ABSTRACT
Transcription of eukaryotic genes requires the cooperative action of the RNA polymerase complex, the general transcription factors (TFIIB, TFIID, TFIIE, TFIIF and TFIIH) and chromatin modifiers. The TFIID complex contributes to transcriptional activation by several mechanisms and has a subunit with associated histone acetyltransferase (HAT) activity. The histone modifier SAGA complex has both HAT and deubiquitylase (DUB) activities. TFIID and SAGA share several TBP-associated factors (TAFs), but not their HAT subunit. Recently, several duplicated TAF proteins have been identified in higher eukaryotes, but their functional diversity has been so far poorly characterized. Here, we report the functional similarities and differences of TAF10 and TAF10b, the two TAF10 orthologs of Drosophila melanogaster. Results from in silico modeling suggest that dTAF10 and dTAF10b have similar secondary structures characterized by the presence of a histone-fold domain. Additionally, dTAF10 and dTAF10b share interaction partners and show similar expression patterns in neuronal tissues. Nonetheless, dTAF10 and dTAF10b seem to have partly distinct functions. To investigate their roles, we generated dTaf10-dTaf10b double-mutants and rescued the mutant flies with transgenes, which allowed the translation of either dTAF10 or dTAF10b protein. We found that the loss of dTAF10b resulted in pupal lethality, while animals lacking dTAF10 were able to form puparium. dTaf10 mutant adults showed distorted eye morphology. During DNA repair, dTAF10 and dTAF10b act redundantly, suggesting that these proteins have distinct but partially overlapping functions.
Introduction
The pre-initiation complex (PIC) consists of the RNA polymerase II (RNAPII), chromatin modifiers, and general transcription factors (TFIIB, TFIID, TFIIE, TFIIF and TFIIH).Citation1 During the well-controlled transcription initiation step, TFIID binds to specific promoters, which facilitates the assembly of PIC.Citation2 TFIID is a multi-protein complex composed of TATA-box-binding protein (TBP) and TBP-associated factors (TAFs).Citation3,4 The TAF protein family consists of 15 members, named TAF1–15 according to their descending molecular weights.Citation3,5 TAF proteins are also present in TAFs containing TBP-free complex (TFTC) and GCN5-containing SAGA complex.Citation6-8 While TFIID contains the whole repertoire of TAF subunits, SAGA consists of the TAF5, TAF6, TAF9, TAF10, and TAF12 proteins.Citation8 The TAF protein composition of TFIID shows tissue-specific alterations, which indicates that TAF proteins can regulate transcription through multiple mechanisms.Citation9-12
Previous reports demonstrated that tissue-specific TAFs and TBP are expressed in various organisms, suggesting the existence of cell-type forms or stage-specific forms of the general transcription factors.Citation11,13 These might be required for the selective activation of specific genes transcribed by RNAPII.Citation14 In addition, tissue-specific Drosophila TAF homologs, such as dTAF4b, dTAF5b, dTAF6b, dTAF8b, and dTAF12b are shown to control expression of genes involved in spermatid differentiation.Citation10,12,15 These data suggest that, in Drosophila, testis-specific TAF homologs might form specific TAF-containing protein complexes to regulate the expression of genes involved in spermatid differentiation.Citation10 These data are also supported by the results of Gazdag and colleagues, as they found that the oocyte-specific TBP homolog TBP2 is required for folliculogenesis, suggesting that TBP2 has a potential role in transcription activation during oocyte growth and maturation.Citation9 This assumes the existence of different TAFs containing TFIID complexes that might be involved in tissue or developmental stage-specific transcriptional programming.
In Drosophila, two TAF proteins (dTAF10 and dTAF10b) show homology with human TAF10. The two dTAF10 proteins show different expression patterns in neuronal tissues during embryonic development and are present in different multiprotein complexes - dSAGA and dTFIID, respectively.Citation16-18 Recently, it has been reported that dTAF10 and dTAF10b have synergistic roles in the regulation of Drosophila morphogenesis and they are required for the activation of Halloween genes involved in ecdysone biosynthesis.Citation18-20 Taken together, these observations support the idea that functionally different TAF-containing complexes participate in gene regulation.
Here, we report that the two Drosophila TAF10 proteins have partially overlapping functions. We show that the absence of both dTaf10 genes results in late larval lethality, while the loss of only dTaf10b causes lethality in the late pupal phase. dTaf10 is not required for morphogenesis, but animals lacking dTaf10 show distorted eye development. These results suggest that, in general, the two Drosophila dTAF10 proteins have partially overlapping functions, having also distinct roles in the regulation of tissue-specific gene expression.
Materials and methods
Fly stocks
Fly stocks were raised at 25°C on standard Drosophila medium. The dTaf10d25 mutant flies were generated by imprecise excision of P(SUPor-P)Taf10KG07031. The mutant flies were maintained over CyO-GFP balancer, which is visible in larval stages. The rest of the fly stocks used in this study were obtained from the Bloomington Drosophila Stock Center, and the RNAi stocks are derived from NIG-Fly center Japan.
Taf10RNAi: http://www.shigen.nig.ac.jp/fly/nigfly/rnaiDetailAction.do?input=srandstockId=2859R-5
P-element remobilization
To mobilize P(SUPor-P)Taf10KG07031 P elements in the 2L(23A5) regions, we crossed P-element-carrying stocks with a CyO P(delta2-3)/Bc transposase source. At F2 progeny, we scored for the loss of the miniwhite (w+mC) marker and identified lethal mutations, which appeared in the absence of complementation of the DF(2L)C144 chromosomal deletion (BL90 stock was used) (http://fly.bio.indiana.edu/). Lethal alleles affecting the dTaf10 and dTaf10b regions were sorted into complementation groups and characterized by molecular methods to find deletions in the dTaf10 and dTaf10b genes. One deletion allele (dTaf10d25) was identified, which removed both the dTaf10 and dTaf10b coding regions ().
RNA isolation and expression quantifications
Total RNAs were isolated from L3 larvae using Trizol (SIGMA) reagent. The homogenized tissues were incubated for 5 minute at 25°C and chloroform was added to each sample. Then, the samples were incubated for 15 minute at 4°C. After the centrifugation step, RNAs were precipitated with isopropyl alcohol and the pellets were washed with 75% ethanol and subsequently dissolved in nuclease-free water. For each experiment, the data obtained from RT-qPCR measurements were calculated by ΔΔCt method.
LOH assay
Loss-of-heterozygosity (LOH) assays were performed by scoring for the appearance of the recessive multiple wing hairs (Mwh) phenotype as reported earlier.Citation21 Every genetic combination was tested in two independent experiments; each involving 15 wings.
Immunostainings
For wing disc immunostaining of L3 larvae, wing discs were fixed in Phosphate Buffer Saline (PBS) buffer containing 4% formaldehyde for 20 minute. To block non-specific staining, wing discs were incubated in 5% BSA-PBST (0.1% (v/v) Tween 20 in PBS) (Sigma-Aldrich) for 2 hour at 4°C. The samples were incubated at 4°C for 1 hour with aniti-Caspase-3 antibody (Cell signaling #9661 1:500) in 1% BSA (Sigma-Aldrich)-PBST (PBS plus 0.1% Tween 20). After several rinses in PBST, wing discs were incubated with secondary antibody (Alexa Fluor 555) for 2 hour at 4°C. Following several washes with PBST, wing discs were mounted in Aqua Poly Mount (Polysciences Inc.) and examined with Olympus BX51 microscope.
Electron microscopy
Drosophila heads were dehydrated by successive incubations in 70%, 80%, and 90% ethanol for 30 minutes, followed by three washing steps in 100% ethanol and one washing in acetone for 30 minutes each. Dehydration was completed by air drying of the coverslips; then the samples were coated with gold. The images were taken using a HITACHI S2400 scanning electron microscope.
Transgene constructions
dTaf10 and dTaf10b transgenes were constructed by inserting genomic DNA fragments into pCaSpeR4.Citation22 The BglII-ScaI (2,177 bp) genomic fragment comprising both dTaf10 and dTaf10b genes was inserted into the pCaSpeR4 vector. Then, the start codon of either dTaf10 or dTaf10b was substituted with TAG, allowing expression of only one of the two Taf10 genes. The c-myc epitope coding fragment CAGATCCTCTTCTGAGATGAGTTTTTGTTC was introduced to the end of coding sequence before the stop codon. The final maps of the plasmids expressing dTAF10 or dTAF10b are given in Fig. S1. DNA was injected into fly stocks, and transgenic flies were obtained using the published protocol.Citation23
Results
Structural characterization of Drosophila TAF10 and TAF10b proteins
Previously, we and others reported that in Drosophila melanogaster two proteins are present that show homology with human TAF10: dTAF10 and dTAF10b (Fig. S2).Citation16,18 The dTAF10 protein consists of 167 amino acids, while the dTAF10b is 146 amino acids long (). Three-dimensional (3D) structure predictions show that although dTAF10 and dTAF10b show only 75% sequence identity, they form similar helix-loop-helix structures that constitute their histone-fold domains (). The 3D protein structure predictions also show that the two proteins differ from each other in their N-terminal regions, which might be involved in the interaction with different sets of proteins. As mentioned above, the most conserved regions of the dTAF10 proteins are the histone-fold domains, in which they show 50% identity and 80% similarity ().
Figure 1. The Drosophila TAF10 and TAF10b proteins have highly similar structures. (A) Schematic representation of the Drosophila TAF10 and TAF10b proteins depicting the localization of the histone-fold domains. (B) Three-dimensional protein structure prediction of dTAF10 and dTAF10b as determined using Swiss-MODEL program.Citation35, 36 (C) Alignment of the amino acid sequences of Drosophila TAF10 proteins. The sequences were downloaded from the NCBI databases, and Gonnet PAM 250 matrix was used to identify the similar regions. The one-letter amino acid code is used. The highly-conserved histone-fold domain is boxed. The star represents the conserved residues, double dots indicates the positions of highly similar amino acids, while single dot marks positions at which weakly similar amino acids are found. (D) The relative position of dTaf10 and dTaf10b genes at the 2L(23A5) position of the D. melanogaster 2nd chromosome based on the data available on Flybase.
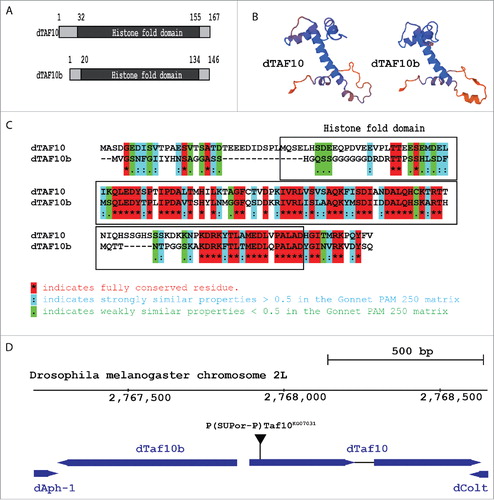
The chromosomal localization of the dTaf10 and dTaf10b genes suggests that the two genes originated from gene duplication event occurred within the Drosophilidae genus. It was previously reported that the dTaf10 and dTaf10b genes partially overlap at their 5′ regions (Fig. S3), but recent data based on RNA-seq results suggest separated coding regions (). This suggests a strict transcriptional control, since dTaf10 and dTaf10b share a 150-bp bidirectional promoter region, from which transcription can start to both directions. Because the orientation of the dTaf10 and dTaf10b genes is opposite to each other, it remains unclear how these two genes are transcriptionally regulated.
dTaf10 and dTaf10b have distinct mRNA profiles in Drosophila tissues
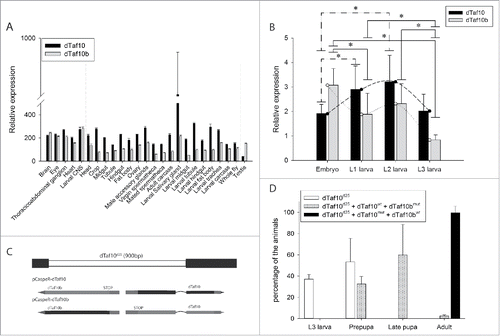
RNA-seq data published by the modENCODE consortia revealed the different expression patterns of the dTaf10 and dTaf10b genes in various tissues ().Citation24 Although the two Drosophila Taf10 genes show similar expression patterns in the nervous system (brain, central nervous system, ganglions, eye), in the rest of larval tissues, the dTaf10 mRNA level is higher than that of dTaf10b (, middle panel). Whereas dTaf10b is highly expressed in the testis. The dTaf10 mRNA level remained fairly constant during larval development, whereas dTaf10b expression is highest at the embryonic stage (). These data are in line with the idea that the dTAF10 and dTAF10b proteins could be equally important in certain tissues, but also play distinct tissue-specific functions in other tissues and developmental stages.
Figure 2. dTaf10 and dTaf10b show alterations in their expression levels in different tissues during Drosophila development. (A) Relative mRNA level of dTaf10 and dTaf10b in various tissues. The data were obtained from the modENCODE and Flyatlas databases. (B) The dTaf10 and dTaf10b mRNAs levels at different developmental stages as determined by quantitative RT-PCR. Each data point represents the average of three replicates. Star indicates statistical significance between the data set (P < 0.03 by Mann-Whitney rank sum test; n = 6). (C) The schematic structure of the dTaf10 null mutant and gene specific rescue constructs. (D) The pupariation and hatching rate of the dTaf10 (dTaf10d25+dTaf10mut+dTaf10bwt) and dTaf10b (dTaf10d25+dTaf10wt+dTaf10bmut) single mutants or dTaf10/dTaf10b (dTaf10d25) double mutant. Mean values and errors were calculated from three independent experiments containing 30 larvae in each category.
dTAF10 and dTAF10b are required for proper Drosophila development
To investigate the functions of dTAF10 and dTAF10b, we deleted the coding region of both dTaf10 genes by P-element remobilization to generate the dTaf10 null mutant (dTaf10d25).Citation18 We also created two transgenes, in which the first methionine encoding triplet of either dTAF10 or dTAF10b was mutated to a stop codon (TAG), allowing the translation of only dTAF10b or dTAF10, respectively. Then, we investigated the rescue effect of the two transgenes in the dTaf10 null animals (). Although, the double mutants showed L3 larval lethality and early pupal lethality, the dTaf10b mutants reached late pupal stage (). dTaf10 was not required for Drosophila larval and pupal development (). In the absence of dTAF10, normal adult tissues could be developed. However, we observed morphology abnormalities in the eye, which might indicate a role in apoptosis (). Based on these results, we concluded that dTaf10b plays an essential role during larval and pupal development, whereas dTaf10 does not have an essential function until the adult stage. Taken together, it seems that although the two dTAF10 proteins are similar, they could regulate distinct biological processes.
Figure 3. The dTAF10 proteins are required for eye development and genome maintenance. (A) Images of eye surface of wild-type and dTaf10 mutant animals obtained by scanning electron microscope. The lack of bristles might indicate elevated level of apoptosis during eye development. (B) The average number of mwh clones detected on the wings of wild-type, dTaf10 (dTaf10d25+dTaf10mut+dTaf10bwt), and dTaf10b (dTaf10d25+dTaf10wt+dTaf10bmut) mutant animals following a low-dose of X-ray irradiation. The inset shows an mwh clone in the wild-type background. Data represent averages of 15 wings in each category. (C) RT-qPCR analysis of dTaf10 and dTaf10b mRNA levels following siRNA silencing. (D) Wing imaginal discs of wild-type and dTaf10 siRNA silenced L3 larvae were stained with anti-Caspase-3 antibody to detect apoptotic cells. The wing morphology of the adults is shown on the lower panel. UAS-GFP; Vg-GAL4 is a negative control for the effect of the tissue-specific driver alone.
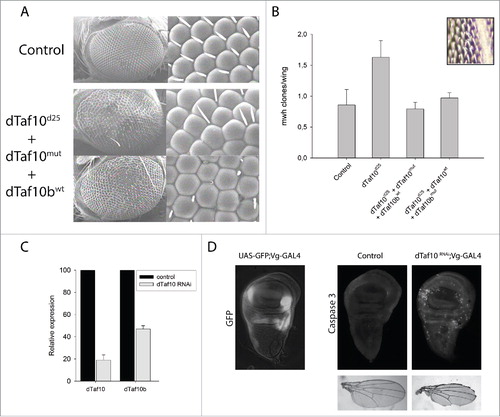
The rough eye phenotype observed in the dTAF10 mutants () could be the consequence of apoptosis-induced cell death. To test whether the loss of dTAF10 proteins indeed caused apoptosis, we silenced dTaf10 with siRNA and immunostained imaginal discs of the larvae with an anti-Caspase-3 antibody. The wing-specific silencing of dTaf10 RNAi lines by vestigial-GAL4 driver led to increased apoptotic cell number at the GFP-positive dTaf10 siRNA expressing regions of the wing margins compared to the control regions, where the dTaf10 mRNA level was not silenced ().
dTAF10 and dTAF10b affect the DNA repair efficiency
The rough eye phenotype observed in dTaf10 mutants could be a result of increased apoptotic rate, DNA damage repair abnormalities, or failure in developmental reprogramming. Earlier it was reported that the dSAGA complex, which also contains dTAF10, is required for the maintenance of genome stability.Citation21 To test whether dTAF10 and dTAF10b are both required for this process, we performed LOH assays to test the contribution of the dTAF10 proteins in preserving genome stability. We performed LOH assays based on the detection of the recessive multiple wing hair (mwh) phenotype revealed by the loss of the wild-type copy of mwh. Late-third-instar larvae heterozygous for the mwh mutation and either wild-type or heterozygous for dTaf10/dTaf10b mutations were irradiated with a low-dose X-ray (25 Gy). Cells that have lost the wild-type copy of mwh display the recessive mwh phenotype, which is easily recognizable and can be scored on the wings. While the dTaf10/dTaf10b double heterozygotes exhibited elevated numbers of mwh cells, the single mutations had no effect on the mwh clone numbers (). This result demonstrates that dTAF10 and dTAF10b have a role in preserving genomic stability in response to DNA damage induced by low-level of X-ray irradiation.
dTAF10 and dTAF10b interacting partners consist of similar but partly different sets of proteins
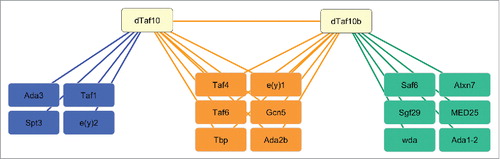
To reveal whether dTAF10 and dTAF10b are incorporated into the same multiprotein complexes, we checked public databases to identify proteins, which interact with either dTAF10 or dTAF10b. It has already been demonstrated that dTAF10 is part of the dSAGA complex, while dTAF10b is a component of the dTFIID complex.Citation16,17,20 We also checked available Drosophila protein-protein interaction databases and collected the interaction partners of dTAF10 and dTAF10b. The results were visualized by using Cytoscape software.Citation25 We found 11 proteins that interact with dTAF10 and 13 proteins that interact with dTAF10b. The interaction map strengthens the view that dTAF10 and dTAF10b interact with each other. Additionally, the dTAF10 and TAF10b proteins interact with dTBP, dTAF4, dTAF6, and dTAF9 (e(y)1), which are components of the dTFIID and the dSAGA complexes. We also found interactions between dTAF10 proteins and additional dSAGA subunits, such as dADA2b and GCN5. dTAF10 interacts with three additional dSAGA subunits (SPT3, ADA3, and e(y)2), whereas dTAF10b interacts with ATAXIN7, SAF6, WDA, ADA1-2, and SGF29. These data also support the notion that dTAF10 and dTAF10b could be incorporated into similar multiprotein complexes sharing the same submodules. Finally, we could identify dTAF10b-dTAF1 and dTAF10-MED25 interactions, which suggests that dTAF10 and dTAF10b could play different roles in the fine-tuning of the tissue-specific transcription activation events.
Discussion
Two orthologs of the human Taf10 gene are present in the Drosophilidae genus: dTaf10 and dTaf10b. The two dTaf10 genes are located at the 23A5 cytological position on the 2nd chromosome of D. melanogaster in a head-to-head orientation to each other, presumably with overlapping transcription regulatory regions. This arrangement suggests an intricate regulation since the two genes share a 150-bp promoter region, from which transcription can initiate to both directions.
It was suggested that the two adjacent dTAF10s have different functions because they show different expression patterns during the Drosophila embryonal development.Citation16,26 Also, the dTaf10 and dTaf10b expression patterns have different stoichiometry, supporting the notion that they might have separated roles in various tissues.Citation24 Although the dTaf10 mRNA level was higher in many tissues than that of dTaf10b, in neuronal tissues the two mRNA levels seem to be equal.
The phenotypic characterization of the dTaf10-dTaf10b double mutants and the results of rescue experiments revealed that dTAF10 and dTAF10b have distinct roles in tissue-specific gene activation. It has been shown that several Drosophila genes have orthologs that are exclusively expressed in the testis.Citation9,10,12,15,27 Additionally, it has been shown that testis-specific dTAF1, dTBP, and dTAF7 subunits of dTFIID are expressed in humanCitation28-30 or in mouse, respectively.Citation9,31 It is possible that for tissue-specific activation of promoters, specific components of the general transcription machinery are required to achieve transcriptional reprogramming occurring in the testis. Our data are in line with this view, since only dTaf10b is expressed in the testis. Accordingly, in a testis-specific gene activation program, TAF homologs could form a different dTFIID complex and regulate the tissue-specific expression of a subset of genes.
The lack of dTAF10 does not result in a lethal phenotype but does produce an abnormal eye bristles phenotypes, which suggests that dTAF10 is not essential for Drosophila development but does have important roles in different tissues. Loss of the dSAGA subunits resulted in photoreceptor axon-targeting defects indicating that the dTAF10 containing dSAGA complex plays a role in neuronal development in the optic lobe.Citation32 We also show that dTaf10 silencing results in increased apoptosis and wing morphology abnormalities.
Although dTAF10 and dTAF10b proteins have similar 3D structures of their histone-fold domain, they differ in their N-terminal variable regions. Consequently, the two proteins could incorporate into similar complexes through their conserved histone-fold domain, while the variable region could facilitate the interaction with different proteins. dTAF10 is a stable component of dTFIID and is essential for transcription activation. dTAF10 plays a role in the nuclear import of the dTFIID subunit dTAF8.Citation33,34 The multisubunit TFIID complex has important roles in the coordination of transcription initiation. dTAF10 protein(s) are also present in the SAGA complex.
The protein-protein interaction map shown in also supports these observations, as dTAF10 and dTAF10b could interact with the core proteins of dTFIID and dSAGA. However, dTAF10 and dTAF10b interact with different dSAGA subunits, suggesting that those proteins might be required for the tissue-specific fine-tuning of transcription. Thus, dTAF10 may help to induce the expression of the general transcription activators, which can activate the transcription of several tissue-specific target genes. It seems probable that dTAF10 and dTAF10b regulate the formation of distinct dTFIID-like structures that could activate different subsets of target genes.
Figure 4. Schematic representations of the interaction network of dTAF10 and dTAF10b. Data were obtained from Flybase and were visualized using Cytoscape software. Left panel indicates sets of identical proteins, which are in interaction with dTAF10. Proteins at middle panel interact with both dTAF10 and dTAF10b, while sets of proteins represented at left panel show an interaction only with dTAF10b.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Author contributions
Conceived and designed the experiments: TP, IMB. Performed the experiments: TP, ZP, BNB, BV, YVS, SGG. Analyzed the data: TP. Contributed reagents/materials/analysis tools: IMB, TP. Wrote the paper: TP, IMB.
1327836_Supplemental_Material.zip
Download Zip (949.1 KB)Acknowledgments
We thank Zsuzsanna Ujfaludi and Gabriella Pankotai-Bodo for helpful discussions and revisions. We also thank Adrienn Csebella-Bakota for maintaining Drosophila strains.
Funding
This work was supported by Hungarian Scientific Research Fund (OTKA PD 112118), the János Bolyai Research Scholarship of the Hungarian Academy of Sciences and the National Research, Development and Innovation Office grant GINOP-2.3.2-15-2016-00032 and GINOP-2.3.2-15-2016-00020. Yulii Shidlovskii was funded by Russian Foundation for Basic Research (16-34-60214).
References
- Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol 2006; 41:105-178; PMID:16858867; https://doi.org/10.1080/10409230600648736
- Morse RH. Transcription factor access to promoter elements. J Cell Biochem 2007; 102:560-570; PMID:17668451; https://doi.org/10.1002/jcb.21493
- Burley SK, Roeder RG. Biochemistry and structural biology of transcription factor IID (TFIID). Annu Rev Biochem 1996; 65:769-799; PMID:8811195; https://doi.org/10.1146/annurev.bi.65.070196.004005
- Juven-Gershon T, Kadonaga JT. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev Biol 2010; 339:225-229; PMID:19682982; https://doi.org/10.1016/j.ydbio.2009.08.009
- Rhee HS, Pugh BF. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature 2012; 483:295-301; PMID:22258509; https://doi.org/10.1038/nature10799
- Demeny MA, Soutoglou E, Nagy Z, Scheer E, Janoshazi A, Richardot M, Argentini M, Kessler P, Tora L. Identification of a small TAF complex and its role in the assembly of TAF-containing complexes. PLoS One 2007; 2:e316; PMID:17375202; https://doi.org/10.1371/journal.pone.0000316
- Grant PA, Schieltz D, Pray-Grant MG, Steger DJ, Reese JC, Yates JR, 3rd, Workman JL. A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell 1998; 94:45-53; PMID:9674426; https://doi.org/10.1016/S0092-8674(00)81220-9
- Timmers HT, Tora L. SAGA unveiled. Trends Biochem Sci 2005; 30:7-10; PMID:15653319; https://doi.org/10.1016/j.tibs.2004.11.007
- Gazdag E, Rajkovic A, Torres-Padilla ME, Tora L. Analysis of TATA-binding protein 2 (TBP2) and TBP expression suggests different roles for the two proteins in regulation of gene expression during oogenesis and early mouse development. Reproduction 2007; 134:51-62; PMID:17641088; https://doi.org/10.1530/REP-06-0337
- Hiller MA, Lin TY, Wood C, Fuller MT. Developmental regulation of transcription by a tissue-specific TAF homolog. Genes Dev 2001; 15:1021-1030; PMID:11316795; https://doi.org/10.1101/gad.869101
- Hochheimer A, Tjian R. Diversified transcription initiation complexes expand promoter selectivity and tissue-specific gene expression. Genes Dev 2003; 17:1309-1320; PMID:12782648; https://doi.org/10.1101/gad.1099903
- Lin TY, Viswanathan S, Wood C, Wilson PG, Wolf N, Fuller MT. Coordinate developmental control of the meiotic cell cycle and spermatid differentiation in Drosophila males. Development 1996; 122:1331-1341; PMID:8620860
- Veenstra GJ, Wolffe AP. Gene-selective developmental roles of general transcription factors. Trends Biochem Sci 2001; 26:665-671; PMID:11701325; https://doi.org/10.1016/S0968-0004(01)01970-3
- Verrijzer CP. Transcription factor IID–not so basal after all. Science 2001; 293:2010-2011; PMID:11557865; https://doi.org/10.1126/science.1064980
- White-Cooper H, Schafer MA, Alphey LS, Fuller MT. Transcriptional and post-transcriptional control mechanisms coordinate the onset of spermatid differentiation with meiosis I in Drosophila. Development 1998; 125:125-134; PMID:9389670
- Georgieva S, Kirschner DB, Jagla T, Nabirochkina E, Hanke S, Schenkel H, de Lorenzo C, Sinha P, Jagla K, Mechler B et al. Two novel Drosophila TAF(II)s have homology with human TAF(II)30 and are differentially regulated during development. Mol Cell Biol 2000; 20:1639-1648; PMID:10669741; https://doi.org/10.1128/MCB.20.5.1639-1648.2000
- Muratoglu S, Georgieva S, Papai G, Scheer E, Enunlu I, Komonyi O, Cserpán I, Lebedeva L, Nabirochkina E, Udvardy A et al. Two different Drosophila ADA2 homologues are present in distinct GCN5 histone acetyltransferase-containing complexes. Mol Cell Biol 2003; 23:306-321; PMID:12482983; https://doi.org/10.1128/MCB.23.1.306-321.2003
- Pahi Z, Kiss Z, Komonyi O, Borsos BN, Tora L, Boros IM, Pankotai T. dTAF10- and dTAF10b-Containing complexes are required for ecdysone-driven larval-pupal morphogenesis in Drosophila melanogaster. PLoS One 2015; 10:e0142226; PMID:26556600; https://doi.org/10.1371/journal.pone.0142226
- Pankotai T, Popescu C, Martin D, Grau B, Zsindely N, Bodai L, Tora L, Ferrús A, Boros I. Genes of the ecdysone biosynthesis pathway are regulated by the dATAC histone acetyltransferase complex in Drosophila. Mol Cell Biol 2010; 30:4254-4266; PMID:20584983; https://doi.org/10.1128/MCB.00142-10
- Weake VM, Swanson SK, Mushegian A, Florens L, Washburn MP, Abmayr SM, Workman JL. A novel histone fold domain-containing protein that replaces TAF6 in Drosophila SAGA is required for SAGA-dependent gene expression. Genes Dev 2009; 23:2818-2823; PMID:20008933; https://doi.org/10.1101/gad.1846409
- Pankotai T, Komonyi O, Bodai L, Ujfaludi Z, Muratoglu S, Ciurciu A, Tora L, Szabad J, Boros I. The homologous Drosophila transcriptional adaptors ADA2a and ADA2b are both required for normal development but have different functions. Mol Cell Biol 2005; 25:8215-8227; PMID:16135810; https://doi.org/10.1128/MCB.25.18.8215-8227.2005
- Le T, Yu M, Williams B, Goel S, Paul SM, Beitel GJ. CaSpeR5, a family of Drosophila transgenesis and shuttle vectors with improved multiple cloning sites. Biotechniques 2007; 42:164-166; PMID:17373479; https://doi.org/10.2144/000112386
- Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science 1982; 218:348-353; PMID:6289436; https://doi.org/10.1126/science.6289436
- The modENCODE Consortium, Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, Landolin JM, Bristow CA, Ma L et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 2010; 330:1787-1797; PMID:21177974; https://doi.org/10.1126/science.1198374
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003; 13:2498-2504; PMID:14597658; https://doi.org/10.1101/gr.1239303
- Hernandez-Hernandez A, Ferrus A. Prodos is a conserved transcriptional regulator that interacts with dTAF(II)16 in Drosophila melanogaster. Mol Cell Biol 2001; 21:614-623; PMID:11134347; https://doi.org/10.1128/MCB.21.2.614-623.2001
- Nakajima T, Takase M, Miura I, Nakamura M. Two isoforms of FTZ-F1 messenger RNA: molecular cloning and their expression in the frog testis. Gene 2000; 248:203-212; PMID:10806365; https://doi.org/10.1016/S0378-1119(00)00119-0
- Ozer J, Moore PA, Lieberman PM. A testis-specific transcription factor IIA (TFIIAtau) stimulates TATA-binding protein-DNA binding and transcription activation. J Biol Chem 2000; 275:122-128; PMID:10617594; https://doi.org/10.1074/jbc.275.1.122
- Upadhyaya AB, Lee SH, DeJong J. Identification of a general transcription factor TFIIAalpha/beta homolog selectively expressed in testis. J Biol Chem 1999; 274:18040-18048; PMID:10364255; https://doi.org/10.1074/jbc.274.25.18040
- Wang PJ, Page DC. Functional substitution for TAF(II)250 by a retroposed homolog that is expressed in human spermatogenesis. Hum Mol Genet 2002; 11:2341-2346; PMID:12217962; https://doi.org/10.1093/hmg/11.19.2341
- Pointud JC, Mengus G, Brancorsini S, Monaco L, Parvinen M, Sassone-Corsi P, Davidson I. The intracellular localisation of TAF7L, a paralogue of transcription factor TFIID subunit TAF7, is developmentally regulated during male germ-cell differentiation. J Cell Sci 2003; 116:1847-1858; PMID:12665565; https://doi.org/10.1242/jcs.00391
- Weake VM, Lee KK, Guelman S, Lin CH, Seidel C, Abmayr SM, Workman JL. SAGA-mediated H2B deubiquitination controls the development of neuronal connectivity in the Drosophila visual system. EMBO J 2008; 27:394-405; PMID:18188155; https://doi.org/10.1038/sj.emboj.7601966
- Soutoglou E, Demeny MA, Scheer E, Fienga G, Sassone-Corsi P, Tora L. The nuclear import of TAF10 is regulated by one of its three histone fold domain-containing interaction partners. Mol Cell Biol 2005; 25:4092-4104; PMID:15870280; https://doi.org/10.1128/MCB.25.10.4092-4104.2005
- Trowitzsch S, Viola C, Scheer E, Conic S, Chavant V, Fournier M, Papai G, Ebong IO, Schaffitzel C, Zou J et al. Cytoplasmic TAF2-TAF8-TAF10 complex provides evidence for nuclear holo-TFIID assembly from preformed submodules. Nat Commun 2015; 6:6011; PMID:25586196; https://doi.org/10.1038/ncomms7011
- Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M, Bordoli L et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 2014; 42:W252-8; PMID:24782522; https://doi.org/10.1093/nar/gku340
- Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res 2009; 37:D387-92; PMID:18931379; https://doi.org/10.1093/nar/gkn750
