ABSTRACT
TFIIH is a 10-subunit complex involved in transcription and DNA repair. It contains several enzymatic activities including a ATP-dependent DNA translocase in XPB and a cyclin-dependent kinase in CDK7. Recently the discovery of several XPB and CDK7 inhibitors with specific impact on the transcriptional addiction of many tumors pinpointed these activities as potential target in cancer chemotherapy. Unexpectedly a basal transcription factor involved in global mRNA expression now emerges a one of the most clinically promising Achilles heels of cancerous cells. These inhibitors also proved to be useful tools to unveil new functions of TFIIH in gene expression.
Introduction
During transcription of mRNA (mRNA), RNA Pol II is recruited to promoters with the support of additional proteins called the general transcription factors (GTFs), forming the basal pre-initiation complex (PIC).Citation1 Initiation takes place following the assembly of the PIC, the opening of the DNA around the transcription start site and the phosphorylation of the carboxyl terminal domain of RPB1 (CTD), the largest RNA Pol II subunitCitation2,3 Downstream from the transcription start site (∼20- to 60-nucleotide), the RNA Pol II remains in a paused state, called promoter proximal pausing, before entering into the elongation step, owing to the action of specific factors.Citation4 Among GTFs, TFIIH plays a peculiar function through its involvement in both transcription initiation and promoter proximal pausing release.Citation5,6 TFIIH is composed of 2 sub-complexes that are bridged by the XPD helicase subunit: the core (including XPB ATPase/Translocase, p62, p52, p44, p34 and TTDA) and the CDK-activating kinase (CAK) (including MNAT1, cyclin H and the CDK7 kinase). TFIIH is also playing a role in RNA Pol I-dependent gene expression although the mechanistic details of its function are not known yet.Citation7 Beyond Pol I and II transcription, TFIIH also participates to nucleotide excision repair (NER),Citation8,9,10 a DNA repair pathway that contributes to the removal of bulky adducts including either UV light or chemotherapeutic agents such as cisplatin.
Beneath the complexity of every cancer cells, their common feature resides in the fact that each of the pathways that constrains the proliferative response in normal situation is dysregulated. Cancer cell hyperproliferation relies on a high level of transcription. Consequently, proteins involved in transcriptional control, including TFIIH, emerge as the attractive targets for the development of a new generation of small chemical molecules that would modulate their enzymatic activity. In addition, the hyperproliferative capacity of cancer cells has been used for a long time in clinics to specifically kill them with chemical compounds interfering with fundamental cellular pathways such as transcription and replication. The cancer cells adapt to these treatments by various means including an increase in DNA repair pathways such as NER.Citation11 Due to its central role in transcription and DNA repair, TFIIH has become a privileged target to exploit in anticancer therapy. Several small molecules have been characterized these last years that inhibit TFIIH enzymatic activities and specifically kill tumor cells, suggesting that they can soon be used in the clinic ().
Figure 1. Drugs targeting the TFIIH transcription/DNA repair complex. TFIIH is a 10-subunit complex composed of a core (In red; XPB, p62, p52, p44, p34 and TTDA) associated to the CAK (In blue; Cdk7, CycH and MAT1) through MAT1 and the XPD subunit (In green). Four enzymatic activities are found in TFIIH; XPB and XPD are 3′ to 5′ and 5′ to 3′ helicases, Cdk7 is a kinase, and p44 has been described as an E3 ubiquitin ligase in yeast. Triptolide (TPL) is a diterpenoid epoxide that is endogenously produced by the thunder god vine, Tripterygium wilfordii and which targets the ATPase activity of XPB. Minnelide is water soluble pro-drug of TPL synthesized at the University of Minnesota (Minnesota and triptolide) by adding a phosphate ester group. Spironolactone is a potassium-sparing diuretic that induced the specific degradation of XPB through the proteasome pathway. THZ1 is phenylaminopyrimidinyl-acrylamide compound that acts as a highly potent, selective, ATP-site directed, and irreversible inhibitor of CDK7
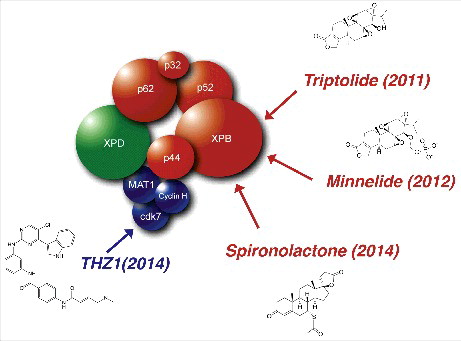
Inhibitors of XPB; impact on transcription and DNA repair
One of the most promising drug for the treatment of many cancer types is triptolide (TPL), a molecule that targets the ATPase activity of XPB. TPL is a diterpen triepoxide extracted from the plant Tripterygium wilfordii. This molecule has a limited solubility in vivo, and a TPL derivative called Minnelide, has been synthetized and was found to be extremely effective in both pancreatic and liver cancer.Citation12
Although other targets have been proposed, TPL shows a very high specificity for XPB and induces Pol I- and II-dependent transcription inhibition but also prevents NER.Citation13,14,15 Consequently, TPL can be used alone to impact the transcriptional program of tumor cells or in combination with platinum-derivatives to increase their anticancer efficiency.Citation15 Indeed, platinum-based compounds exert their antineoplastic effect through their binding to DNA and the saturation of several DNA repair mechanisms, including NER, that they induce. Chemosensitization of cancer cells, for instance through the inhibition of DNA repair pathways, constitutes a strategy with important clinical implications since it may minimize resistance and ultimately decrease the possibility of negative side effects.Citation11
Along this line, Spironolactone (SP), an antagonist of aldosterone, proved to strongly inhibit NER through the specific degradation of XPB. Consequently, combination of SP with Cisplatin or Oxaliplatin chemo-sensitizes human carcinoma cells.16
One could have imagined that these 2 molecules would have the same impact on transcription since they target the same TFIIH subunit. Surprisingly, we recently demonstrated that it was not the case and that mRNA expression could take place in the absence of XPB after SP treatment while it was impairer when XPB ATPase activity was inhibited by TPL. These results, together with biochemical studies, prompted us to propose a new model for the function of XPB in transcription initiation in which the recruitment of TFIIH to the PIC induces the XPB-dependent block of DNA unwinding that is overcome by its ATP-dependent translocation along the DNA, followed by promoter melting achieved with the use of binding energy alone.Citation17 Satisfactory enough, this model unified the 3 mechanisms of DNA melting in class I, II, and III gene expression wherein DNA unwinding at the promoter only operates through the energy released that is generated upon the creation of the PIC. These results also show that TFIIH inhibitors could be very useful tools to unveil the function of this complex in fundamental cellular pathways. More broadly, these inhibitors could be used in genome-wide gene expression studies to reveal unexpected gene behavior such as the existence of a small group of genes that show a stably paused Pol II after TPL inhibition.Citation18
Inhibitor of CDK7; affecting super-enhancer dependent gene transcription
Another important target in TFIIH is the CDK7 kinase that is part of the sub complex CAK with cyclin H and MNAT1. CDK7 controls transcription initiation through the phosphorylation of the serine 5 of the carboxyl-terminal domain of the RPB1 subunit of RNA Pol II.Citation19 In 2014, Kwiatkowski et al., first described THZ1, a small molecule able to irreversibly bind CDK7 with high affinity and specificity among other CDKs.Citation20 This specificity relies on that fact that the residue that is targeted by THZ1, the cysteine 312, is located outside of the canonical kinase domain. This specificity was useful to analyze the role of CDK7 in gene expression and to reveal new roles for this kinase in co-transcriptional capping and pausing.Citation21 The same authors also showed that the inhibition of CDK7 could unexpectedly leads to the disruption of a specific transcription program fundamental for the T-cell acute lymphoblastic leukemia homeostasis without major effect on basal transcription. Several papers subsequently extended the class of tumor cells that display sensitive to THZ1 vs. wild-type cells at the low nanomolar range and CDK7 kinase activity quickly appeared as a druggable target in cancer chemotherapy due to its involvement in super-enhancer dependent gene transcription.Citation22,23
Normal enhancers are DNA sequence of 200–500bp base-pairs able to increase the transcription of one or more genes by providing transcription factors to the promoters that trigger the formation of PIC.Citation24 Enhancer-localized proteins and the interactions between enhancers and promoters, contribute to define the cooperative regulatory environment in a phase-separation fashion.Citation25 Recently, a subclass of hundreds of enhancers, named super-enhancers (SEs), has been described through genome-wide methods with a stronger enrichment for the binding of master transcription factors (i.e OCT4 in ESCs), coactivators (such as the Mediator) and specific chromatin marks such as H3K27 acetylation or H3K4 mono-methylation. SEs can be seen as “very strong enhancers” that are of more than 12Kb and that are enriched near highly transcribed genes encoding important regulators.Citation26
Cancer cells are enriched in aberrant SEs at the level of specific oncogenes highly transcribed and involved in several cellular functions that established the cancer dysregulated transcriptional program. The maintenance of this transcriptional program, and the genes that sustain it, are vital for the cancer cells which introduce the concept of “transcriptional addiction” described above, strictly correlated with oncogene addiction.Citation27 In a mechanism that is not completely understood, THZ1 treatment at low nanomolecular range affects mainly transcription of genes under the dependency of SEs and as thus specifically kills tumor cells vs. wild-type. The hypothesis of such CDK7 SEs vulnerability may depend on the high concentration of CDK7 in SEs vs. normal enhancers together with others coactivators such as BRD4.Citation28 Thus, depriving SEs of CDK7 kinase activity would decrease the transactivation of oncogenes (i.e MYC) and the progression from initiation to elongation state of RNA pol II affecting not only mRNAs, but also ncRNAs which regulate the environment of SEs.Citation29 It is however still not known why SEs are specifically vulnerable to CDK7 kinase inhibition and whether CAK is present as an independent entity from TFIIH in SEs or if all TFIIH compounds are recruited there. Indeed, CAK can be found associated to TFIIH or free in the cell and the function of the free complex is not clearly defined.Citation30 A comparison of the sensitivity of cancerous cells to THZ1 vs. TPL, and a complete analyze of the SEs occupancy after treatment with these drugs in these cells should provide some important answers to these questions and bring new insights into the role of TFIIH in transcription regulation.
Conclusion
Many human syndromes, other than cancer, imply an uncontrolled cellular proliferation. Although these pathologies are diverse and heterogeneous, the different pathways that guide their progression imply an increased level of cellular division and an increased level of transcriptional activity by the RNA Pol I and II to sustain the high metabolism of hyperproliferative cells. Importantly, ribosome biogenesis by RNA Pol I is one of the energetically most expensive molecular processes for the cell and the capacity of proliferation of the cell is closely related to the level of production of ribosomal particles.Citation31 The importance of transcriptional activity during proliferation is best highlighted by the fact than many molecules interfering with RNA metabolism have been used to contain these syndromes. For instance, psoriasis is contained using bethamethasone and cyclosporine A, both are well known molecular transcription inhibitors. As a basal transcription factor, TFIIH intervenes in the process of basal transcription by the RNA Pol I and II and is thus an interesting biologic target in many human syndromes, other than cancer, that imply a control of the transcriptional activity. Consequently, the social and scientific impact of studies on TFIIH can be high in the future as more and more disorders will be characterized as transcription syndromes and will require “transcription therapy.”
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- Buratowski S, Hahn S, Guarente L, Sharp PA. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell 1989; 56:549-61; PMID:2917366; https://doi.org/10.1016/0092-8674(89)90578-3
- Grunberg S, Hahn S. Structural insights into transcription initiation by RNA polymerase II. Trends Biochem Sci 2013; 38(12):603-11; PMID:24120742; https://doi.org/10.1016/j.tibs.2013.09.002
- Sainsbury S, Bernecky C, Cramer P. Structural basis of transcription initiation by RNA polymerase II. Nat Rev Mol Cell Biol 2015; 16(3):129-43; PMID:25693126; https://doi.org/10.1038/nrm3952
- Jonkers I, Lis JT. Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol 2015; 16(3):167-77; PMID:25693130; https://doi.org/10.1038/nrm3953
- Schwartz BE, Larochelle S, Suter B, Lis JT. Cdk7 is required for full activation of Drosophila heat shock genes and RNA polymerase II phosphorylation in vivo. Mol Cell Biol 2003; 23(19):6876-86; PMID:12972606; https://doi.org/10.1128/MCB.23.19.6876-6886.2003
- Glover-Cutter K, Larochelle S, Erickson B, Zhang C, Shokat K, Fisher RP, Bentley DL. TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter-proximal pausing, and termination by RNA polymerase II. Mol Cell Biol 2009; 29(20):5455-64; PMID:19667075; https://doi.org/10.1128/MCB.00637-09
- Iben S, Tschochner H, Bier M, Hoogstraten D, Hozák P, Egly JM, Grummt I. TFIIH plays an essential role in RNA polymerase I transcription. Cell 2002; 109(3):297-306; PMID:12015980; https://doi.org/10.1016/S0092-8674(02)00729-8
- Drapkin R, Reardon JT, Ansari A, Huang JC, Zawel L, Ahn K, Sancar A, Reinberg D. Dual role of TFIIH in DNA excision repair and in transcription by RNA polymerase II. Nature 1994; 368:769-72; PMID:8152490; https://doi.org/10.1038/368769a0
- Feaver WJ, Svejstrup JQ, Bardwell L, Bardwell AJ, Buratowski S, Gulyas KD, Donahue TF, Friedberg EC, Kornberg RD. Dual roles of a multiprotein complex from S. cerevisiae in transcription and DNA repair. Cell 1993; 75(7):1379-87; PMID:8269516; https://doi.org/10.1016/0092-8674(93)90624-Y
- Schaeffer L, Roy R, Humbert S, Moncollin V, Vermeulen W, Hoeijmakers JH, Chambon P, Egly JM. DNA repair helicase: a component of BTF2 (TFIIH) basic transcription factor. Science 1993; 260(5104):58-63; PMID:8465201; https://doi.org/10.1126/science.8465201
- Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer 2007; 7(8):573-84; PMID:17625587; https://doi.org/10.1038/nrc2167
- Chugh R, Sangwan V, Patil SP, Dudeja V, Dawra RK, Banerjee S, Schumacher RJ, Blazar BR, Georg GI, Vickers SM, et al. A preclinical evaluation of Minnelide as a therapeutic agent against pancreatic cancer. Sci Transl Med 2012; 4(156):156ra139; PMID:23076356; https://doi.org/10.1126/scitranslmed.3004334
- Titov DV, Gilman B, He QL, Bhat S, Low WK, Dang Y, Smeaton M, Demain AL, Miller PS, Kugel JF, et al. XPB, a subunit of TFIIH, is a target of the natural product triptolide. Nat Chem Biol 2011; 7(3):182-8; PMID:21278739; https://doi.org/10.1038/nchembio.522
- Nonnekens J, Perez-Fernandez J, Theil AF, Gadal O, Bonnart C, Giglia-Mari G. Mutations in TFIIH causing trichothiodystrophy are responsible for defects in ribosomal RNA production and processing. Hum Mol Genet 2013; 22(14):2881-93; PMID:23562818; https://doi.org/10.1093/hmg/ddt143
- Wang G, Wang X, Xu X. Triptolide potentiates lung cancer cells to cisplatin-induced apoptosis by selectively inhibiting the NER activity. Biomark Res 2015; 3:17; PMID:26161259; https://doi.org/10.1186/s40364-015-0043-2
- Alekseev S, Ayadi M, Brino L, Egly JM, Larsen AK, Coin F. A small molecule screen identifies an inhibitor of dna repair inducing the degradation of TFIIH and the chemosensitization of tumor cells to platinum. Chem Biol 2014; 21(3):398-407; PMID:24508195; https://doi.org/10.1016/j.chembiol.2013.12.014
- Alekseev S, Nagy Z, Sandoz J, Weiss A, Egly JM, Le May N, Coin F. Transcription without XPB establishes a Unified Helicase-Independent mechanism of promoter opening in eukaryotic gene expression. Mol Cell 2017; 65(3):504-514 e4; PMID:28157507; https://doi.org/10.1016/j.molcel.2017.01.012
- Chen F, Gao X, Shilatifard A. Stably paused genes revealed through inhibition of transcription initiation by the TFIIH inhibitor triptolide. Genes Dev 2015; 29(1):39-47; PMID:25561494; https://doi.org/10.1101/gad.246173.114
- Roy R, Adamczewski JP, Seroz T, Vermeulen W, Tassan JP, Schaeffer L, Nigg EA, Hoeijmakers JH, Egly JM. The MO15 cell cycle kinase is associated with the TFIIH transcription-DNA repair factor. Cell 1994; 79:1093-101; PMID:8001135; https://doi.org/10.1016/0092-8674(94)90039-6
- Kwiatkowski N, Zhang T, Rahl PB, Abraham BJ, Reddy J, Ficarro SB, Dastur A, Amzallag A, Ramaswamy S, Tesar B, et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature 2014; 511(7511):616-20; PMID:25043025; https://doi.org/10.1038/nature13393
- Nilson KA, Guo J, Turek ME, Brogie JE, Delaney E, Luse DS, Price DH. THZ1 reveals roles for Cdk7 in Co-transcriptional capping and pausing. Mol Cell 2015; 59(4):576-87; PMID:26257281; https://doi.org/10.1016/j.molcel.2015.06.032
- Christensen CL, Kwiatkowski N, Abraham BJ, Carretero J, Al-Shahrour F, Zhang T, Chipumuro E, Herter-Sprie GS, Akbay EA, Altabef A, et al. Targeting transcriptional addictions in small cell lung cancer with a covalent CDK7 inhibitor. Cancer Cell 2014; 26(6):909-22; PMID:25490451; https://doi.org/10.1016/j.ccell.2014.10.019
- Chipumuro E, Marco E, Christensen CL, Kwiatkowski N, Zhang T, Hatheway CM, Abraham BJ, Sharma B, Yeung C, Altabef A, et al. CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell 2014; 159(5):1126-39; PMID:25416950; https://doi.org/10.1016/j.cell.2014.10.024
- Calo E, Wysocka J. Modification of enhancer chromatin: what, how, and why? Mol Cell 2013; 49(5):825-37; PMID:23473601; https://doi.org/10.1016/j.molcel.2013.01.038
- Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. A phase separation model for transcriptional control. Cell 2017; 169(1):13-23; PMID:28340338; https://doi.org/10.1016/j.cell.2017.02.007
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell 2013; 155(4):934-47; PMID:24119843; https://doi.org/10.1016/j.cell.2013.09.053
- Bradner JE, Hnisz D, Young RA. Transcriptional addiction in cancer. Cell 2017; 168(4):629-43; PMID:28187285; https://doi.org/10.1016/j.cell.2016.12.013
- Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 2013; 153(2):320-34; PMID:23582323; https://doi.org/10.1016/j.cell.2013.03.036
- Pefanis E, Wang J, Rothschild G, Lim J, Kazadi D, Sun J, Federation A, Chao J, Elliott O, Liu ZP, et al. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell 2015; 161(4):774-89; PMID:25957685; https://doi.org/10.1016/j.cell.2015.04.034
- Compe E, Egly JM. Nucleotide excision repair and transcriptional regulation: TFIIH and Beyond. Annu Rev Biochem 2016; 85:265-90; PMID:27294439; https://doi.org/10.1146/annurev-biochem-060815-014857
- Freed EF, Bleichert F, Dutca LM, Baserga SJ. When ribosomes go bad: diseases of ribosome biogenesis. Mol Biosyst 2010; 6(3):481-93; PMID:20174677; https://doi.org/10.1039/b919670f
