ABSTRACT
Sub1 was initially identified as a coactivator factor with a role during transcription initiation. However, over the last years, many evidences showed that it influences processes downstream during mRNA biogenesis, such as elongation, termination, and RNAPII phosphorylation. The recent discover that Sub1 directly interacts with the RNAPII stalk adds new insights into how it achieves all these tasks.
Introduction
In eukaryotes, transcription by RNA polymerase II (RNAPII) is strictly regulated by general transcription factors, regulators, cofactors, and chromatin regulators that include histone modifiers and chromatin remodeling complexes.Citation1,2 Additionally, the interactions of the polymerase is tightly regulated by post-transcriptional modifications of the carboxy-terminal domain (CTD) of its largest subunit, Rpb1. Being, for instance, the regulation of Rpb1-CTD phosphorylation crucial for the biogenesis of mRNAs.Citation3,4
Eukaryotic RNAPII contains 12 subunits, Rpb1 to Rpb12, that in Saccharomyces cerevisiae dissociate into a 10-subunit core and a Rpb4/Rpb7 heterodimer that forms the stalk domain.Citation5 Rpb4 and Rpb7 are conserved from yeast to humans, and orthologs found in archaea also function in transcription.Citation6 Rpb4/7 participates in a broad range of activities under a variety of conditions.Citation7,8 In S. cerevisiae, Rpb4/7 is required for promoter-dependent transcription in vitro, is involved in elongation and termination,Citation8 and is important for co-transcriptional recruitment of factors required for 3′-end formation of mRNA and snoRNA genes.Citation9 Rpb4/7 may also function in mRNA quality control and translation, where it is thought to bind co-transcriptionally to nascent transcripts and promote nuclear export. It was proposed that, once in the cytoplasm, it stimulates translation initiation and subsequent deadenylation and mRNA decay.Citation10,11 Moreover, we showed that Rpb4/7 plays a key role in modulating Rpb1-CTD phosphorylation.
Among RNAPII regulators, Sub1, in S. cerevisiae, was originally characterized as a coactivator due to its homology with human Positive Coactivator 4 (PC4), its genetic and physical interaction with the general transcription factor IIB, and its capacity to activate transcription.Citation12,13 Later, in vitro assays demonstrated that in fact Sub1 is a functional component of the Preinitiation complex (PIC),Citation14 and has a role in the selection of transcription start site (TSS).Citation15 Accordingly, Sub1 is predominantly associated with gene promoters,Citation14,16 and therefore, its function has been primarily linked to processes that take place at the 5′-end region of coding genes. Nevertheless, a number of evidences showed that Sub1 also plays a significant role in transcription elongation.Citation17,18 It can be localized within coding regions in a transcription-dependent manner and affects the levels of total RNAPII associated to genes during the entire transcription cycle. Furthermore, it co-purifies with the elongation factor Spt5, influences the elongation rate and promotes splicing.Citation17 Likewise, Sub1 has been found to play an anti-terminator role at 3′-end regions, while interacting with the termination factor Rna15.Citation16,19,20 More recently, we have shown that Sub1 directly interacts with the RNAPII stalk domain formed by the Rpb4 and Rpb7 subunits, and is genetically related to the polymerase clamp domain.Citation21 This may help to explain how this multifaceted factor may influence different processes during the whole transcription cycle in addition to globally modulating Rpb1-CTD phosphorylation.Citation18 Thus, in the course of the transcription cycle, Sub1 could exert its function at different stages through the association with specific factors that directly interact with Rpb4/7 and/or the clamp domain.
Sub1 has been also described as a regulator of RNAPIII transcription. It stimulates transcription initiation and reinitiation in vitro, and it is required for optimal transcription by RNAPIII in exponentially growing cells.Citation22,23 Similarly, PC4 stimulates human RNAPIII transcription.Citation24,25 Sub1 and PC4, as ssDNA binding proteins, have been also implicated in DNA-dependent processes other than transcription, such as DNA repair, replication and in the maintenance of genome stability.Citation26-31 Here, the focus will be on the implications of Sub1 interaction with Rpb4/7 during the biogenesis of mRNAs.
Some features of Sub1 and Rpb4, keys to understand the current working model
The Rpb4/7 heterodimer is a stalk-like protrusion extending from the main body of the RNAPII complex.Citation32 Although Rpb4 is required for stable interaction of Rpb7 with the RNAPII core, most contacts between Rpb4/7 and the core complex occur between Rpb7 and the Rpb1 and Rpb6 subunits.Citation32,33 The stalk is located near the RNA exit channel and the Rpb1-CTD. Indeed, two studies showed that the nascent RNA exits making contacts with the Rpb7 subunit Citation34,35 Then, this location suggests that the stalk might play a role in the recruitment of factors important for RNA biogenesis Citation9 and/or CTD modification.Citation36
Sub1 is a polypeptide of 292 amino acids that shows strong similarity to PC4 (127 amino acids) over a 64-residue region (amino acids 41–105) that includes a ssDNA binding domain (DBD) and sequences essential for co-activator function.Citation37,38 Although Sub1 is highly related to PC4, yeast Sub1 is much larger. Specifically, Sub1 has an extra carboxy-terminal (CT) region of approximately 190 amino acids with no functions assigned up to date, but suggesting that Sub1 might have functional differences due to this additional region. Like PC4, Sub1 has the capacity to tightly bind melted DNA and ssDNA in vitro.Citation12 The DBD of PC4 and Sub1 are involved in transcriptional activationCitation12,39,40 and repression,Citation38,41 and it has been proposed that the CT region is dispensable for the functions of Sub1 related to its DNA binding capacity, such as regulation of IMD2 transcription and response to DNA damage.Citation27,41 Regardless, our studies suggest the CT region is necessary for the stability of Sub1 and it could regulate its DNA binding capacity, while interacting with Rpb4/7.Citation21
On the other hand, it has been proposed that PC4 phosphorylation negatively impacts its dsDNA binding capacity, thereby promoting its release from promoters.Citation42 Yeast Sub1 can be also phosphorylated in vitro, binding more weakly to DNA than unphosphorylated Sub1.Citation12 Interestingly, Sub1 has been identified as a phosphoprotein in proteome-wide studies, being three specific amino acids of the CT sequence the target of these phosphorylations.Citation43 If the phosphorylated form has a role in elongation, this would explain why lower levels of Sub1 associated with coding regions are usually detected.Citation14,17 Although it is unknown if Sub1 binding capacity is modulated by phosphorylation in vivo, our results indicate that it may be regulated, at least, by its extra CT region, where phosphorylated residues are placed.Citation43
Sub1 at the initiation stage
Our more recent work presents the first demonstration that Sub1 directly interacts with RNAPII through the Rpb4/7 stalk, and most likely via Rpb7, because Sub1 binds RNAPII in the absence of Rpb4. However, a fully functional Rpb4/7 heterodimer is necessary for Sub1 to stably associate with chromatin after RNAPII recruitment to the PIC.Citation21 We have proposed a model, where Sub1 is recruited to RNAPII through interaction with Rpb4/7, TFIIB, and DNA.Citation12-14,16 One hypothesis is that once at the PIC, Sub1 interacts with Rpb4/7 via its CT region to keep associated to RNAPII and chromatin (). This interaction would help to maintain Sub1 associated to gene promoters until the next step in transcription. Supporting this idea, we identified a specific genetic interaction between sub1ΔCT and rpb4Δ, but not with a mutation altering Sub1 DNA binding, and showed that the association of Sub1ΔCT with gene promoters is significantly reduced in the absence of RPB4.Citation21 This is the first evidence for a role of the Sub1-CT region.
Figure 1. Schematic model showing the hypothetical localization of Sub1 during transcription initiation. Sub1 is bound to the promoter by interacting with upstream DNA at the junction between single- and double-stranded DNACitation14 through its DNA Binding Domain (Sub1 DBD) Citation21. The proposed localization of Sub1 in this model explains the reported physical and genetic interaction of Sub1 with TFIIBCitation13,55, as well with TFIIE and TFIIH.Citation14 The intrinsically-disordered CT region of Sub1 (Sub1 CT) may extend to directly interact with the Rpb4/7.Citation21 During initiation, Rpb7 contacts with the nascent RNACitation34,35. The genetic interaction between Sub1 and Rpb1 clamp is also illustrated. In addition, the model shows the connections of Fcp1 phosphatase with Rpb4 and Sub1 revealed by our studiesCitation16,21,36that could occur during the initiation–elongation transition, and consistent with the structural dataCitation56.
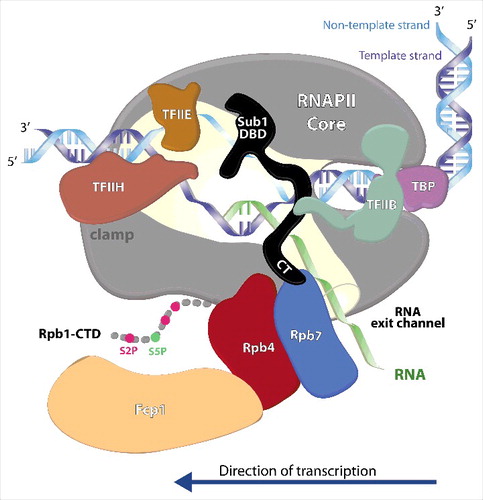
With respect to components of the PIC, a direct interaction between Sub1 and TFIIB were described and showed that Sub1 inhibited the formation of the TATA binding protein (TBP)-TFIIB-promoter complexes in vitro.Citation13 Accordingly, TFIIB is required for Sub1 recruitment to the promoters of constitutively transcribed genes,Citation40 and Sub1 mainly localizes to the promoter region in a manner dependent upon TBP.Citation14 A model proposed by S. Buratowski and collaborators Citation14 is that Sub1 is first recruited to the PIC by interactions with transcription factors, likely with TFIIB, and at that point both factors would cooperate in promoter melting. Moreover, they showed that Sub1 is located near the leading edge of the HIS4 transcription bubble. Hence, in this model, Sub1, upon promoter melting, can interact with the non-template strand or perhaps both strands at the upstream junction between single- and double-stranded DNA. Agreeing with this model, the intrinsically-disordered nature of the Sub1 CT could allow the protein to span the distance between the bubble upstream junction and the RNAPII stalk (). Additionally, genetic data also suggest that Sub1, bound to gene promoters, could help TFIIE and TFIIH to maintain the PIC in a stable, but inactive conformation in the open complex.Citation14 Comparably, it has been shown that PC4 directly associates with various transcriptional activators, for instance TFIIB and TFIIH, and weakly with TBP. In particular, PC4 participates in the initiation–elongation transition by binding to melted DNA in collaboration with TFIIE.Citation44
Considering more ideas and hypotheses, one possibility is that Sub1 would have effects over the stability of factor interactions established with the clamp domain during initiation and early elongation; for instance cooperating with TFIIE at initiation stage and with Spt5 during elongation. This hypothesis is supported by the fact that SUB1 deletion causes lethality when combined with the rpb1-L1397S mutation localized within the clamp domain,Citation21,45 and that both TFIIE and Spt5 directly interact with this domain.Citation46-49 In that sense, it is worthy to mention that during initiation, TFIIE contributes to the formation of the open complex and to the maintenance of the stability of the transcription bubble, thanks to its interaction with the clamp, whose mobility it aids to modulate.Citation46,47 In Archeae, it has been proposed that the TFIIE homologue TFE could be recruited to the transcription complex via the E/F polymerase subunits (Rpb4/7), and/or TFE could modify the conformation and function of the polymerase via E/F.Citation6 Furthermore, in these organisms, TFE competes with Spt4/5 for the access to the clamp during the initiation, but after clearance of TFE, Spt4/5, together with E/F, will contribute to the processivity of the polymerase.Citation6 Sub1 could participate in the replacement of TFIIE by Spt4/5, contributing this way to the initiation–elongation transition.Citation17
Sub1 as an elongation factor
In the course of transcription initiation, the clamp controls the entry of dsDNA to the polymerase active center and, throughout elongation it contributes to the separation of the RNA–DNA hybrid at the end of the transcription bubble. For this purpose, the clamp adopts two distinct structural configurations: open, where dsDNA accesses to the active center, and closed, where ssDNA is seated in the active center and facilitates the processivity of the polymerase.Citation50 Curiously, Rpb7 associates with the polymerase through the interaction with the clamp, restricting its configuration to the closed state.Citation32 In S. cerevisiae, Spt5 extensively interacts with the stalk, and with three other domains formed by Rpb1 and Rpb2: the clamp, protrusion, and wall.Citation48,49 These interactions may fasten the transcribing DNA onto the central cleft of the RNAPII, as the clamp is in a closed configuration. Thus, Spt5 has been proposed to be crucial for holding the RNAPII complex in a closed conformation that is highly competent for transcription elongation, favoring processivity.Citation48 In this situation, Sub1 would be recruited to the chromatin in a Rpb4/7 dependent manner, and associate with both complexes, Spt4/5 and Rpb4/7.Citation17,21
During transcription initiation, it has been proposed that changes in Sub1, such as phosphorylationCitation12,42 could reduce its DNA-binding capacity and facilitate promoter clearance.Citation12-14 During the transition to elongation, Sub1, most likely via its CT, would remain attached to RNAPII through Rpb7,Citation21 thus joining the elongation complex to influence transcription elongation.Citation17 This is likely achieved by stabilizing Spt5-Rpb1 association with DNA, because in the absence of Sub1, Spt5-Rpb1 interaction decreases.Citation17 Accordingly, as mentioned above, sub1Δ is synthetically lethal with rpb1-L1397S, Citation21 and also enhances growth phenotypes of the spt5–194 mutant that alters Spt5 binding to the clamp. In contrast, SUB1 overexpression suppresses these phenotypes.Citation17 Thus, we proposed that Sub1 plays a role in the stabilization of the clamp domain, and it is essential when the function of the clamp is altered, as in the rpb1-L1397S and spt5–194 mutants.Citation17,21 In fact, deletion of SUB1 has no effect on Spt5 association with chromatin,Citation17 while RPB4 deletion affects both Sub1Citation21 and Spt5 occupancy (unpublished data). Hence, Sub1 interactions with Rpb4/7 and Spt4/5 may concurrently promote closed complex conformation and thus facilitating processivity and transcription elongation ().
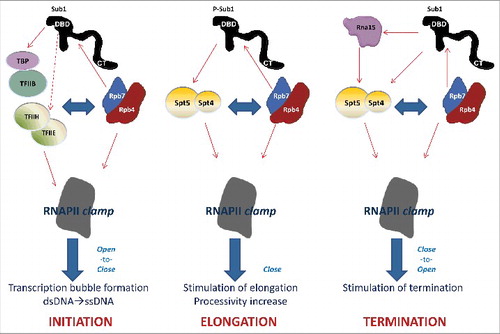
Figure 2. A working model: Sub1 function is linked to the stalk and clamp domains, assisting transcription and processing factors during the whole transcription cycle. See the text for detail. Continuous-line arrows indicate physical interaction/co-purification while discontinuous lines indicate genetic interaction.
A controversial point: Initiation versus elongation
So far, we have focused on the idea that the biological meaning of rpb1-L1397S sub1Δ synthetic lethality is directly linked to participation of Sub1 in elongation. It is instead possible this genetic interaction could be explained as a direct consequence of Sub1 role in initiation. In fact, Sub1 is involved in TSS selection,Citation15 and its association with coding region significantly decreased with respect to promoters. Besides, in rpb1-L1397S cells, IMD2, and URA2 are upregulated, which is a hallmark of a defect on TSS selection.Citation45 However, several facts support that Sub1s role in elongation is linked to the clamp function: (1) the clamp domain is essential for RNAPII processivity, and genome wide association of RNAPII with coding regions is reduced in rpb1-L1397S cells.Citation45 (2) Sub1 promotes elongation rate and splicing, in association with Spt5,Citation15,16 which in turn interacts with the clamp.Citation48,49 Indeed, Sub1 effect on splicing was corroborated in a nice study that also showed that Sub1 is involved TSS selection.Citation15 In this study, two groups of RNAPII mutants were identified: one that preferentially initiates upstream and exhibits an increased rate of transcription and other initiating downstream and transcribing slowly. They showed that sub1Δ exacerbated slow RNAPII alleles and partially suppressed fast RNAPII alleles. They revealed that fast RNAPII mutations resulted in upstream transcription start and diminished splicing, whereas slow mutations or SUB1 deletion gave rise to downstream transcription start and enhanced splicing. More interestingly, they showed that only deletion of SUB1, and not the sua7–3 mutation, that also exhibits defects on TSS selection, correlates with splicing defects, as do slow and fast RNAPII mutants. Thus, the catalytic rate of RNAPII has effects on TSS selection. Therefore, Sub1 effects on TSS and elongation could be also due to its role influencing RNAPII rate,Citation17 which is closed related to the clamp domain function. Consistently, both, the speed of the polymerase and Sub1, impact on the selection of the TSS, splicing, and transcription elongationCitation15,17. Furthermore, we have also shown that, though in sub1Δ the IMD2 gene is upregulated, there is also a decrease in de novo synthesis of IMD2 transcript due to defects on elongation Citation17 and this may be the case for rpb1-L1397S. In fact, this mutant is sensitive to 6AU.Citation45 In any case, we cannot discard the possibility that the lethality of the sub1Δ rpb1-L1397S double mutant is a consequence of defects on transcription start site selection.
Sub1 participates in transcription termination
Rpb4 contributes to co-transcriptional recruitment of 3′-end processing factors,Citation9 and in Archea, E/F stimulates transcription termination dependent on U-rich sequences.Citation6 The location of Rpb4/7 near the CTD Citation32 and the fact that Rpb7 contact the emerging RNA Citation34,35 let us and othersCitation9,36 to propose that the stalk is in a perfect position to stabilize interactions with 3′-end processing factors, the CTD and the polyadenylation site sequences within the nascent transcript. Again, S. Buratowski studiesCitation9 suggested that such stabilization could be mediated by a direct interaction of Rpb4/7 with Rna15, a component of the cleavage factor I (CFI), involved in termination and polyadenylation, with the RNA and with an intermediary factor. Could be this intermediary factor Sub1? Accordingly, on one hand, Sub1 interacts with Rna15, and this interaction is evolutionary conserved in human cells. Strong evidences suggested that Sub1 could exert an anti-terminator function, avoiding premature termination of pre-mRNAs while inhibiting Rna15 function.Citation19 Furthermore, Sub1 directly interacts with Rpb7,Citation51 which in turns contacts the nascent RNA.Citation34,35 Hence, Sub1 is properly placed to influence pre-mRNA processing, while interacting with the stalk. On the other hand, Rpb4 and Spt4/5 are required for Rna15 recruitment.Citation9,19,20,52 Because in the absence of Sub1, the Spt5-RNAPII interaction is unstable, it might be that the recruitment and function of the CFI complex would be altered. Then, we can visualize a model for Sub1 role during transcription termination that could imply a new change in the conformation of Sub1 and its localization within the transcription complex or even Sub1 exit. Thus, for instance, its dephosphorylation, coincident with RNAPII dephosphorylation by Fcp1, could relieve the inhibition over Rna15, allowing termination.Citation19 Simultaneously, this dephosphorylation could promote Sub1 exit from the complex, destabilizing Spt4/5-RNAPII interaction and bring about transcription termination.Citation52 Hence, Sub1 would not only contribute to the initiation–elongation transition, but also to the elongation–termination transition, participating in transcription regulation from PIC formation to transcription termination.
New insights to understand how Sub1 modulates Rpb1-CTD phosphorylation
The location of Rpb4/7 within the RNAPII complex near Rpb1-CTD hinted that it may play a role in the modulation of its modifications.Citation32 Indeed, we earlier demonstrated that Rpb4/7, as well as Sub1, have a role in regulating RNAPII CTD phosphorylation levels.Citation18,36 Rpb4 modulates the functionality of Fcp1,Citation36 and Sub1 associates with Fcp1, and is required to maintain proper Fcp1 protein levels.Citation16 Interestingly, we know now that a full length CTD is a requisite for Sub1 association with RNAPII and with chromatin.Citation21 Altogether, our data provide significant new insight into the relationship among Sub1, Fcp1, Rpb4/7, and the modulation of Rpb1-CTD phosphorylation at least during elongation and termination, which crucially regulates the biogenesis of mRNAs, and RNAPII recycling.Citation3,4
A model for Sub1 role during the whole transcription cycle
The data accumulated over the years about Sub1 indicate that Sub1 binds DNA through its rminal DBD, while is stably maintained in the RNAPII-DNA complex by binding to Rpb4/7 via its CT region (). In addition, Sub1 interaction with the stalk and polymerase clamp domains may help to explain how it may influence TSS selectionCitation15 and transcription elongation rate.Citation17 Similarly, Sub1 association with Rpb4/7 and Spt4/5 offers an insight into Sub1 role during transcription termination; and its localization within RNAPII helps to understand how it may influence Rpb1-CTD phosphorylation. Consistent with all that data, a working model for Sub1 function, linked to Rpb4/7 and the configuration of the clamp (), could be the following: During initiation, Sub1 is recruited to promoters by TFIIB and Rpb4/7, where it helps to maintain the PIC in a stable but inactive conformation (open), for instance collaborating with TFIIE and TFIIH.Citation14,18 Here, Sub1 binds DNA through its N-terminal DBD, while is stably maintained in the RNAPII-DNA complex by binding to Rpb4/7 via its CT region (). In this situation, Sub1 could be near the emerging RNA, which make contacts with Rpb7.Citation34,35 After PIC activation, the transcription complex undergoes a conformational change that allows TFIIE exit, and its replacement by Spt4/5, which binds to the clamp and Rpb4/7, assuring the polymerase closed conformation and promoting processivity. At this point, RNAPII is being phosphorylated and transcription starts.Citation3,4 Then, during elongation Sub1 would interact with Spt4/5 (maybe both as phosphoproteinsCitation42,43,53), stabilizing Spt5-Rpb1 association with DNA. This could help to decrease the time that this complex is paused immediately after the initiation–elongation transition and will positively influence the transcription elongation rate.Citation17 Moreover, extensive and specific phosphorylation events targeting RNAPII,Citation3,4 Spt5,Citation53 and Sub1 (P-Sub1, ) Citation12,42,43,54 could take place simultaneously and ultimately regulate the initiation–elongation transition and, therefore, transcription elongation. Finally, during termination, Sub1 and Spt4/5 would promote, together with Rpb4/7 the activity of termination and 3′-end processing factors, such as CFI (for instance, Rna15),Citation9,16,20,52 as well as clamp opening and transcription termination. One possibility is that Sub1 dephosphorylation and/or Sub1-associated factors would trigger its dissociation from Spt4/5 and Rna15, favoring transcription termination and 3′-end processing.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
I thank all people from my lab that contribute to the knowledge of Sub1 functions, and especially to M. Garavís. I also thank C. Fernández-Tornero for its support over the last years; and I would like to express my gratitude to S. Buratowski for reading this manuscript, for his comments, and for his always invaluable help.
Funding
O.C. acknowledges the Spanish Ministry of Economy and Competitiveness (MINECO; [BFU2013–48374-P]) for funding.
References
- Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol 2006; 41:105-178; PMID:16858867; https://doi.org/10.1080/10409230600648736
- Hahn S, Young ET. Transcriptional regulation in Saccharomyces cerevisiae: transcription factor regulation and function, mechanisms of initiation, and roles of activators and coactivators. Genetics 2011; 189:705-736; PMID:22084422; https://doi.org/10.1534/genetics.111.127019
- Buratowski S. The CTD code. Nat Struct Biol 2003; 10:679-680; PMID:12942140; https://doi.org/10.1038/nsb0903-679
- Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell 2009; 36:541-546; PMID:19941815; https://doi.org/10.1016/j.molcel.2009.10.019
- Cramer P. RNA polymerase II structure: from core to functional complexes. Curr Opin Genet Dev 2004; 14:218-226; PMID:15196470; https://doi.org/10.1016/j.gde.2004.01.003
- Grohmann D, Werner F. Cycling through transcription with the RNA polymerase F/E (RPB4/7) complex: structure, function and evolution of archaeal RNA polymerase. Res Microbiol 2011; 162:10-18; PMID:20863887; https://doi.org/10.1016/j.resmic.2010.09.002
- Choder M. Rpb4 and Rpb7: subunits of RNA polymerase II and beyond. Trends Biochem Sci 2004; 29:674-681; PMID:15544954; https://doi.org/10.1016/j.tibs.2004.10.007
- Sharma N, Kumari R. Rpb4 and Rpb7: multifunctional subunits of RNA polymerase II. Crit Rev Microbiol 2013; 39:362-372; PMID:22917057; https://doi.org/10.3109/1040841X.2012.711742
- Runner VM, Podolny V, Buratowski S. The Rpb4 subunit of RNA polymerase II contributes to cotranscriptional recruitment of 3′ processing factors. Mol Cell Biol 2008; 28:1883-1891; PMID:18195044; https://doi.org/10.1128/MCB.01714-07
- Dahan N, Choder M. The eukaryotic transcriptional machinery regulates mRNA translation and decay in the cytoplasm. Biochim Biophys Acta 2013; 1829:169-173; PMID:22982191; https://doi.org/10.1016/j.bbagrm.2012.08.004
- Harel-Sharvit L, Eldad N, Haimovich G, Barkai O, Duek L, Choder M. RNA polymerase II subunits link transcription and mRNA decay to translation. Cell 2010; 143:552-563; PMID:21074047; https://doi.org/10.1016/j.cell.2010.10.033
- Henry NL, Bushnell DA, Kornberg RD. A yeast transcriptional stimulatory protein similar to human PC4. J Biol Chem 1996; 271:21842-21847; PMID:8702984; https://doi.org/10.1074/jbc.271.36.21842
- Knaus R, Pollock R, Guarente L. Yeast SUB1 is a suppressor of TFIIB mutations and has homology to the human co-activator PC4. EMBO J 1996; 15:1933-1940; PMID:8617240
- Sikorski TW, Ficarro SB, Holik J, Kim T, Rando OJ, Marto JA, Buratowski S. Sub1 and RPA associate with RNA polymerase II at different stages of transcription. Mol Cell 2011; 44:397-409; PMID:22055186; https://doi.org/10.1016/j.molcel.2011.09.013
- Braberg H, Jin H, Moehle EA, Chan YA, Wang S, Shales M, Benschop JJ, Morris JH, Qiu C, Hu F, et al. From structure to systems: high-resolution, quantitative genetic analysis of RNA polymerase II. Cell 2013; 154:775-788; PMID:23932120; https://doi.org/10.1016/j.cell.2013.07.033
- Calvo O, Manley JL. The transcriptional coactivator PC4/Sub1 has multiple functions in RNA polymerase II transcription. EMBO J 2005; 24:1009-1020; PMID:15692559; https://doi.org/10.1038/sj.emboj.7600575
- Garcia A, Collin A, Calvo O. Sub1 associates with Spt5 and influences RNA polymerase II transcription elongation rate. Mol Biol Cell 2012; 23:4297-4312; PMID:22973055; https://doi.org/10.1091/mbc.E12-04-0331
- Garcia A, Rosonina E, Manley JL, Calvo O. Sub1 globally regulates RNA polymerase II C-terminal domain phosphorylation. Mol Cell Biol 2010; 30:5180-5193; PMID:20823273; https://doi.org/10.1128/MCB.00819-10
- Calvo O, Manley JL. Evolutionarily conserved interaction between CstF-64 and PC4 links transcription, polyadenylation, and termination. Mol Cell 2001; 7:1013-1023; PMID:11389848; https://doi.org/10.1016/S1097-2765(01)00236-2
- Calvo O, Manley JL. Strange bedfellows: polyadenylation factors at the promoter. Genes Dev 2003; 17:1321-1327; PMID:12782649; https://doi.org/10.1101/gad.1093603
- Garavis M, Gonzalez-Polo N, Allepuz-Fuster P, Louro JA, Fernandez-Tornero C, Calvo O. Sub1 contacts the RNA polymerase II stalk to modulate mRNA synthesis. Nucleic Acids Res 2017; 45:2458-2471; PMID:27924005; https://doi.org/10.1093/nar/gkw1206
- Tavenet A, Suleau A, Dubreuil G, Ferrari R, Ducrot C, Michaut M, Aude JC, Dieci G, Lefebvre O, Conesa C, et al. Genome-wide location analysis reveals a role for Sub1 in RNA polymerase III transcription. Proc Natl Acad Sci USA 2009; 106:14265-14270; PMID:19706510; https://doi.org/10.1073/pnas.0900162106
- Acker J, Nguyen NT, Vandamme M, Tavenet A, Briand-Suleau A, Conesa C. Sub1 and Maf1, two effectors of RNA polymerase III, are involved in the yeast quiescence cycle. PLoS One 2014; 9:e114587; PMID:25531541; https://doi.org/10.1371/journal.pone.0114587
- Wang Z, Roeder RG. DNA topoisomerase I and PC4 can interact with human TFIIIC to promote both accurate termination and transcription reinitiation by RNA polymerase III. Mol Cell 1998; 1:749-757; PMID:9660958; https://doi.org/10.1016/S1097-2765(00)80074-X
- Mertens C, Roeder RG. Different functional modes of p300 in activation of RNA polymerase III transcription from chromatin templates. Mol Cell Biol 2008; 28:5764-5776; PMID:18644873; https://doi.org/10.1128/MCB.01262-07
- Mortusewicz O, Evers B, Helleday T. PC4 promotes genome stability and DNA repair through binding of ssDNA at DNA damage sites. Oncogene 2016; 35:761-770; PMID:25961912; https://doi.org/10.1038/onc.2015.135
- Yu L, Ma H, Ji X, Volkert MR. The Sub1 nuclear protein protects DNA from oxidative damage. Mol Cell Biochem 2016; 412:165-171; PMID:26708217; https://doi.org/10.1007/s11010-015-2621-x
- Lopez CR, Singh S, Hambarde S, Griffin WC, Gao J, Chib S, Yu Y, Ira G, Raney KD, Kim N. Yeast Sub1 and human PC4 are G-quadruplex binding proteins that suppress genome instability at co-transcriptionally formed G4 DNA. Nucleic Acids Res 2017; 45:5850–5862; PMID:28369605; https://doi.org/10.1093/nar/gkx201
- Gao J, Zybailov BL, Byrd AK, Griffin WC, Chib S, Mackintosh SG, Tackett AJ, Raney KD. Yeast transcription co-activator Sub1 and its human homolog PC4 preferentially bind to G-quadruplex DNA. Chem Commun 2015; 51:7242-7244; PMID:25813861; https://doi.org/10.1039/C5CC00742A
- Lada AG, Kliver SF, Dhar A, Polev DE, Masharsky AE, Rogozin IB, Pavlov YI. Disruption of transcriptional coactivator sub1 leads to genome-wide re-distribution of clustered mutations induced by APOBEC in active yeast genes. PLoS Genet 2015; 11:e1005217; PMID:25941824; https://doi.org/10.1371/journal.pgen.1005217
- Yu L, Volkert MR. Differential requirement for SUB1 in chromosomal and plasmid double-strand DNA break repair. PLoS One 2013; 8:e58015; PMID:23554872; https://doi.org/10.1371/journal.pone.0058015
- Armache KJ, Mitterweger S, Meinhart A, Cramer P. Structures of complete RNA polymerase II and its subcomplex, Rpb4/7. J Biol Chem 2005; 280:7131-7134; PMID:15591044; https://doi.org/10.1074/jbc.M413038200
- Bushnell DA, Kornberg RD. Complete, 12-subunit RNA polymerase II at 4.1-A resolution: implications for the initiation of transcription. Proc Natl Acad Sci USA 2003; 100:6969-6973; PMID:12746498; https://doi.org/10.1073/pnas.1130601100
- Chen CY, Chang CC, Yen CF, Chiu MT, Chang WH. Mapping RNA exit channel on transcribing RNA polymerase II by FRET analysis. Proc Natl Acad Sci USA 2009; 106:127-132; PMID:19109435; https://doi.org/10.1073/pnas.0811689106
- Ujvari A, Luse DS. RNA emerging from the active site of RNA polymerase II interacts with the Rpb7 subunit. Nat Struct Mol Biol 2006; 13:49-54; PMID:16327806; https://doi.org/10.1038/nsmb1026
- Allepuz-Fuster P, Martinez-Fernandez V, Garrido-Godino AI, Alonso-Aguado S, Hanes SD, Navarro F, Calvo O. Rpb4/7 facilitates RNA polymerase II CTD dephosphorylation. Nucleic Acids Res 2014; 42:13674-13688; PMID:25416796; https://doi.org/10.1093/nar/gku1227
- Brandsen J, Werten S, van der Vliet PC, Meisterernst M, Kroon J, Gros P. C-terminal domain of transcription cofactor PC4 reveals dimeric ssDNA binding site. Nat Struct Biol 1997; 4:900-903; PMID:9360603; https://doi.org/10.1038/nsb1197-900
- Werten S, Stelzer G, Goppelt A, Langen FM, Gros P, Timmers HT, Van der Vliet PC, Meisterernst M. Interaction of PC4 with melted DNA inhibits transcription. EMBO J 1998; 17:5103-5111; PMID:9724646; https://doi.org/10.1093/emboj/17.17.5103
- Ge H, Roeder RG. Purification, cloning, and characterization of a human coactivator, PC4, that mediates transcriptional activation of class II genes. Cell 1994; 78:513-523; PMID:8062391; https://doi.org/10.1016/0092-8674(94)90428-6
- Rosonina E, Willis IM, Manley JL. Sub1 functions in osmoregulation and in transcription by both RNA polymerases II and III. Mol Cell Biol 2009; 29:2308-2321; PMID:19204085; https://doi.org/10.1128/MCB.01841-08
- Koyama H, Sumiya E, Nagata M, Ito T, Sekimizu K. Transcriptional repression of the IMD2 gene mediated by the transcriptional co-activator Sub1. Genes Cells 2008; 13:1113-1126; PMID:18823333; https://doi.org/10.1111/j.1365-2443.2008.01229.x
- Jonker HR, Wechselberger RW, Pinkse M, Kaptein R, Folkers GE. Gradual phosphorylation regulates PC4 coactivator function. FEBS J 2006; 273:1430-1444; PMID:16689930; https://doi.org/10.1111/j.1742-4658.2006.05165.x
- Smolka MB, Albuquerque CP, Chen SH, Zhou H. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc Natl Acad Sci USA 2007; 104:10364-10369; PMID:17563356; https://doi.org/10.1073/pnas.0701622104
- Akimoto Y, Yamamoto S, Iida S, Hirose Y, Tanaka A, Hanaoka F, Ohkuma Y. Transcription cofactor PC4 plays essential roles in collaboration with the small subunit of general transcription factor TFIIE. Genes Cells 2014; 19:879-890; PMID:25308091; https://doi.org/10.1111/gtc.12187
- Kwapisz M, Wery M, Despres D, Ghavi-Helm Y, Soutourina J, Thuriaux P, Lacroute F. Mutations of RNA polymerase II activate key genes of the nucleoside triphosphate biosynthetic pathways. EMBO J 2008; 27:2411-2421; PMID:18716630; https://doi.org/10.1038/emboj.2008.165
- Chen HT, Warfield L, Hahn S. The positions of TFIIF and TFIIE in the RNA polymerase II transcription preinitiation complex. Nat Struct Mol Biol 2007; 14:696-703; PMID:17632521; https://doi.org/10.1038/nsmb1272
- Naji S, Grunberg S, Thomm M. The RPB7 orthologue E' is required for transcriptional activity of a reconstituted archaeal core enzyme at low temperatures and stimulates open complex formation. J Biol Chem 2007; 282:11047-11057; PMID:17311916; https://doi.org/10.1074/jbc.M611674200
- Li W, Giles C, Li S. Insights into how Spt5 functions in transcription elongation and repressing transcription coupled DNA repair. Nucleic Acids Res 2014; 42:7069-7083; PMID:24813444; https://doi.org/10.1093/nar/gku333
- Martinez-Rucobo FW, Sainsbury S, Cheung AC, Cramer P. Architecture of the RNA polymerase-Spt4/5 complex and basis of universal transcription processivity. EMBO J 2011; 30:1302-1310; PMID:21386817; https://doi.org/10.1038/emboj.2011.64
- Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 Å resolution. Science 2001; 292:1863-1876; PMID:11313498; https://doi.org/10.1126/science.1059493
- Garavis M, Gonzalez-Polo N, Allepuz-Fuster P, Louro JA, Fernandez-Tornero C, Calvo O. Sub1 contacts the RNA polymerase II stalk to modulate mRNA synthesis. Nucleic Acids Res 2017; 45:2458-2471; PMID:27924005; https://doi.org/10.1093/nar/gkw1206
- Mayer A, Schreieck A, Lidschreiber M, Leike K, Martin DE, Cramer P. The spt5 C-terminal region recruits yeast 3′ RNA cleavage factor I. Mol Cell Biol 2012; 32:1321-1331; PMID:22290438; https://doi.org/10.1128/MCB.06310-11
- iu Y, Warfield L, Zhang C, Luo J, Allen J, Lang WH, Ranish J, Shokat KM, Hahn S. Phosphorylation of the transcription elongation factor Spt5 by yeast Bur1 kinase stimulates recruitment of the PAF complex. Mol Cell Biol 2009; 29:4852-4863; PMID:19581288; https://doi.org/10.1128/MCB.00609-09
- Malik S, Guermah M, Roeder RG. A dynamic model for PC4 coactivator function in RNA polymerase II transcription. Proc Natl Acad Sci USA 1998; 95:2192-2197; PMID:9482861; https://doi.org/10.1073/pnas.95.5.2192
- Wu WH, Pinto I, Chen BS, Hampsey M. Mutational analysis of yeast TFIIB. A functional relationship between Ssu72 and Sub1/Tsp1 defined by allele-specific interactions with TFIIB. Genetics 1999; 153:643-652; PMID:10511545
- Kamenski T, Heilmeier S, Meinhart A, Cramer P. Structure and mechanism of RNA polymerase II CTD phosphatases. Mol Cell 2004; 15:399-407; PMID:15304220; https://doi.org/10.1016/j.molcel.2004.06.035
