ABSTRACT
While structures of the RNA polymerase (Pol) II initiation complex have been resolved and extensively studied, the Pol I initiation complex remained elusive. Here, we review the recent structural analyses of the yeast Pol I transcription initiation complex that reveal several unique and unexpected Pol I-specific properties.
Introduction
Eukaryotes encode at least three multisubunit RNA polymerases (Pols I-III) that synthesize RNA from DNA templates [Citation1]. Pol I is the most specialized as it exclusively transcribes ribosomal DNA (rDNA) into ribosomal RNA (rRNA) that is processed into the structural and catalytic components of the ribosome [Citation1,Citation2]. High-resolution structures of the large and dynamic Pol II initiation complexes have been solved with the help of technical advances in cryo-EM, but until recently, similar structures of the Pol I initiation complex remained elusive. Given that the Pols share an evolutionarily conserved core and a set of conserved initiation factors, it was anticipated that the Pol I initiation complex may resemble that of the Pol II system [Citation1]. However, these recent Pol I structures reveal an entirely new paradigm for eukaryotic Pol initiation factor architecture that breaks the mold of how initiation complexes assemble and interact with promoter DNA [Citation3–Citation5]. In this point of view, we summarize recent progress in understanding the molecular architecture of the yeast Pol I initiation complex.
Core Factor structure
Core Factor (CF) is a central player at the heart of Pol I transcription and recent structures reveal a novel architecture that is directly involved in many key steps of the initiation process. Transcription initiation can be subdivided into the formation of a pre-initiation complex (PIC), closed complex (CC), open complex (OC), and initially transcribing complex (ITC) [Citation6]. Efficient transcription of rDNA by Pol I relies on the recruitment of transcription factors to form the PIC that includes upstream activating factor (UAF), CF, TATA-box binding protein (TBP) and Rrn3 [Citation2,Citation7]. Once formed, the PIC transitions into the early initiation phases starting with the CC where the rDNA is double stranded to the OC where DNA near the transcription start site is melted, and finally the ITC where nascent RNA is first synthesized before entering the elongation phase [Citation6,Citation8].
CF is a heterotrimer comprised of two Pol I-specific subunits, Rrn6 and Rrn11, and a TFIIB-like factor, Rrn7 [Citation9,Citation10]. Each CF subunit contains unique structural elements. Rrn6 contains a WD40 β-propeller domain, known for coordinating protein complex assembly, and a helical headlock domain followed by a lysine-rich CTD [Citation3,Citation9]. Rrn11 contains a disordered, yet essential, N-terminal domain (NTD) and a tetratricopeptide repeat (TPR) domain that is also known for mediating protein-protein interactions [Citation3,Citation9]. Finally, Rrn7 contains an N-terminal zinc ribbon (ZR) domain and linker region followed by two cyclin fold domains where a unique insertion is found in the C-terminal cyclin fold [Citation9,Citation10]. The crystal structure of the CF complex is subdivided into two modules linked by a central hinge region [Citation3]. Module I includes Rrn11 and the Rrn6 β-propeller domain whereas module II includes Rrn7 and the Rrn6 C-terminal helical headlock domain. Surprisingly, the Rrn7 cyclin folds, although similar in structure, have an altered rotational axis compared to TFIIB that is likely due to its unique interaction with the rDNA promoter [Citation3–Citation5].
Polymerase interaction
TFIIB makes two important contact points with Pol II [Citation11]. For instance in Pol II, the TFIIB ZR and N-terminal cyclin domain contact the dock domain and the periphery of the wall domain, respectively [Citation11]. In the Pol I system, Rrn7 seems to have a minor role in Pol I interaction compared to TFIIB and Brf1. CF contains three Pol I interacting regions (PIRs) with Pol I-Rrn3 () [Citation3]. The Rrn7 N-terminal ZR domain, designated PIR I, contacts the Pol I dock domain, similar to TFIIB, in addition to making contact with Rrn3 [Citation3,Citation5]. PIR II consists of the essential Rrn11 C-terminal TPR domain, which contacts the Pol I clamp and protrusion [Citation3–Citation5]. Finally, PIR III is found in the Rrn7 C-terminal cyclin fold insertion domain, which contacts the Pol I wall domain of A135 [Citation3,Citation4]. Interestingly, previous mutational analysis showed that this region in Rrn7 is not essential for yeast growth [Citation9], so it is possible that PIR III is less important for CF-Pol I interaction. However, the Rrn11 TPR domain that encompasses PIR II is essential for cell viability [Citation9], likely due its importance in Rrn6 and Pol I interaction [Citation3,Citation5]. Contact between CF and Pol I is mainly mediated by a small and essential interaction surface of CF subunit Rrn11 [Citation3–Citation5,Citation9]. Additional studies will be necessary to understand the contribution of each PIR to understand their individual and collective importance in Pol I binding and initiation.
Figure 1. CF contacts with Pol I and the rDNA promoter. CF-rDNA (cyan and blue) interactions are primarily mediated through backbone interactions of Rrn7 cyclin folds and the Rrn11 NTD with the promoter from positions -27 to -20 and -24 to -16, respectively. Rrn6 acts as a scaffold for CF complex. PIR I: Rrn7 N-terminal zinc ribbon domain contacts the Pol I dock and within Rrn3 HEAT repeats 4–5, residues 1–39. PIR II: C-terminal TPR domain of Rrn11 contacts the Pol I clamp and protrusion, residues 265–440. PIR III: Rrn7 insertion domain contacts the Pol I wall of A135, residues 405–415.
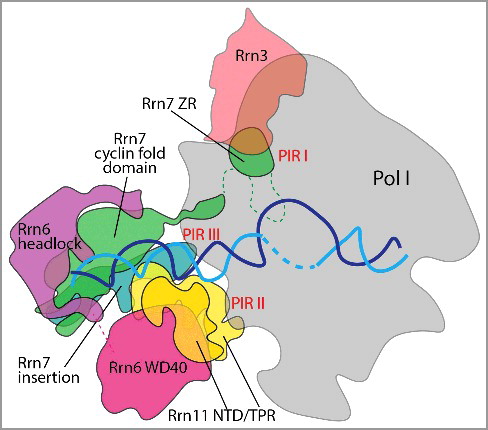
Rrn7 has unique characteristics that set it apart from its TFIIB paralog. While the Rrn7 ZR occupies a similar position as the TFIIB B-reader, the cyclin domains adopt a different position [Citation3–Citation5]. The Rrn7 N-terminal cyclin domain is buried within module II of CF, relatively far away from Pol I, whereas its Pol II counterpart TFIIB contacts the wall domain of the polymerase [Citation3]. The Rrn7 cyclin domains lack an interface with Pol I, different from their TFIIB counterparts, but engage the promoter DNA [Citation3–Citation5].
The Rrn6 C-terminus, the majority of which is not resolved in the CF crystal structure, is necessary for Rrn3 interaction in yeast two-hybrid studies [Citation3,Citation12]. A striking feature of the Pol I PIC structures is the distance between CF and Rrn3. It is noted in negative stain reconstructions of the Pol I-Rrn3-CF structure that additional density is present that lies between CF and Rrn3 [Citation3]. It is possible that this density may represent the lysine-rich Rrn6 CTD that is non-essential for yeast growth or the disordered Rrn3 N-terminus [Citation9].
In the absence of promoter DNA, CF module I contacts the upstream end of the Pol I cleft while module II projects outward and is mobile as demonstrated by its ability to adopt various positions in EM reconstructions [Citation3]. Given the positioning of CF in the structure, as it is shown to block the upstream end of the Pol I cleft, major structural changes must occur for DNA to be loaded into the Pol I active center. Indeed, when a nucleic acid scaffold was added to the cryo-EM experiments, CF adopted a different position than when observed with just the Pol I-Rrn3-CF complex [Citation3]. A movement of 90Å and rotation of up to 60° allows the CF structure to open and contact Pol I between the protrusion and wall domains [Citation3]. The rotation of CF facilitates loading of the promoter DNA into the Pol I cleft while PIRs are maintained [Citation3]. This is consistent with other recent work that describes the movement of CF during PIC formation [Citation4].
In the presence of promoter DNA, CF and DNA were found to have a rotational movement up to 10° that allow them to adopt a variety of conformational states on Pol I where State 1, 2, and 3 depict distinct orientations of CF during Pol I transcription initiation [Citation4]. State 1 reveals CF and DNA rotated towards the back of Pol I placing the upstream DNA away from the active site. In State 2, the stabilization of the Rrn7 ZR on Pol I corresponds to the movement of CF and DNA [Citation4]. CF adopts a more intermediate position in State 3 where strong density was observed near the Pol I wall domain corresponding to the A49 tandem winged helix (tWH) domain [Citation4].
A49 forms heterodimer with A34.5 that act as a built-in elongation factor related to TFIIF [Citation13]. Resolving the A49 linker domain structure, which connects the A49/A34.5 dimerization interface to the A49 tWH, has been attempted with little success [Citation13]. In recent structures, both the A49 tWH and the A49 linker domain were resolved and shown to interact with template DNA and Pol I, respectively [Citation4]. The A49 tWH contacts upstream DNA while the linker domain spans the active site cleft and contacts the Pol I clamp coiled-coil [Citation4]. Strikingly, the position of the Rrn7 linker domain and the A49 tWH clash between State 2 and 3 [Citation4]. Together, these structures suggest that A49 might displace the Rrn7 linker, opening the RNA exit channel and CF pre-conditions Pol I in an elongation-competent state during initiation [Citation4]. Although A49 is non-essential, its deletion drastically impairs cell growth [Citation14], and furthermore, mutations within the tWH exhibit 6-azauracil and mycophenolic acid sensitivity that are indicative of defects in transcription elongation [Citation15]. A49 tWH mutants also show reduced Pol I occupancy at the rDNA locus and less recruitment of Pol I and Rrn3 at the promoter further supporting a direct role for A49 in transcription initiation [Citation15].
Promoter recognition
The rDNA promoter region is bipartite comprising an upstream activation sequence and core element in which UAF and CF bind, respectively [Citation2,Citation7]. Collectively, recent structures demonstrate that CF primarily engages the rDNA promoter via two subunits Rrn7 and Rrn11, while Rrn6 has more of a scaffolding role in assembly of the complex and the PIC [Citation3–Citation5]. CF bound to rDNA is thought to resemble a right hand holding the rDNA between the palm and fingers [Citation4]. In this context, the thumb pointing towards Pol I is composed of the Rrn11 C-terminus, the palm consists of the N-terminal regions of Rrn11 and Rrn6, the fingers are composed of Rrn7, and knuckles are the Rrn6 C-terminus [Citation4].
Rrn7 and Rrn11 mediate rDNA promoter interactions mainly through backbone contacts from positions -27 to -20 and -24 to -16, respectively [Citation3–Citation5]. The Rrn7 C-terminal cyclin fold domain contacts the major groove at position -27, which is likely a critical interaction as the PIC fails to assemble with DNA template lacking the -27 nucleotide base [Citation4].
Rrn11 uses three of the four helices within the NTD and helice 5 of the TPR domain to interact with the DNA backbone near position -16 [Citation3–Citation5]. It is worth noting that majority the Rrn11 TPR domain is important for cell viability and complex integrity [Citation9]. More importantly, mutation of the Rrn11 NTD is lethal yet CF complex integrity is retained, consistent with its role in promoter engagement [Citation9]. Pol I also makes contacts with the rDNA sequence downstream of the promoter via Rpb5, the lobe domain of A135 and the clamp head, cleft and jaw domains of A190, similar to DNA contacts found in Pol II initiation complexes [Citation3–Citation5]. However, Pol I contacts the promoter near position -10 where the DNA lies between the wall and protrusion, which is different from Pol II where DNA is located above the cleft [Citation3–Citation5].
The path of the rDNA promoter is fundamentally different from the path of DNA in the Pol II system. Two kinks were identified in the rDNA at position -21 of ∼35° and position -16 of ∼45° that presumably form as a result of protein-DNA interactions [Citation3–Citation5]. In the Pol II system, TBP is responsible for a 90° bend that projects upstream DNA toward the side of Pol II [Citation16]. In the Pol I PIC, the orientation of the rDNA promoter more closely resembles the path of DNA found in the elongating Pol I structures, which further pre-conditions Pol I for a more efficient transition from initiation to elongation [Citation17,Citation18].
An insightful observation noted that the rDNA promoter naturally bends within the core element [Citation3,Citation19]. Intriguingly, DNA bending prediction analysis of the rDNA promoter shows that the essential -27 position lies at the apex of the DNA bend [Citation3,Citation19,Citation20]. It is speculated that CF may recognize DNA structure rather than a specific sequence, which is in line with its primarily backbone mechanism of rDNA interaction [Citation3,Citation19]. Furthermore, this may help explain the enigmatic observation that rDNA promoters lack sequence conservation across species, and are relatively unaffected by single base substitutions [Citation3,Citation21]. However, it is also noted that Pol I factors evolve at a faster rate compared to their Pol II and III counterparts, which may also contribute to the lack of conservation of the rDNA promoter and the Pol I factors that bind them [Citation22]. Given that multiple domains of CF and Pol I contact DNA, the importance of specific sequences in Pol I promoter interaction may be masked and could be revealed when domains are analyzed in isolation.
Promoter opening
Unlike the Pol II system which contains TFIIH, an ATP-helicase that assists with promoter opening, Pol I transcription initiation is independent of ATP [Citation23]. The identification of three distinct States of CF bound to rDNA lead to the hypothesis that the intrinsic mobility of CF may act as a ratchet in promoter opening [Citation4]. Within the three States, two mobile domains play active roles in establishing the PIC that include the Rrn7 ZR/linker and A49 tWH/linker region [Citation4]. Movement of CF-rDNA in State 2 correlates with the stabilization of the Rrn7 ZR/linker region on Pol I near the single-stranded template DNA [Citation4]. In the absence of the ZR, CF is still recruited to the promoter in vivo, which would suggest the Rrn7 ZR or linker region plays a post-recruitment role [Citation9].
Previous domain swap studies showed that the TFIIB and Rrn7 B-linker domains are interchangeable, pointing to a possible shared mechanism for the TFIIB-related factors in the stabilization of OC DNA [Citation10]. In combination with the intrinsic mobility of CF-rDNA, the recent structural studies suggest that Rrn7 ZR and linker domain play a direct role in promoter opening, which agrees with importance of the Rrn7 ZR and possibly linker domain for melting promoter DNA [Citation4]. One caveat to these studies is the absence of Rrn3, which may help position the Rrn7 ZR in the Pol I dock domain, although the linker domain would still be able to localize to the open DNA [Citation3,Citation5]. Further studies will be necessary to understand the importance of the Rrn7 ZR and linker positioning during Pol I initiation.
Recent results with human TFIIH showed that XPB helicase ATPase activity is dispensable for Pol II transcription as global initiation is relatively unaffected upon XPB deletion [Citation24]. In agreement with this, the yeast Pol II CC shows signs of DNA melting in the absence of TFIIH [Citation24]. This led to the hypothesis that all three eukaryotic Pols use a similar mechanism of promoter opening where the Pols and initiation factors have an intrinsic ability to melt DNA without ATP [Citation24]. This fits well with the model presented for Pol I, which has likely evolved an efficient means to melt Pol I promoter DNA without the help of ATP-dependent translocases [Citation3–Citation5]. It will be exciting to elucidate the ATP-independent promoter opening mechanism for Pol III, which remains unsolved.
A different role for TBP in Pol I transcription?
TBP may occupy a different location in the Pol I PIC than once previously thought. TBP most strongly interacts with the CF subunit Rrn6, leading to the possibility that one of Rrn6 domains, such as the β-propeller or headlock domain that lie on the periphery of the CF complex, may provide a platform for TBP interaction [Citation5,Citation25,Citation26]. Although TBP was added to the experimental conditions in the Han et al. reconstitutions, it was not resolved in the cryo-EM studies [Citation4]. Clearly, the mechanism of CF-TBP interaction remains unclear and warrants further attention.
Previous studies using Acanthamoeba castellanii postulated that TBP's role in the Pol I system is fundamentally different [Citation27]. This was partially based on the lack of sensitivity of Pol I transcription to TATA-containing oligonucleotides which successfully competed away Pol II and III activity [Citation27]. Additional site directed DNA-protein photo-crosslinking studies also illuminated differences in Pol I PIC architecture, where TBP appears to crosslink further upstream of the transcription start site compared to the Pol II system [Citation28]. Recent Pol I initiation structures leave little room for TBP to adopt its typical Pol II position [Citation3–Citation5]. It is also clear from in vitro studies that TBP stimulates yeast Pol I transcription activity [Citation29], indicating that TBP acts more like a coactivator in the Pol I system rather than a basal initiation factor. Given that TBP is a stable component of human Selectivity Factor 1 (SL1) unlike CF [Citation30,Citation31], this also leaves open the possibility that TBP may be utilized differently between yeast and human Pol I systems.
Conclusions and further perspectives
These most recent structural and biochemical efforts have elucidated reconstructions that greatly enhance our understanding of the Pol I transcription machinery [Citation3–Citation5]. Overall, it appears that Pol I initiation machinery is very different from the Pol II system. The key findings of these studies show that the intrinsic mobility of CF allows the complex to adopt a variety of interaction surfaces, which suggests CF has both recruitment and post-recruitment roles in transcription initiation including promoter opening. In addition, these studies show that DNA takes a different path through Pol I than in Pol II [Citation3–Citation5].
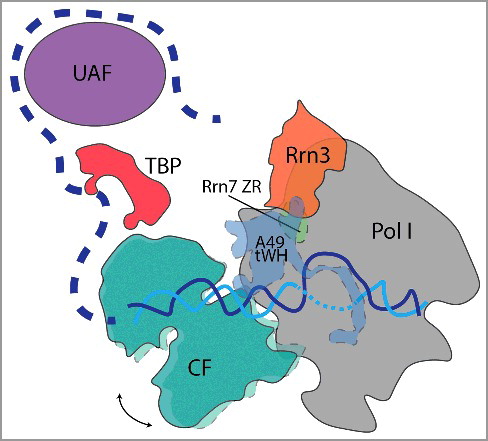
These recent structural efforts have taken huge strides to provide high-resolution structures of CF and near-atomic resolution structures of the Pol I PIC [Citation3–Citation5]. Future studies are poised to expand upon these structural analyses to include UAF and TBP with the ultimate goal to complete the entire yeast Pol I PIC (). It will be exciting to see if these features are conserved in the human Pol I PIC and if the Rrn7 human ortholog, TAF1B of the SL1 complex, retains similar recruitment and post-recruitment properties in transcription initiation [Citation10,Citation32]. It is also noteworthy that TAF1B contains a large insertion within the N-terminal cyclin domain that is rich in modifiable residues that could possibly serve as a regulatory protein-protein interaction surface or have additional functions in SL1 recruitment, DNA engagement, or polymerase interaction given its placement within the structure [Citation7,Citation10]. We are hopeful that these new structures will help mold new strategies to specifically target the Pol I transcription machinery that is often upregulated in cancer [Citation33–Citation35]. More broadly, these studies may also be used to help further characterize the Pol III PIC structure that remains unresolved.
Figure 2. Yeast Pol I PIC highlighting possible upstream location of TBP and UAF. CF (teal) moves in the three States of initiation acting like a ratchet that opens DNA (cyan and blue) using an ATP-independent mechanism. An arrow depicts CF rotational movement. Dashed outline denotes CF, A49 tWH, and Rrn7 ZR mobility during initiation. Previous biochemical studies, in combination with the recent Pol I PIC structures, suggest that TBP (red) may occupy a position more upstream between UAF (purple) and CF. Pol I and Rrn3 are depicted in grey and orange, respectively. Upstream DNA is depicted by a dash blue line.
For more information, we direct readers to two other excellent recent reviews on this subject that complements the work presented above [Citation36,Citation37].
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgements
We apologize for those whose work was not cited due to space limitations.
Additional information
Funding
References
- Vannini A, Cramer P. Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol Cell. 2012;45(4):439–446. doi:10.1016/j.molcel.2012.01.023. PMID:22365827
- Schneider D. RNA polymerase I activity is regulated at multiple steps in the transcription cycle: recent insights into factors that influence transcription elongation. Gene. 2012;493(2):176–184. doi:10.1016/j.gene.2011.08.006. PMID:21893173
- Engel C, Gubbey T, Neyer S, et al. Structural basis of RNA polymerase I transcription initiation. Cell. 2017;169(1):120–131, e22. doi:10.1016/j.cell.2017.03.003. PMID:28340337
- Han Y, Yan C, Nguyen THD, et al. Structural mechanism of ATP-independent transcription initiation by RNA polymerase I. Elife. 2017;6. doi:10.7554/eLife.27414.
- Sadian Y, Tafur L, Kosinski J, et al. Structural insights into transcription initiation by yeast RNA polymerase I. Embo j. 2017;36(18):2698–2709. doi:10.15252/embj.201796958. PMID:28739580
- Holstege F, Fiedler U, Timmers H. Three transitions in the RNA polymerase II transcription complex during initiation. Embo j. 1997;16(24):7468–7480. doi:10.1093/emboj/16.24.7468. PMID:9405375
- Knutson B, Hahn S. TFIIB-related factors in RNA polymerase I transcription. Biochim Biophys Acta. 2013;1829(3–4):265–273. doi:10.1016/j.bbagrm.2012.08.003. PMID:22960599
- Sainsbury S, Bernecky C, Cramer P. Structural basis of transcription initiation by RNA polymerase II. Nat Rev Mol Cell Biol. 2015;16(3):129–143. doi:10.1038/nrm3952. PMID:25693126
- Knutson B, Luo J, Ranish J, et al. Architecture of the Saccharomyces cerevisiae RNA polymerase I Core Factor complex. Nat Struct Mol Biol. 2014;21(9):810–816. doi:10.1038/nsmb.2873. PMID:25132180
- Knutson B, Hahn S. Yeast Rrn7 and human TAF1B are TFIIB-related RNA polymerase I general transcription factors. Science. 2011;333(6049):1637–1640. doi:10.1126/science.1207699. PMID:21921198
- Kostrewa D, Zeller ME, Armache KJ, et al. RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature. 2009;462(7271):323–330. doi:10.1038/nature08548. PMID:19820686
- Peyroche G, Milkereit P, Bischler N, et al. The recruitment of RNA polymerase I on rDNA is mediated by the interaction of the A43 subunit with Rrn3. Embo j. 2000;19(20):5473–5482. doi:10.1093/emboj/19.20.5473. PMID:11032814
- Geiger S, Lorenzen K, Schreieck A, et al. RNA polymerase I contains a TFIIF-related DNA-binding subcomplex. Mol Cell. 2010;39(4):583–594. doi:10.1016/j.molcel.2010.07.028. PMID:20797630
- Liljelund P, Mariotte S, Buhler JM, et al. Characterization and mutagenesis of the gene encoding the A49 subunit of RNA polymerase A in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1992;89(19):9302–9305. doi:10.1073/pnas.89.19.9302. PMID:1409638
- Beckouet F, Labarre-Mariotte S, Albert B, et al. Two RNA polymerase I subunits control the binding and release of Rrn3 during transcription. Mol Cell Biol. 2008;28(5):1596–1605. doi:10.1128/MCB.01464-07. PMID:18086878
- Burley S, Roeder R. Biochemistry and structural biology of transcription factor IID (TFIID). Annu Rev Biochem. 1996;65:769–799. doi:10.1146/annurev.bi.65.070196.004005. PMID:8811195
- Neyer S, Kunz M, Geiss C, et al. Structure of RNA polymerase I transcribing ribosomal DNA genes. Nature. 2016;540(7634):607–610. doi:10.1038/nature20561. PMID:27842382
- Tafur L, Sadian Y, Hoffmann NA, et al. Molecular structures of transcribing RNA polymerase I. Mol Cell. 2016;64(6):1135–1143. doi:10.1016/j.molcel.2016.11.013. PMID:27867008
- Marilley M, Radebaugh CA, Geiss GK, et al. DNA structural variation affects complex formation and promoter melting in ribosomal RNA transcription. Mol Genet Genomics. 2002;267(6):781–791. doi:10.1007/s00438-002-0708-z. PMID:12207225
- Smircich P, Duhagon M, Garat B. Conserved curvature of RNA polymerase I core promoter beyond rRNA genes: the case of the tritryps. Genomics Proteomics Bioinformatics. 2015;13(6):355–363. doi:10.1016/j.gpb.2015.09.005. PMID:26718450
- Kownin P, Bateman E, Paule M, Effects of single-base substitutions within the Acanthamoeba castellanii rRNA promoter on transcription and on binding of transcription initiation factor and RNA polymerase I. Mol Cell Biol. 1988;8(2):747–753.
- Carter R, Drouin G. The evolutionary rates of eukaryotic RNA polymerases and of their transcription factors are affected by the level of concerted evolution of the genes they transcribe. Mol Biol Evol. 2009;26(11):2515–2520. doi:10.1093/molbev/msp164. PMID:19633229
- Lofquist A, Li H, Imboden MA, et al. Promoter opening (melting) and transcription initiation by RNA polymerase I requires neither nucleotide beta,gamma hydrolysis nor protein phosphorylation. Nucleic Acids Res. 1993;21(14):3233–3238. doi:10.1093/nar/21.14.3233. PMID:7688114
- Alekseev S, Nagy Z, Sandoz J, et al. Transcription without XPB establishes a unified helicase-independent mechanism of promoter opening in eukaryotic gene expression. Mol Cell. 2017;65(3):504–514, e4. doi:10.1016/j.molcel.2017.01.012. PMID:28157507
- Steffan J, Keys DA, Dodd JA, et al. The role of TBP in rDNA transcription by RNA polymerase I in Saccharomyces cerevisiae: TBP is required for upstream activation factor-dependent recruitment of core factor. Genes Dev. 1996;10(20):2551–2563. doi:10.1101/gad.10.20.2551. PMID:8895657
- Steffan J, Keys DA, Vu L, et al. Interaction of TATA-binding protein with upstream activation factor is required for activated transcription of ribosomal DNA by RNA polymerase I in Saccharomyces cerevisiae in vivo. Mol Cell Biol. 1998;18(7):3752–3761. doi:10.1128/MCB.18.7.3752. PMID:9632758
- Radebaugh C, Matthews JL, Geiss GK, et al. TATA box-binding protein (TBP) is a constituent of the polymerase I-specific transcription initiation factor TIF-IB (SL1) bound to the rRNA promoter and shows differential sensitivity to TBP-directed reagents in polymerase I, II, and III transcription factors. Mol Cell Biol. 1994;14(1):597–605. doi:10.1128/MCB.14.1.597. PMID:8264628
- Gong X, Radebaugh CA, Geiss GK, et al. Site-directed photo-cross-linking of rRNA transcription initiation complexes. Mol Cell Biol. 1995;15(9):4956–4963. doi:10.1128/MCB.15.9.4956. PMID:7651413
- Aprikian P, Moorefield B, Reeder R. TATA binding protein can stimulate core-directed transcription by yeast RNA polymerase I. Mol Cell Biol. 2000;20(14):5269–5275. doi:10.1128/MCB.20.14.5269-5275.2000. PMID:10866683
- Comai L, Tanese N, Tjian R. The TATA-binding protein and associated factors are integral components of the RNA polymerase I transcription factor, SL1. Cell. 1992;68(5):965–976. doi:10.1016/0092-8674(92)90039-F. PMID:1547496
- Comai L, Zomerdijk JC, Beckmann H, et al. Reconstitution of transcription factor SL1: exclusive binding of TBP by SL1 or TFIID subunits. Science. 1994;266(5193):1966–1972. doi:10.1126/science.7801123. PMID:7801123
- Naidu S, Friedrich JK, Russell J, et al. TAF1B is a TFIIB-like component of the basal transcription machinery for RNA polymerase I. Science. 2011;333(6049):1640–1642. doi:10.1126/science.1207656. PMID:21921199
- Williamson D, Lu YJ, Fang C, et al. Nascent pre-rRNA overexpression correlates with an adverse prognosis in alveolar rhabdomyosarcoma. Genes Chromosom Cancer. 2006;45(9):839–845. doi:10.1002/gcc.20347. PMID:16770781
- Hannan K, Sanij E, Rothblum LI, et al. Dysregulation of RNA polymerase I transcription during disease. Biochim Biophys Acta. 2013;1829(3–4):342–360. doi:10.1016/j.bbagrm.2012.10.014. PMID:23153826
- Bywater M, Pearson RB, McArthur GA, et al. Dysregulation of the basal RNA polymerase transcription apparatus in cancer. Nat Rev Cancer. 2013;13(5):299–314. doi:10.1038/nrc3496. PMID:23612459
- Jochem L, Ramsay E, Vannini A. RNA polymerase I, bending the rules? Embo j. 2017;36(18):2664–2666. doi:10.15252/embj.201797924. PMID:28842442
- Khatter H, Vorlander M, Muller C. RNA polymerase I and III: similar yet unique. Curr Opin Struct Biol. 2017;47:88–94. doi:10.1016/j.sbi.2017.05.008. PMID:28743025
