ABSTRACT
Hexim1 acts as a tumor suppressor and is involved in the regulation of innate immunity. It was initially described as a non-coding RNA-dependent regulator of transcription. Here, we detail how 7SK RNA binds to Hexim1 and turns it into an inhibitor of the positive transcription elongation factor (P-TEFb). In addition to its action on P-TEFb, it plays a role in a variety of different mechanisms: it controls the stability of transcription factor components and assists binding of transcription factors to their targets.
Introduction
Hexim stands for “HEXamethylene-bis-acetamide-Inducible protein in vascular smooth Muscle cells” [Citation1]. Other names are Cardiac Lineage-associated Protein (CLP-1) [Citation2], Ménage A Quatre (MAQ-1) [Citation3] or Estrogen Down-regulated Gene (EDG-1) [Citation4]. Hexim1 synthesis is induced by HEXamethylene-bis-acetamide (HMBA) and analogs [Citation5], histone deacetylase inhibitors [Citation6] and nucleotide depletion [Citation7]. Hexim1 is degraded by autophagy in some circumstances [Citation8]. Hexim proteins are evolutionarily conserved proteins found in vertebrates, mollusks, insects and nematodes [Citation9]. Drosophila melanogaster has only one dHexim gene. Its knock-down generates multiple developmental defects and homozygous knock-outs flies could not be obtained [Citation10]. Mammalian genomes generally, but not always, have two adjacent genes coding for two paralogue proteins, Hexim1 and Hexim2. In contrast to Hexim2 genes, Hexim1 genes do not have introns. Homozygous Hexim1 knock-out mouse embryos develop but die at birth [Citation11]. Hexim2 can compensate for the loss of Hexim1 in knock-down cells [Citation12]. These proteins share similar properties with subtle variations in affinity for partners [Citation13]. Hexim1 regulates heart physiology and has strong links to cancer, differentiation, development and inflammation [Citation14–Citation18]. It acts as an anti-tumor and pro-differentiation factor. Most studies are on human Hexim1 protein. This review focuses on its biochemical properties. Amino acid numbers refer to human Hexim1 (1 to 359).
Hexim1 domain architecture
Six regions can be distinguished in Hexim1 ((A)). An unstructured N-terminal domain (a.a. 1 to 255) comprises a poorly conserved sequence (a.a. 1 to 149) followed by a stretch of conserved basic amino acids, mostly lysines and arginines (a.a. 150 to 163) [Citation19,Citation20]. This basic region (BR) is sometimes called the Arginine-Rich Motif (ARM) by analogy with a similar motif in the HIV Tat protein. It overlaps a bipartite nuclear localization sequence responsible for its nuclear localization [Citation3,Citation21]. Hexim1 is sometimes described as a cytoplasmic protein as it is present in low salt “cytosolic” extracts [Citation22]. However, many nuclear proteins are found in such extracts, and localization cannot be deduced from biochemical extractions. As a matter of fact, immunofluorescence clearly shows that Hexim1 is nuclear [Citation3,Citation21]. Furthermore, Hexim1 binds strongly in vitro to Kpna2/Importin alpha1 of the Ran-mediated nuclear import pathway [Citation13].
Figure 1. – (A) Hexim domain architecture. Amino-acid numbers delimiting the basic region (BR), the central region (CR), the PYNT motif, the acidic regions (AR1 and AR2) and the C-terminal helices (α1, α2 and α3) are indicated. (B) 3-D structure of Hexim1 C-terminal coiled-coil domain deduced from NMR (pdb – 2gd7). Hexim1 forms dimers through its C-terminal domain. Helices α2 and α3 are separated by a GGD a.a. sequence. (C) The basic region (BR), the PYNT motif and acidic regions (AR1 and AR2) are schematically positioned. The BR and AR1 regions interact within “free” Hexim1 dimers. (D) Binding of Hexim1 to 7SK RNA releases the BR/AR interaction leading to Hexim1 sequences between a.a. 260 and 310 binding to Cyclin T1 (CycT1) and the PYNT sequence binding to Cdk9 catalytic cleft. resulting in inhibition of Cdk9/Cyclin T1 (P-TEFb) kinase activity. MePCE and LaRP7 proteins bind to 7SK 5’ and 3’ extremities, respectively. The same color codes are used for Hexim 1 regions in A, B, C and D.
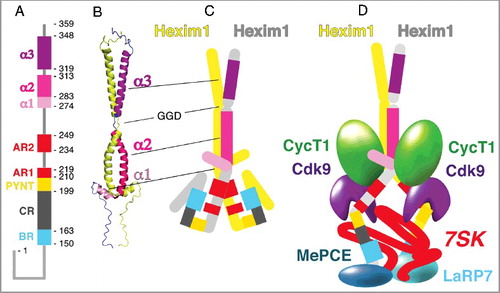
A “central region” (a.a. 164 to 198) follows the BR and continues with the so-called “PYNT” sequence. This sequence (PVAPYNTTQFLM – a.a. 199 to 210) is the most conserved one among all Hexim proteins [Citation9]. The “PYNT” sequence is followed by two sequences rich in aspartic and glutamic acids, the acidic regions, AR1 (a.a. 211 to 219) and AR2 (a.a. 234 to 249).
The Hexim1 C-terminal domain (a.a. 255 to 359) folds into three alpha-helices as shown by NMR [Citation23] ((B)). The first helix, α1 spans from a.a. 274 to 283. The two C-terminal alpha-helices, α2 (a.a. 284 to 313) and α3 (a.a. 319 to 348), are entangled in a 9.5 nm long bipartite coiled-coil leucine-zipper [Citation24]. Hence, Hexim1 proteins form highly anisotropic dimers [Citation12,Citation25–Citation27]. Although the last helix is not essential, the flexibility brought by a stretch of three residues (GGD) between helices α2 and α3 is critical for Hexim1 function [Citation24]. Apart from its three C-terminal helices, Hexim1 is poorly structured. It might be speculated that it is structured upon interaction with its binding partners, like p21 when associated to Cdk2/Cyclin A [Citation28].
7SK RNA binding turns Hexim1 into a P-TEFb inhibitor
Hexim1 has been isolated from cell extracts bound to the inactive positive transcription factor (P-TEFb) complex and 7SK RNA [Citation20]. P-TEFb is required to release RNA polymerase II pausing to achieve productive transcription of most Pol II genes [Citation29–Citation31]. Hence, Hexim1 and 7SK RNA act as transcription repressors. Human 7SK RNA is a small (331 bases), abundant nuclear RNA polymerase III transcript [Citation29,Citation32–Citation34]. 7SK RNA forms a stable core 7SK snRNP particle with MePCE and LaRP7 proteins.
In vitro, full-length Hexim1 requires 7SK RNA to associate with and inhibit P-TEFb, but MePCE and LARP7 proteins are not necessary in this process [Citation19,Citation20]. Drosophila dHexim also associates with the fly 7SK RNA homologue to inactivate fly P-TEFb [Citation35]. The C.elegans proteins, ceHexim, and cit-1.2 (cyclin T1/T2 homologue) interact in a yeast two-hybrid assay [Citation36]. But, although a putative 7SK RNA homologue has been identified in this species, its association with ceHexim and P-TEFb has not been demonstrated [Citation9].
A Hexim1 dimer binds a single 7SK RNA molecule. Two Hexim1 binding sites have been identified in a 7SK 5’ hairpin [Citation37], one Hexim monomer for each site [Citation26,Citation38]. The main Hexim1 binding site consists of an inverted GAUC double-strand repeat within the hairpin. Chemical foot printing data suggest that 7SK RNA undergoes important conformational changes when bound to Hexim1 and P-TEFb [Citation39]. An NMR study of the 7SK 5’ hairpin shows that binding of a peptide corresponding to the Hexim1 BR induces the opening of a GAUC repeat as well as a hydrogen-bond network involving two uridines next to the GAUC repeat [Citation40]. Interestingly, the crystal structure of the 7SK 5’ hairpin indicates that the same uridines can adopt two very distinct conformations [Citation41] and in vivo RNA tethering assay suggests that these two uridines indeed contribute to Hexim1 binding [Citation42].
The Hexim1 basic region (BR) is involved in 7SK RNA binding. Removal of 4 of its 12 basic residues suppresses its binding capacity [Citation20]. However, the BR is unlikely involved in 7SK recognition due to the low complexity of its amino-acid composition. Indeed, an in vivo RNA tethering assay suggests that Hexim1 sequences overlapping the “central region”, the “PYNT” sequence and a small part of the acidic region (AR1) (a.a. 178 to 220) are required for specific recognition [Citation42]. Furthermore, a 7SK 5’ hairpin with a thiouridine substitution of uridine U30, cross-links to a Hexim1 peptide positioned just after the “PYNT” sequence, corresponding to the N-terminal acid region (AR1) (a.a. 211–219) [Citation43].
Hexim1 dimerization is essential for P-TEFb binding and inhibition [Citation26,Citation27]. One Hexim1 dimer binds one 7SK RNA molecule and two core P-TEFb modules. 7SK binding to Hexim1 triggers formation of an inactive P-TEFb/Hexim1/7SK complex. However, RNA binding to Hexim1 per se is not required in this process. When the N-terminal part of Hexim1 lacking the BR but keeping the PYNT sequence is deleted, the resulting truncated protein binds to and inhibits P-TEFb in absence of RNA [Citation20,Citation44]. According to a very elegant model proposed by the Peterlin lab and confirmed by others, including the Geyer lab, the positive charges of the BR (a.a. 150–164) attract the negative charges of the acidic region (AR1) (a.a. 211–219) [Citation21,Citation44] ((C)). 7SK RNA neutralizes the positive charges of the BR that subsequently releases the AR1. The likely resulting Hexim1 conformational change unmasks a P-TEFb binding surface ((D)).
The Hexim1 “PYNT” sequence masks the Cdk9 catalytic site and inhibits its kinase activity
Core P-TEFb comprises two subunits, Cdk9 and Cyclin T1 or T2. Yeast two-hybrid and in vitro binding assays demonstrate that Hexim1 C-terminal domain contacts Cyclin T1 or T2 cyclin box [Citation20,Citation36,Citation45]. Several point mutations (F262L, F267L, H275, E295A) disrupt Hexim1-Cyclin T1 binding in vitro or in yeast two hybrid assays and Hexim1-P-TEFb binding in vivo. These conserved residues are localized next or within helices α1 and α2 in the C-terminal domain (between a.a. 262 and 300) ((B)). NMR data further support the identification of this region as the Cyclin T binding domain (TBD) [Citation23]. Homozygous mice expressing a Hexim1 protein lacking helix α3 are viable thereby indicating that it has little functional importance [Citation46].
A C-terminal Hexim1 fragment (255 to 359) binds Cyclin T1 in vitro but does not inhibit P-TEFb. In contrast, a longer Hexim1 fragment (200 to 359) is almost as efficient as the wild-type full-length protein in inhibiting P-TEFb in vitro [Citation20,Citation44]. Importantly, 7SK RNA is required for binding and inhibition by the full-length Hexim1 but not by the (200 to 359) fragment. Furthermore, removing just the PYNT sequence as in the (207 to 359) fragment, generates a protein that is inactive for P-TEFb inhibition, although it still binds P-TEFb. Mutations in residues of the “PYNT” sequence (Pro202, Thr205 or Phe208) in a full-length Hexim1 abolish P-TEFb binding in vivo and in vitro [Citation20,Citation47]. Photocross-linking experiments suggest close contacts between Cdk9 and residues Trp164, Tyr167, Phe178 and Phe194 in the Hexim1 central region, Phe208 in the “PYNT” sequence and residues Phe267, Tyr271 and Tyr274 in helix α1 that precedes the coiled-coil leucine zipper [Citation47]. Mass spectrometry of cross-links further demonstrates that residue Phe208 in the “PYNT” sequence contacts a Cdk9 peptide at the border of its catalytic cleft. This peptide is nearby the “T-loop” that controls access to the Cdk9 catalytic site. By homology with the 3-D structure of Cdk2/Cyclin A, this Cdk9 peptide likely contacts its peptide substrate. Like all Cyclin Dependent Kinases, Cdk9 activity requires phosphorylation of its T-loop threonine Thr186. In Cdk2/Cyclin A, the homologous T-loop phosphothreonine interacts with a model peptide substrate [Citation47]. As Cdk9 Thr186 phosphorylation is required for P-TEFb-Hexim1 binding [Citation26,Citation48], it might be hypothesized that phospho-Thr186 is directly involved in Cdk9/Hexim1 interaction. In conclusion, the Hexim1 “PYNT” sequence masks the Cdk9 catalytic site and occupies the substrate-binding site, resulting in Cdk9 inactivation. These findings confirm initial observations of the importance of the “PYNT” sequence for P-TEFb inhibition [Citation20].
HIV Tat and TAR RNA compete with Hexim1 and 7SK RNA for P-TEFb
Transcription of the human immunodeficiency viral (HIV) genome is controlled by recognition by the viral Tat protein of the TAR hairpin that forms at the 5’ end of the viral transcript [Citation49]. The TAR.Tat complex recruits active P-TEFb to release RNA polymerase II paused after an initial transcription of 50–80 nt. A Tat.Cyclin T1 contact surface overlapping the two cyclin box subdomains has been identified by X-ray crystallography [Citation50]. Cyclin T1 mutations abolishing Hexim1 interaction have been identified in a yeast two-hybrid screen [Citation36]. They are located on the Tat.Cyclin T1 contact surface, suggesting Hexim1 and Tat bind to the same Cyclin T1 region. Importantly, a Cyclin T1 point mutation in the middle of this surface abolishes both Tat and Hexim1 binding to P-TEFb without affecting P-TEFb activity. Not surprisingly, Tat and Hexim1 binding to P-TEFb are mutually exclusive [Citation45]. Like Hexim1, Tat contacts Cdk9 [Citation50] at the T-loop but in contrast to Hexim1, Tat does not mask its catalytic site. Thus, in contrast to the Hexim1.7SK complex, Tat.TAR binding keeps P-TEFb in an active state [Citation44].
Tat's TAR binding site is an arginine-rich motif (ARM) that resembles the Hexim1 BR [Citation19,Citation20]. Like 7SK RNA, TAR RNA folds into a hairpin with a bulge [Citation51]. Not surprisingly, both Tat and Hexim1 bind either 7SK or TAR RNA. Importantly, any RNA that binds Hexim1 promotes P-TEFb inhibition [Citation51,Citation52]. Thus, Hexim1 binding to TAR recruits an inactive P-TEFb.Hexim1 complex to the nascent viral transcript [Citation53]. Removal of Hexim1.7SK is required to activate P-TEFb [Citation54] and the Tat.TAR RNA complex competes out Hexim1.7SK in a following step, thus activating P-TEFb and releasing paused RNA polymerase [Citation51,Citation55]. On the other hand, as Hexim1 inactivates P-TEFb when bound to TAR, it may contribute to HIV silencing in absence of Tat through either P-TEFb or TAR hairpin trapping [Citation56].
Post-translational modifications regulate P-TEFb.Hexim1.7SK snRNP complex formation
Hexim1 inhibition of P-TEFb is regulated. Several mechanisms concur to control Hexim1 inhibition of P-TEFb. Other feed-back loops involving 7SK snRNP trapping, RNA helicases targeting 7SK RNA have been discussed [Citation6,Citation57–Citation62]. Here, we focus on post-translational modifications of Hexim1 or P-TEFb subunits.
Protein kinase cascades control P-TEFb.Hexim1.7SK snRNP assembly. Cdk9 phosphorylation is required for both P-TEFb activity and binding to Hexim1 [Citation26,Citation48]. Thr186 dephosphorylation, for example in response to UV irradiation, coincides with dissociation from Hexim1, at the same time, Cdk9 substrates cannot bind either. Cdk9 is rephosphorylated within a few hours after UV irradiation. When CDK9 is dephosphorylated, transient transcription arrest takes place, suggesting that Cdk9 dephosphorylation might contribute to this process.
Phosphorylation is directly involved in P-TEFb.Hexim1.7SK snRNP disruption. Indeed, mass spectrometry indicates that Hexim1 Thr270, Tyr271, Tyr274 and Ser278, at the tip of helix α1 are phosphorylated [Citation63,Citation64]. The phosphomimetic mutations T270D, S278D and Y271E/Y274E disrupt the assembly of P-TEFb and Hexim1 into a 7SK snRNP complex. By contrast, preventing phosphorylation of these residues (in particular the Y271F mutation) impairs release of active P-TEFb from the 7SK snRNP complex in response to T-cell activating signals. Phosphorylation of Thr270 and Ser278 relies on the PI3/Akt pathway [Citation63] and Tyr271 on the Erk1 pathway [Citation65]. PKC phosphorylates Ser158 in the middle of the basic region (BR) and neutralizes one of the charges in this region [Citation66]. Ser158 phosphorylation impairs 7SK RNA binding to Hexim1, and therefore impairs Hexim1 association with P-TEFb. Treatments leading to heart hypertrophy promote dissociation of P-TEFb.Hexim1.7SK snRNP [Citation67]. The Jak/STAT signal transduction pathway is involved.
Several protein phosphatases have also been proposed to control P-TEFb.Hexim1.7SK snRNP assembly. Indeed, following stimulations such as UV irradiation, the PP1α, PPM1A/PP2Cα and PPM1G phosphatases dephosphorylate CDK9 on Thr186 [Citation68–Citation70], thus inactivating kinase activity. PP2B (Ca++-dependent, calmodulin-stimulated phosphatase) facilitates direct dephosphorylation of Cdk9 Thr186 by PP1α [Citation68].
Cdk9 and Cyclin T1 acetylation play a role in P-TEFb.Hexim1.7SK snRNP assembly. Deacetylation of Cdk9 Lys48 by the sirtuin deacetylases SIRT2 [Citation71] or SIRT7 [Citation72] promotes P-TEFb release from Hexim1.7SK. Hexim1 associates with the MyoD transcription factor and Histone deacetylases HDAC1, HDAC3 and HDAC5 to inhibit MyoD-dependent gene expression, suggesting that Hexim1 might as well recruit deacetylases to P-TEFb [Citation73].
In conclusion, major signaling pathways affecting global gene activity converge on P-TEFb.Hexim1.7SK RNP and play a role in its activity.
Hexim1 partners recruit P-TEFb on target genes
Transcription factor-assisted recruitment of P-TEFb.Hexim1.7SK snRNP and subsequent disassembly of the complex at promoters might be a general mechanism [Citation74–Citation76]. The KAP1/TRIM28/TIF1B transcription intermediary factor recruits P-TEFb.Hexim1.7SK snRNP on promoters through binding of the Larp7 core 7SK snRNP subunit [Citation76]. The RNA binding protein SR protein SRSF2/SC35 also binds and recruits the P-TEFb.Hexim1.7SK RNP complex to promoters [Citation77]. Transcription factor NF-κB recruits P-TEFb to activate elongation at inflammatory-responsive genes. NF-κB interacts with both Hexim1 and P-TEFb [Citation74,Citation78]. PPM1G/PP2Cγ phosphatase might further assist in recruitment as it is recruited to promoters and interacts directly with NF-κB, Hexim1 and 7SK RNA [Citation70]. PPM1G binds both 7SK RNA and Hexim1. PPM1G binds Hexim1 through the C-terminal helix α2 (a.a. 286–314) that has also P-TEFb binding sites. Thus, binding of PPM1G and P-TEFb to Hexim1 is mutually exclusive. It is proposed that PPM1G dephosphorylates Thr186 in the CDK9 T-loop leading to P-TEFb.Hexim1.7SK disassembly. Furthermore, PPM1G would trap Hexim1.7SK RNA while Thr186 would be rephosphorylated later for in situ reactivation.
Steroid receptors directly recruit active P-TEFb on their target genes [Citation79–Citation82]. They interact with Hexim1 basic region (BR). Thus, they compete with 7SK RNA and favor formation of the active P-TEFb form. The androgen receptor recruits PP1α phosphatase, also resulting in P-TEFb detachment from Hexim1 and 7SK RNA, though, it is not known whether Thr186 dephosphorylation is involved in this event [Citation83]. Nevertheless, Hexim1 acts as tumor suppressor for breast and prostate cancers [Citation84]. Overexpression of full-length wild-type Hexim1 in mice represses estrogen receptor recruitment to a target promoter, whereas overexpression of a truncated protein lacking helix α3 (313 to 359 deletion) leads to an increased recruitment [Citation46]. This finding is a rare example that proposes a function for this helix.
Transcription of many genes involves a competition mechanism by chromatin-associated proteins. Brd4 is a histone-associated protein that interacts with active P-TEFb and might contribute in its recruitment to promoters [Citation15,Citation85]. Yet, a recent study suggests that Brd4 might not be directly involved in the recruitment of P-TEFb [Citation86]. Anyhow, Brd4 directly interacts with Cyclin T1 through two binding domains [Citation87]. Although such competition was not observed in an in vitro assay [Citation88], the Brd4 C-terminal domain might compete out Hexim1 binding for P-TEFb in living cells. Furthermore, Hexim1 expression enhances Brd4 sensitivity to its inhibitor, JQ1 [Citation8]. In contrast to Brd4, Hexim1 antagonizes cancer progression and stem cell reprogramming [Citation15,Citation89].
Hexim1 and NEAT1 ncRNA contribute to the innate immune response
Although P-TEFb and 7SK RNP are major Hexim1 partners, both have independent functions on their own. Several papers have reviewed the involvement of 7SK RNA in transcription control [Citation29,Citation33,Citation34,Citation90]. Here we focus on Hexim1 binding to other proteins and other RNAs with important physiological consequences.
For example, Hexim1 binds nucleophosmin, a nucleolar phosphoprotein involved in ribosome biogenesis and acting as a proto-oncogene [Citation91]. Nucleophosmin stimulates Hexim1 degradation and might lead to its cytoplasmic mislocalization in some leukemias. Hexim1 also binds and stabilizes the p53 tumor suppressor protein preventing its degradation by the ubiquitination pathway [Citation92]. In contrast, Hexim1 interacts with the hypoxia-inducible factor α (HIF-1α) and stimulates its degradation through a ubiquitination pathway [Citation93]. Both effects may contribute to Hexim1's anti oncogenic properties.
A series of cellular mRNAs that bind Hexim1 and are stabilized as such have been identified by RNA UV cross-linking (eCLIP) [Citation7]. Some of these mRNAs code for antitumor proteins and may contribute to the tumor suppressor function of Hexim1, for example in melanomas that respond to nucleotide stress. A SELEX study also identified three mRNAs (coding for Cad, Brd4 and Tcf3) binding to Hexim1 [Citation94]. The presence of a bulge within a double-stranded structure is required for their affinity for Hexim1. Noteworthy, Cad and Brd4 mRNAs also came out in the above-mentioned eCLIP screen.
Hexim1 was recently shown to bind the NEAT1 non-coding nuclear RNA [Citation95]. NEAT1 is a long (22 kb) abundant nuclear RNA polymerase II transcript required for the formation of nuclear paraspeckles [Citation96]. Hexim1 associated with NEAT1 serves as a scaffold for a multiprotein complex involved in innate immunity signaling [Citation95]. This complex comprises two protein kinases, DNA-PKc and TBK1, among other components and has been named Hexim1-DNA-PK-paraspeckle components-ribonucleoprotein complex (HDP-RNP). The NEAT1 RNA anchors this complex within the paraspeckles. Upon DNA-dependent activation of DNA-PK and cGAMP synthesis, the NEAT1-containing HDP-RNP complex detaches from paraspeckles. The STING transcription factor is released from the endoplasmic reticulum and recruited onto HDP-RNP to promoters to activate innate immunity response genes [Citation97]. There might be cooperation with NF-κB, as mentioned above, to recruit Hexim1 and P-TEFb and activate elongation at inflammatory-responsive genes in a stimulus-dependent manner [Citation74]. The Hexim1 BR sequence is involved in HDP-RNP assembly [Citation95]. The HDP-RNP binding sites are located in the “central region” (CR) of Hexim1. This is in contrast with the major P-TEFb binding sites that are located in the “PYNT” sequence and in the C-terminal helices [Citation32,Citation47]. Although distinct binding sites are involved, P-TEFb and HDP-RNP binding is mutually exclusive [Citation95]. The Hexim1 BR sequence is involved in NEAT1 RNA recognition. However, the NEAT1 elements targeted by Hexim1 are unknown. NEAT1 is essential for formation of the HDP-RNP complex. 7SK RNA promotes P-TEFb binding to Hexim1; it is remarkable that the RNA species that binds Hexim1 determines the complex that assembles on it. It will be highly interesting to identify other RNA-Hexim1 complexes and unravel their functions.
Concluding remarks
There is no protein family related to Hexim1, the Hexim family is unique. Hexim1 combines an extended coiled-coil C-terminal domain responsible for its anisotropy with the capacity to specifically bind some non-coding RNAs, thus forming different RNA.Hexim.protein complexes with different functions. At least one of these complexes, P-TEFb.Hexim1.7SK snRNP, is dynamic and its activity is fine tuned in response to the environment. Remarkably, the RNA species that binds Hexim1 determines the protein complex that assembles on it. Therefore, Hexim1 serves as an RNA-dependent regulatory protein hub. Single mutations due to single nucleotide variations in Hexim1 and in Hexim2 are associated with human cancers (High-performance Integrated Virtual Environment (HIVE) at George Washington University). It is so far unknown whether some of these mutations affect Hexim1 functions.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by grants ANR 10-LABX-54 MEMOLIFE and ANR-11-IDEX-0001‐02 PSL* Research University.
Additional information
Funding
References
- Kusuhara M, Nagasaki K, Kimura K, et al. Cloning of hexamethylene-bis-acetamide-inducible transcript, HEXIM1, in human vascular smooth muscle cells. Biomed Res. 1999;20:273–279. doi:10.2220/biomedres.20.273.
- Ghatpande S, Goswami S, Mathew S, et al. Identification of a novel cardiac lineage-associated protein(cCLP-1): a candidate regulator of cardiogenesis. Dev Biol. 1999;208:210–221. doi:10.1006/dbio.1998.9180. PMID:10075853
- Michels AA, Nguyen VT, Fraldi A, et al. MAQ1 and 7SK RNA interact with CDK9/cyclin T complexes in a transcription-dependent manner. Mol Cell Biol. 2003;23(14):4859–4869. doi:10.1128/MCB.23.14.4859-4869.2003. PMID:12832472
- Wittmann BM, Wang N, Montano MM. Identification of a novel inhibitor of breast cell growth that is down-regulated by estrogens and decreased in breast tumors. Cancer Res. 2003;63(16):5151–5158. PMID:12941847
- Ketchart W, Yeh IJ, Zhou H, et al. Induction of HEXIM1 activities by HMBA derivative 4a1: functional consequences and mechanism. Cancer Lett. 2016;379(1):60–69. doi:10.1016/j.canlet.2016.05.029. PMID:27238569
- Liu P, Xiang Y, Fujinaga K, et al. Release of positive transcription elongation factor b (P-TEFb) from 7SK small nuclear ribonucleoprotein (snRNP) activates hexamethylene bisacetamide-inducible protein (HEXIM1) transcription. J Biol Chem. 2014;289(14):9918–9925. doi:10.1074/jbc.M113.539015. PMID:24515107
- Tan JL, Zhao Q, Liang X, et al. Stress from nucleotide depletion activates the transcriptional regulator HEXIM1 to suppress melanoma. Mol Cell. 2016;62(1):34–46. doi:10.1016/j.molcel.2016.03.013. PMID:27058786
- Han L, Zhao Q, Liang X, et al. Ubenimex enhances Brd4 inhibition by suppressing HEXIM1 autophagic degradation and suppressing the Akt pathway in glioma cells. Oncotarget. 2017;8(28):45643–45655. doi:10.18632/oncotarget.17314. PMID:28484091
- Marz M, Donath A, Verstraete N, et al. Evolution of 7SK RNA and its protein partners in metazoa. Mol Biol Evol. 2009;26:2821–2830. doi:10.1093/molbev/msp198. PMID:19734296
- Nguyen D, Fayol O, Buisine N, et al. Functional interaction between HEXIM and hedgehog signaling during drosophila wing development. PLoS One. 2016;11(5):e0155438. doi:10.1371/journal.pone.0155438. PMID:27176767
- Huang F, Wagner M, Siddiqui MA. Ablation of the CLP-1 gene leads to down-regulation of the HAND1 gene and abnormality of the left ventricle of the heart and fetal death. Mech Dev. 2004;121(6):559–572. doi:10.1016/j.mod.2004.04.012. PMID:15172687
- Yik JH, Chen R, Pezda AC, et al. Compensatory contributions of HEXIM1 and HEXIM2 in maintaining the balance of active and inactive P-TEFb complexes for control of transcription. J Biol Chem. 2005;280:16368–16376. doi:10.1074/jbc.M500912200. PMID:15713661
- Czudnochowski N, Vollmuth F, Baumann S, et al. Specificity of Hexim1 and Hexim2 complex formation with cyclin T1/T2, importin alpha and 7SK snRNA. J Mol Biol. 2010;395(1):28–41. doi:10.1016/j.jmb.2009.10.055. PMID:19883659
- Badrock A, Hurlstone A. A hexIM1 on your melanocytes: transcription elongation – the Achilles' heel of melanoma? Pigment Cell Melanoma Res. 2016;29(5):496–497. doi:10.1111/pcmr.12509. PMID:27399276
- Chen R, Yik JH, Lew QJ, et al. Brd4 and HEXIM1: multiple roles in P-TEFb regulation and cancer. Biomed Res Int. 2014;2014:232870. PMID:24592384
- Catalucci D, Condorelli G. HEXIM1: a new player in myocardial hypertrophy? Cardiovasc Res. 2013;99(1):1–3. doi:10.1093/cvr/cvt134. PMID:23720275
- Dey A, Chao SH, Lane DP. HEXIM1 and the control of transcription elongation: from cancer and inflammation to AIDS and cardiac hypertrophy. Cell Cycle. 2007;6(15):1856–1863. doi:10.4161/cc.6.15.4556. PMID:17671421
- Asakura A. Vascular endothelial growth factor gene regulation by HEXIM1 in heart. Circ Res. 2008;102(4):398–400. doi:10.1161/CIRCRESAHA.108.172114. PMID:18309107
- Yik JH, Chen R, Pezda AC, et al. A human immunodeficiency virus type 1 Tat-like arginine-rich RNA-binding domain is essential for HEXIM1 to inhibit RNA polymerase II transcription through 7SK snRNA-mediated inactivation of P-TEFb. Mol Cell Biol. 2004;24(12):5094–5105. doi:10.1128/MCB.24.12.5094-5105.2004. PMID:15169877
- Michels AA, Fraldi A, Li Q, et al. Binding of the 7SK snRNA turns the HEXIM1 protein into a P-TEFb (CDK9/cyclin T) inhibitor. Embo J. 2004;23(13):2608–2619. doi:10.1038/sj.emboj.7600275. PMID:15201869
- Barboric M, Kohoutek J, Price JP, et al. Interplay between 7SK snRNA and oppositely charged regions in HEXIM1 direct the inhibition of P-TEFb. Embo J. 2005;24(24):4291–4303. doi:10.1038/sj.emboj.7600883. PMID:16362050
- Biglione S, Byers SA, Price JP, et al. Inhibition of HIV-1 replication by P-TEFb inhibitors DRB, seliciclib and flavopiridol correlates with release of free P-TEFb from the large, inactive form of the complex. Retrovirology. 2007;4:47. doi:10.1186/1742-4690-4-47. PMID:17625008
- Dames SA, Schönichen A, Schulte A, et al. Structure of the Cyclin T binding domain of Hexim1 and molecular basis for its recognition of P-TEFb. Proc Natl Acad Sci U S A. 2007;104:14312–14317. doi:10.1073/pnas.0701848104. PMID:17724342
- Schonichen A, Bigalke JM, Urbanke C, et al. A flexible bipartite coiled coil structure is required for the interaction of Hexim1 with the P-TEFB subunit cyclin T1. Biochemistry. 2010;49:3083–3091. doi:10.1021/bi902072f. PMID:20210365
- Byers SA, Price JP, Cooper JJ, et al. HEXIM2, A HEXIM1 related protein, regulates P-TEFb through association with 7SK. J Biol Chem. 2005;280:16360–16367. doi:10.1074/jbc.M500424200. PMID:15713662
- Li Q, Price JP, Byers SA, et al. Analysis of the large inactive P-TEFb complex indicates that it contains one 7SK molecule, a dimer of HEXIM1 or HEXIM2, and two P-TEFb molecules containing Cdk9 phosphorylated at threonine 186. J Biol Chem. 2005;280:28819–28826. doi:10.1074/jbc.M502712200. PMID:15965233
- Dulac C, Michels AA, Fraldi A, et al. Transcription-dependent association of multiple P-TEFb units to a HEXIM multimer. J Biol Chem. 2005;280:30619–30629. doi:10.1074/jbc.M502471200. PMID:15994294
- Mitrea DM, Yoon MK, Ou L, et al. Disorder-function relationships for the cell cycle regulatory proteins p21 and p27. Biol Chem. 2012;393(4):259–274. doi:10.1515/hsz-2011-0254. PMID:23029651
- Zhou Q, Li T, Price DH. RNA Polymerase II Elongation Control. Annu Rev Biochem. 2012;81:119–143. doi:10.1146/annurev-biochem-052610-095910. PMID:22404626
- Jonkers I, Lis JT. Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol. 2015;16(3):167–177. doi:10.1038/nrm3953. PMID:25693130
- Li Y, Liu M, Chen LF, et al. P-TEFb: finding its ways to release promoter-proximally paused RNA Polymerase II. Transcription. 2017;19:1–7. doi:10.1080/21541264.2017.1281864.
- Diribarne G, Bensaude O. 7SK RNA, a non-coding RNA regulating P-TEFb, a general transcription factor. RNA Biol. 2009;6:122–128. doi:10.4161/rna.6.2.8115. PMID:19246988
- McNamara RP, Bacon CW, D'Orso I. Transcription elongation control by the 7SK snRNP complex: releasing the pause. Cell Cycle. 2016;15(16):2115–2123. doi:10.1080/15384101.2016.1181241. PMID:27152730
- Quaresma AJ, Bugai A, Barboric M. Cracking the control of RNA polymerase II elongation by 7SK snRNP and P-TEFb. Nucleic Acids Res. 2016;44(16):7527–7539. doi:10.1093/nar/gkw585. PMID:27369380
- Nguyen D, Krueger BJ, Sedore SC, et al. The Drosophila 7SK snRNP and the essential role of dHEXIM in development. Nucleic Acids Res. 2012;40(12):5283–5297. doi:10.1093/nar/gks191.
- Verstraete N, Kuzmina A, Diribarne G, et al. A cyclin T1 point mutation that abolishes positive transcription elongation factor (P-TEFb) binding to HEXIM1 and HIV tat. Retrovirology. 2014;11(50): doi:10.1186/1742-4690-11-50.
- Egloff S, Van Herreweghe E, Kiss T. Regulation of polymerase II transcription by 7SK snRNA: two distinct RNA elements direct P-TEFb and HEXIM1 binding. Mol Cell Biol. 2006;26(2):630–642. doi:10.1128/MCB.26.2.630-642.2006. PMID:16382153
- Martinez-Zapien D, Saliou JM, Han X, et al. Intermolecular recognition of the non-coding RNA 7SK and HEXIM protein in perspective. Biochimie. 2015;117:63–71. doi:10.1016/j.biochi.2015.03.020. PMID:25863285
- Brogie JE, Price DH. Reconstitution of a functional 7SK snRNP. Nucleic Acids Res. 2017;45(11):6864–6880. doi:10.1093/nar/gkx262. PMID:28431135
- Lebars I, Martinez-Zapien D, Durand A, et al. HEXIM1 targets a repeated GAUC motif in the riboregulator of transcription 7SK and promotes base pair rearrangements. Nucleic Acids Res. 2010;38:7749–7763. doi:10.1093/nar/gkq660. PMID:20675720
- Martinez-Zapien D, Legrand P, McEwen AG, et al. The crystal structure of the 5 functional domain of the transcription riboregulator 7SK. Nucleic Acids Res. 2017;45(6):3568–3579. PMID:28082395
- Fujinaga K, Luo Z, Peterlin BM. Genetic analysis of the structure and function of 7SK small nuclear ribonucleoprotein (snRNP) in cells. J Biol Chem. 2014;289(30):21181–21190. doi:10.1074/jbc.M114.557751. PMID:24917669
- Bélanger F, Baigude H, Rana TM. U30 of 7SK RNA Forms a Specific Photo-cross-link with Hexim1 in the Context of Both a Minimal RNA-binding,site and a Fully Reconstituted 7SK/Hexim1/P-TEFb,Ribonucleoprotein Complex. J Mol Biol. 2009;386:1094–1107. doi:10.1016/j.jmb.2009.01.015. PMID:19244621
- Czudnochowski N, Bosken CA, Geyer M. Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition. Nat Commun. 2012;3:842. doi:10.1038/ncomms1846. PMID:22588304
- Schulte A, Czudnochowski N, Barboric M, et al. Identification of a cyclin T-binding domain in Hexim1 and biochemical analysis of its binding competition with HIV-1 Tat. J Biol Chem. 2005;280:24968–24977. doi:10.1074/jbc.M501431200. PMID:15855166
- Ogba N, Doughman YQ, Chaplin LJ, et al. HEXIM1 modulates vascular endothelial growth factor expression and function in breast epithelial cells and mammary gland. Oncogene. 2010;29:3639–3649. doi:10.1038/onc.2010.110. PMID:20453883
- Kobbi L, Demey-Thomas E, Braye F, et al. An evolutionary conserved Hexim1 peptide binds to the Cdk9 catalytic site to inhibit P-TEFb. Proc Natl Acad Sci USA. 2016; 201612331. doi:10.1073/pnas.1612331113. PMID:27791144
- Chen R, Yang Z, Zhou Q. Phosphorylated P-TEFb is tagged for inhibition through association with 7SK snRNA. J Biol Chem. 2004;279:4153–4160. doi:10.1074/jbc.M310044200. PMID:14627702
- Ott M, Geyer M, Zhou Q. The control of HIV transcription: keeping RNA polymerase II on track. Cell Host Microbe. 2011;10(5):426–435. doi:10.1016/j.chom.2011.11.002. PMID:22100159
- Tahirov TH, Babayeva ND, Varzavand K, et al. Crystal structure of HIV-1 Tat complexed with human P-TEFb. Nature. 2010;465:747–751. doi:10.1038/nature09131. PMID:20535204
- Sedore SC, Byers SA, Biglione S, et al. Manipulation of P-TEFb control machinery by HIV: recruitment of P-TEFb from the large form by Tat and binding of HEXIM1 to TAR. Nucleic Acids Res. 2007;35:4347–4358. doi:10.1093/nar/gkm443. PMID:17576689
- Li Q, Cooper JJ, Altwerger GH, et al. HEXIM1 is a promiscuous double-stranded RNA-binding protein and interacts with RNAs in addition to 7SK in cultured cells. Nucleic Acids Res. 2007;35:2503–2512. doi:10.1093/nar/gkm150. PMID:17395637
- D'Orso I, Jang GM, Pastuszak AW, et al. Transition step during assembly of HIV Tat:P-TEFb transcription complexes and transfer to TAR RNA. Mol Cell Biol. 2012;32(23):4780–4793. doi:10.1128/MCB.00206-12. PMID:23007159
- Mizutani T, Ishizaka A, Suzuki Y, et al. 7SK small nuclear ribonucleoprotein complex is recruited to the HIV-1 promoter via short viral transcripts. FEBS Lett. 2014;588(9):1630–1636. doi:10.1016/j.febslet.2014.01.067. PMID:24607481
- Barboric M, Yik JH, Czudnochowski N, et al. Tat competes with HEXIM1 to increase the active pool of P-TEFb for HIV-1 transcription. Nucleic Acids Res. 2007;35:2003–2012. doi:10.1093/nar/gkm063. PMID:17341462
- Fraldi A, Varrone F, Napolitano G, et al. Inhibition of Tat activity by the HEXIM1 protein. Retrovirology. 2005;2(1):42. doi:10.1186/1742-4690-2-42. PMID:15992410
- Muck F, Bracharz S, Marschalek R. DDX6 transfers P-TEFb kinase to the AF4/AF4N (AFF1) super elongation complex. Am J Blood Res. 2016;6(3):28–45. PMID:27679741
- Nguyen VT, Kiss T, Michels AA, et al. 7SK snRNA binds to and inhibits the activity of Cdk9/cyclin T complexes. Nature. 2001;414:322–325. doi:10.1038/35104581. PMID:11713533
- Yang Z, Zhu Q, Luo K, et al. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature. 2001;414:317–322. doi:10.1038/35104575. PMID:11713532
- Barrandon C, Bonnet F, Nguyen VT, et al. The transcription-dependent dissociation of P-TEFb.HEXIM1.7SK RNA relies upon formation of hnRNP.7SK RNA complexes. Mol Cell Biol. 2007;27:6996–7006. doi:10.1128/MCB.00975-07. PMID:17709395
- Van Herreweghe E, Egloff S, Goiffon I, et al. Dynamic remodelling of human 7SK snRNP controls the nuclear level of active P-TEFb. EMBO J. 2007;26(15):3570–3580. doi:10.1038/sj.emboj.7601783. PMID:17611602
- Lemieux B, Blanchette M, Monette A, et al. A Function for the hnRNP A1/A2 proteins in transcription elongation. PLoS One. 2015;10(5):e0126654. doi:10.1371/journal.pone.0126654. PMID:26011126
- Contreras X, Barboric M, Lenasi T, et al. HMBA releases P-TEFb from HEXIM1 and 7SK snRNA via PI3K/Akt and activates HIV transcription. PLoS Pathog. 2007;3(10):1459–1469. doi:10.1371/journal.ppat.0030146. PMID:17937499
- Mbonye UR, Wang B, Gokulrangan G, et al. Phosphorylation of HEXIM1 at Tyr271 and Tyr274 Promotes Release of P-TEFb from the 7SK snRNP Complex and Enhances Proviral HIV Gene Expression. Proteomics. 2015;15(12):2078–2086. doi:10.1002/pmic.201500038. PMID:25900325
- Kim YK, Mbonye U, Hokello J, et al. T-cell receptor signaling enhances transcriptional elongation from latent HIV proviruses by activating P-TEFb through an ERK-dependent pathway. J Mol Biol. 2011;410:896–916. doi:10.1016/j.jmb.2011.03.054. PMID:21763495
- Fujinaga K, Barboric M, Li Q, et al. PKC phosphorylates HEXIM1 and regulates P-TEFb activity. Nucleic Acids Res. 2012;40(18):9160–9170. doi:10.1093/nar/gks682. PMID:22821562
- Espinoza-Derout J, Wagner M, Shahmiri K, et al. Pivotal role of cardiac lineage protein-1 (CLP-1) in transcriptional elongation factor P-TEFb complex formation in cardiac hypertrophy. Cardiovasc Res. 2007;75:129–138. doi:10.1016/j.cardiores.2007.03.019. PMID:17459355
- Chen R, Liu M, Li H, et al. PP2B and PP1cooperatively disrupt 7SK snRNP to release P-TEFb for transcription in response to Ca 2+ signaling Genes Dev. 2008;22:1356–1368. doi:10.1101/gad.1636008. PMID:18483222
- Wang Y, Dow EC, Liang YY, et al. Phosphatase PPM1A regulates phosphorylation of Thr-186 in the Cdk9 T-loop. J Biol Chem. 2008;283(48):33578–33584. doi:10.1074/jbc.M807495200. PMID:18829461
- Gudipaty SA, D'Orso I. Functional interplay between PPM1G and the transcription elongation machinery. RNA Dis. 2016;3(1): PMID:27088130
- Zhang H, Park SH, Pantazides BG, et al. SIRT2 directs the replication stress response through CDK9 deacetylation. Proc Natl Acad Sci USA. 2013;110(33):13546–13551. doi:10.1073/pnas.1301463110. PMID:23898190
- Blank MF, Chen S, Poetz F, et al. SIRT7-dependent deacetylation of CDK9 activates RNA polymerase II transcription. Nucleic Acids Res. 2017;45(5):2675–2686. doi:10.1093/nar/gkx053. PMID:28426094
- Galatioto J, Mascareno E, Siddiqui MA. CLP-1 associates with MyoD and HDAC to restore skeletal muscle cell regeneration. J Cell Sci. 2010;123(Pt 21):3789–3795. doi:10.1242/jcs.073387. PMID:20940258
- McNamara RP, McCann JL, Gudipaty SA, et al. Transcription factors mediate the enzymatic disassembly of promoter-bound 7SK snRNP to locally recruit P-TEFb for transcription elongation. Cell Rep. 2013;5:1256–1268. doi:10.1016/j.celrep.2013.11.003. PMID:24316072
- Gudipaty SA, McNamara RP, Morton EL, et al. PPM1G binds 7SK RNA and Hexim1 to block P-TEFb assembly into the 7SK snRNP and sustain transcription elongation. Mol Cell Biol. 2015;35(22):3810–3828. doi:10.1128/MCB.00226-15. PMID:26324325
- D'Orso I. 7SKiing on chromatin: move globally, act locally. RNA Biol. 2016;13(6):545–553. doi:10.1080/15476286.2016.1181254. PMID:27128603
- Ji X, Zhou Y, Pandit S, et al. SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase. Cell. 2013;153(4):855–868. doi:10.1016/j.cell.2013.04.028. PMID:23663783
- Ouchida R, Kusuhara M, Shimizu N, et al. Suppression of NF-kappaB-dependent gene expression by a hexamethylene bisacetamide-inducible protein HEXIM1 in human vascular smooth muscle cells. Genes Cells. 2003;8(2):95–107. doi:10.1046/j.1365-2443.2003.00618.x. PMID:12581153
- Ketchart W, Ogba N, Kresak A, et al. HEXIM1 is a critical determinant of the response to tamoxifen. Oncogene. 2011;30:3563–3569. doi:10.1038/onc.2011.76. PMID:21423213
- Wittmann BM, Fujinaga K, Deng H, et al. The breast cell growth inhibitor, estrogen down regulated gene 1, modulates a novel functional interaction between estrogen receptor alpha and transcriptional elongation factor cyclin T1. Oncogene. 2005;24: doi:10.1038/sj.onc.1208728. PMID:15940264
- Yeh IJ, Song K, Wittmann BM, et al. HEXIM1 plays a critical role in the inhibition of the androgen receptor by anti-androgens. Biochem J. 2014;462(2):315–327. doi:10.1042/BJ20140174. PMID:24844355
- Shimizu N, Ouchida R, Yoshikawa N, et al. HEXIM1 forms a transcriptionally abortive complex with glucocorticoid receptor without involving 7SK RNA and positive transcription elongation factor b. Proc Natl Acad Sci USA. 2005;102:8555–8560. doi:10.1073/pnas.0409863102. PMID:15941832
- Liu X, Gao Y, Ye H, et al. Positive feedback loop mediated by protein phosphatase 1alpha mobilization of P-TEFb and basal CDK1 drives androgen receptor in prostate cancer. Nucleic Acids Res. 2017;45(7):3738–3751. PMID:28062857
- Lama R, Gan C, Idippily N, et al. HMBA is a putative HSP70 activator stimulating HEXIM1 expression that is down-regulated by estrogen. J Steroid Biochem Mol Biol. 2017;168:91–101. doi:10.1016/j.jsbmb.2017.02.008. PMID:28213333
- Kanno T, Kanno Y, LeRoy G, et al. BRD4 assists elongation of both coding and enhancer RNAs by interacting with acetylated histones. Nat Struct Mol Biol. 2014;21(12):1047–1057. doi:10.1038/nsmb.2912.
- Winter GE, Mayer A, Buckley DL, et al. BET bromodomain proteins function as master transcription elongation factors independent of CDK9 recruitment. Mol Cell. 2017;67(1):5–18, e19. doi:10.1016/j.molcel.2017.06.004. PMID:28673542
- Schroder S, Cho S, Zeng L, et al. Two-pronged binding with bromodomain-containing protein 4 liberates positive transcription elongation factor b from inactive ribonucleoprotein complexes. J Biol Chem. 2012;287(2):1090–1099. doi:10.1074/jbc.M111.282855. PMID:22084242
- Itzen F, Greifenberg AK, Bosken CA, et al. Brd4 activates P-TEFb for RNA polymerase II CTD phosphorylation. Nucleic Acids Res. 2014;42(12):7577–7590. doi:10.1093/nar/gku449. PMID:24860166
- Liu L, Xu Y, He M, et al. Transcriptional pause release is a rate-limiting step for somatic cell reprogramming. Cell Stem Cell. 2014;15(5):574–588. doi:10.1016/j.stem.2014.09.018. PMID:25312495
- Egloff S, Studniarek C, Kiss T. 7SK small nuclear RNA, a multifunctional transcriptional regulatory RNA with gene-specific features. Transcription. 2017;18:1–7. doi:10.1080/21541264.2017.1344346.
- Gurumurthy M, Tan CH, Ng R, et al. Nucleophosmin interacts with HEXIM1 and regulates RNA polymerase II transcription. J Mol Biol.2008;378(2):302–317. doi:10.1016/j.jmb.2008.02.055. PMID:18371977
- Lew QJ, Chu KL, Chia YL, et al. HEXIM1, a New Player in the p53 Pathway. Cancers (Basel). 2013;5(3):838–856. doi:10.3390/cancers5030838. PMID:24202322
- Yeh IJ, Ogba N, Bensigner H, et al. HEXIM1 down-regulates hypoxia-inducible factor-1alpha protein stability. Biochem J. 2013;456(2):195–204. doi:10.1042/BJ20130592. PMID:24015760
- Fujimoto Y, Nakamura Y, Ohuchi S. HEXIM1-binding elements on mRNAs identified through transcriptomic SELEX and computational screening. Biochimie. 2012;94(9):1900–1909. doi:10.1016/j.biochi.2012.05.003. PMID:22609015
- Morchikh M, Cribier A, Raffel R, et al. HEXIM1 and NEAT1 Long Non-coding RNA form a multi-subunit complex that regulates DNA-mediated innate immune response. Mol Cell. 2017;67:387–399. doi:10.1016/j.molcel.2017.06.020. PMID:28712728
- Adriaens C, Marine JC. NEAT1-containing paraspeckles: Central hubs in stress response and tumor formation. Cell Cycle. 2017;16(2):137–138. doi:10.1080/15384101.2016.1235847. PMID:27700225
- Kato K, Omura H, Ishitani R, et al. Cyclic GMP-AMP as an endogenous second messenger in innate immune signaling by cytosolic DNA. Annu Rev Biochem. 2017;86:541–566. doi:10.1146/annurev-biochem-061516-044813. PMID:28399655
