ABSTRACT
RNA modifications are prevalent among all the classes of RNA, regulate diverse biological processes, and have emerged as a key regulatory mechanism in post-transcriptional control of gene expression. They are subjected to precise spatial and temporal control and shown to be critical for the maintenance of normal development and physiology. For example, m6A modification of mRNA affects stability, recruitment of RNA binding protein (RBP), translation, and splicing. The deposition of m6A on the RNA happens co-transcriptionally, allowing the tight coupling between the transcription and RNA modification machinery. The m6A modification is affected by transcriptional dynamics, but recent insights also suggest that m6A machinery impacts transcription and chromatin signature.
Keywords:
1. Introduction
Like DNA and proteins, RNAs can be modified extensively. These RNA modifications, collectively known as epitranscriptome, have emerged in the last decade as an important additional regulatory layer of gene expression [Citation1]. To date, more than 170 distinct RNA modifications have been identified across different classes of RNA [Citation2]. RNA modifications can control many aspects of RNA metabolism, and their disruption has been associated with a wide range of physiological alterations and many diseases including neurological disease as well as various cancers. The development of genomic approaches over the past decade, which can map their asymmetric distribution, has facilitated the identification of diverse modifications on mRNA molecules [Citation3,Citation4]. These modifications are subjected to strict temporal and spatial regulation, suggesting their critical role in regulation of mRNA fate. Indeed, they are dynamic in nature, asymmetric in distribution, and demonstrate enrichment for developmental genes or genes sensitive to environmental stresses. Subsequently, their role in developmental control and stress regulated gene expression has been highlighted by several studies [Citation1,Citation5–7].
However, owing to the extensive role, it plays in the regulation of mRNA fate, N6-methyladenosine (m6A) modification is the most well-studied epitranscriptomic signature [Citation8,Citation9]. The critical role of the m6A in gene expression regulation is also reflected by the high degree of conservation across the metazoans, including among mammals. Not surprisingly, intensive research into this modification in recent years has implicated m6A in a wide range of processes, from early development, immunological response, and to cancers [Citation2,Citation10].
On the basis of transcriptome-wide mapping approaches: pseudouridine (Ψ), Inosine, N1- methyladenosine (m1A), ribose methylations (Nm), or 5-methylcytidine (m5C), are also among the most prevalent internal modifications on mRNA, but their precise role in the regulation of gene expression remains relatively poorly studied, so far. In this review, we will specifically describe recent findings which highlights the cross-talk between RNA modifications and transcription regulation with a special focus on the most studied one, m6A.
2. m6A mRNA modification process
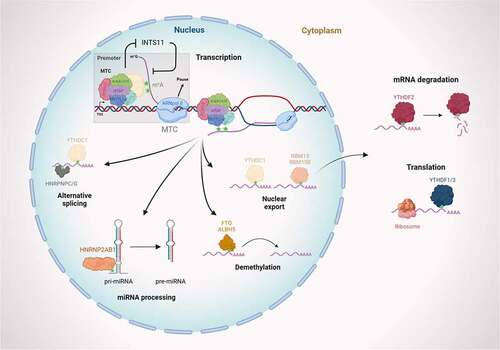
The heterodimeric complex of methyltransferases METTL3 and METTL14 often referred to as “writers” is responsible for the deposition of the m6A modification to target RNAs, through the catalytic domain of METTL3. Two additional regulatory members of the complex, WTAP and KIAA1429, are required for the targeting and formation of an active m6A methyltransferase complex (MTC) () [Citation11]. RBM15 and RBM15B bind the MTC and recruit it to specific sites in mRNA [Citation12]. Notably, both RBM15 and RBM15B have been implicated in mRNA export control [Citation13,Citation14]. Finally, the most recently discovered member of the MTC, ZC3H13, bridges RBM15 to WTAP to maintain the nuclear localization of the m6A MTC [Citation15]. In addition to writers, the literature suggests the existence of m6A demethylases also called erasers. Among them, we find fat mass- and obesity-associated protein (FTO) and ALKBH5, a ALKB subfamily member of the superfamily of Fe(II)/2-oxoglutarate dioxygenases with demethylase activity preferences for single stranded RNA () [Citation16,Citation17, Citation18]. Finally, the last category consists of “reader proteins” which recognize and bind the m6A modification. They include cytoplasmic YTHDF1-3 [Citation19,Citation24] nuclear YTHDC1,2 [Citation18], and several hnRNP proteins including HNRNPC/G which alters splicing decisions [Citation20,Citation21] and HNRNPA2B1 which regulates microRNA processing events () [Citation22]. The cytoplasmic reader proteins play roles in translation and mRNA stability [Citation23,Citation24]. The nuclear reader protein, YTHDC1, is also implicated in splicing control [Citation25] and in mRNA export in conjunction with SRSF3 [Citation26]. Many more processes have so far been described as m6A dependent, but it is only very recently that a role of direct feedback on transcription has been demonstrated.
m6A methylation is catalyzed by the writer complex including METTL3, METTL14, WTAP, KIAA1429, and ZC3H13. The m6A modification is erased by demethylases including FTO and ALKBH5. The m6A-modified RNA reader proteins include YTHDC1/2, YTHDF1/2/3, and RBM15/15B HNRNPC/G/2AB1. m6A modification modulates transcription, miRNA biogenesis, RNA translocation, pre-mRNA splicing, RNA translation, RNA decay, and RNA stability (created with BioRender.com).
3. m6A: A context dependent transcription regulator?
The deposition of m6A modification occurs co-transcriptionally, and transcription efficiency has been shown to impact the m6A methyltransferase catalytic activity on mRNA. Slow transcription rate will allow high methylation on mRNA while rapid transcriptional activity of RNAP II will decrease methylation. These divergent methylation profiles then impact the translation efficiency of these mRNAs bridging transcription and translation [Citation27]. Through this mechanism the transcriptional control of translation was demonstrated, whereby m6A on the RNA acts as key regulatory module that negatively correlated with translational efficiency. Interestingly, recent findings have also shown that m6A methyl transferase complex, together with nuclear reader protein Ythdc1, can regulate the transcription itself. These findings combined with the transcriptional control of m6A deposition points toward extensive cross talk between the transcription and RNA modification machinery. We will describe below some of the studies which establishes this link and show different kind of transcriptional effect, viz, activation or repression, mediated by m6A MTC.
3.1 m6A and transcriptional repression
The transcriptional regulation is context dependent often involving a precise interplay between activation or repression to maintain cellular functions and homeostasis. Heat shock regulation is a prime example of transcriptional response underlying the maintenance of homeostasis. In mouse, heat-shock induced lncRNA, HEAT, acts as an attenuating factor for heat-shock response. The lncRNA HEAT, which is m6A modified, directly binds to the heat shock transcription factor 1 (HSF1) and functions in trans to negatively regulate heat shock genes [Citation28]. Although, dispensable for HSF1 binding, m6A modification is required for formation of Ythdc1-mediated transcriptional silencer complex. Another interesting example of m6A-directed transcriptional repression is the long non-coding RNA X-inactive specific transcript (XIST) mediated repression. In human cells, XIST is heavily methylated which recruits RBM15 and RBM15B to maintain XIST-mediated transcriptional silencing [Citation12]. Furthermore, the knockdown of Mettl3 leads to the loss of XIST-mediated repression and tethering of Ythdc1 is sufficient to rescue the repression, demonstrating the role of m6A and the nuclear reader Ythdc1 in this repression.
Finally, an indirect mechanism of transcriptional control involving the degradation of chromatin associated RNAs (carRNAs), mediated by Ythdc1 has also been reported in mESCs. This mechanism involves the nuclear exosome targeting complex (NEXT) recruited by Ythdc1 on m6A-methylated carRNAs, including LINE-1, that promotes their degradation resulting in a repressed chromatin context, which is non-permissive to transcription () [Citation29]. However, effects of m6A methylation on carRNAs could vary and m6A may stabilize modified carRNAs in different cell types.
3.2 m6A eRNA and transcription condensates
In two recent reports, it was shown that enhancer RNAs can be m6A methylated and the extent of methylation modulate their own transcription as well as of their downstream target genes [Citation29,Citation30]. Furthermore, long eRNA seems to be the most extensively methylated, which recruits Ythdc1, leading to the phase separation and facilitation of coactivator condensate formation [Citation30]. This function seems to be mediated by the C-terminal domain of the Ythdc1 protein that contains the Intrinsically Disordered Regions (IDR2) and arginine residues. The interaction of Ythdc1 and eRNA allows phase transition to form biomolecular condensates, which are dynamically modulated by RNAs. These domains form liquid-like droplets within the nucleus and co-mixes and facilitates the formation of co-activator condensates with BRD4, which correlates with strong enhancer activity and target gene transcriptional induction () [Citation30]. Transcriptional condensate represents a new paradigm in gene regulation that can explain the presence of high concentrations of transcription factors, cofactors, and RNA Pol II at specific genomic regions and in particular at super-enhancers [Citation31]. The recent evidence points toward an exciting role m6A modification play in the regulation of this paradigm. Additionally, Ythdc1 has been also shown to localize in the nucleus in foci called YT bodies that are in close contact to other subnuclear structures such as speckles and coiled bodies [Citation32]. YT bodies are formed during S-phase of the cell cycle and represent dynamic active transcription focal sites. Evidence to date is lacking whether these two mechanisms are similar and co-exist or if they are dependent on specific cellular contexts. Nevertheless, together they point toward an interesting mechanism where m6A modification plays a critical role in sub-cellular compartmentalization of biological processes.
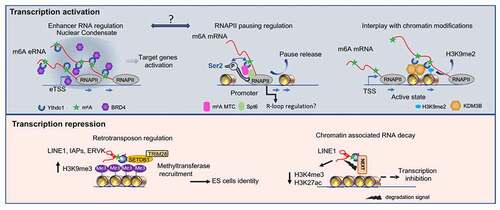
The top panel shows different mechanisms leading to the activation of transcription.
Left: m6A-modified enhancer RNAs recruit Ythdc1 which physically interacts with the BRD4 coactivator via their IDR domain leading to a transcriptionally active nuclear condensate.
The middle part represents a model of m6A-mediated RNAPII pause release involving the dual interaction between promoter and nascent RNA via m6A MTC and Ythdc1. Release to the elongation form of RNAPII (Ser2P) involves physical interaction with the elongation factor Spt6. It would be interesting in the future to study the potential implication of the m6A RNA on the interaction between enhancer (RNA) and promoter. The link between transcription activation, m6A, and regulation of the R loop at the promoter level also needs to be further investigated.
The last schematic representation shows the recruitment by Ythdc1 of the H3K9me2 demethylase leading to modifications of the chromatin and to the activation of transcription.
Lower panel: m6A-mediated transcriptional repression. MTC can methylate retrotransposons like LINE1, intracisternal A particles (IAP), or ERVK, leading to Ythdc1-dependent recruitment of the methyltransferase SETDB1 and its cofactor TRIM28. This will lead to an increase in H3K9me3 and the silencing of retrotransposons.
The right part is a model showing how the m6A methylation of LINE1 leads to its degradation through the Ythdc1 dependent recruitment of the nuclear exosome targeting-mediated nuclear degradation complex (NEXT). This mechanism leads to changes in the chromatin state with loss of active histone marks and inhibition of transcription.
3.3 m6A modulates RNAPII pause release
Transcription comprises three different and highly regulated steps: initiation, elongation, and termination. Following initiation, RNA polymerase II (RNA Pol II) enters a key rate-limiting stage during which it accumulates downstream of the transcription start site, known as promoter-proximal pausing of RNA Pol II. This RNA Pol II pausing mechanism is particularly important for developmental genes or signal activated genes, under stimuli control. This checkpoint allows tight coupling between the transcription and RNA processing events.
Control of the pause step is related to the phosphorylation state of the C-terminal domain (CTD) of the larger RNA Pol II subunit [Citation33]. Specifically, the phosphorylation of Serine at position 2 of the CTD domain corresponds to the elongation form of RNA Pol II [Citation34,Citation35]. The transition from pause to elongation is critical for gene expression in higher metazoans and thus subject to tight regulation [Citation36–38].
In a recent study, researchers found that the m6A MTC directly regulates the release of RNA Pol II from its paused state () [Citation39]. In Drosophila cells, the complex is primarily recruited to gene promoters. This recruitment dependent on active transcription and showed enrichment for genes which are subjected to higher level of pausing. Additionally, this effect was not observed in a mutant of the catalytic domain of Mettl3 methyl transferase strongly suggesting that this regulation is dependent on m6A modification on the RNA. In addition, tethering Mettl3 to the promoter of heterologous gene is sufficient to stimulate the release of RNA Pol II from the promoter into productive elongation. By measuring the amount of RNA produced in a defined time-window, through DRB 4SU-Seq, the authors also showed an overall decrease in transcription when components of m6A MTC were depleted [Citation39].
One of the outstanding questions in the field is regarding the specificity of the m6A deposition. Recent study in mammalian cells showed that the m6A deposition on the RNA is guided by H3K36me3 [Citation40]. However, few evidences rule out this being an exclusive mechanism. First, the H3K36me3 deposition is primarily on exons which cannot explain the m6A deposition on the 5’ UTR. Second, the deposition of m6A in different species, cell types, and physiological conditions are different whose specificity cannot be explained by H3K36me3 pattern. Indeed, the recent study in Drosophila cells showed that m6A deposition is not guided by H3K36me3. The authors by using machine learning algorithm identified the RNA Pol II, chromatin, and pause factors binding as primary determinants of m6A deposition. Although, the matrix of chromatin and transcription associated factors was not exhaustive, yet the involvement of these factors in the recruitment of m6A MTC at the selected promoters could also be validated [Citation39]. In addition, the authors provide evidence that components of the m6A complex interact with a histone chaperone protein, Spt6, previously shown to participate in the initiation and elongation of transcription [Citation39].
Taken together, these findings support a model that m6A methylation, via m6A MTC, acts directly on the transcription machinery by inducing the release of RNA Pol II from the paused state. This highlights a novel positive feedback mechanism mediated via the epitranscriptome to promote the expression of genes.
3.4 Additional m6A regulators in transcription control
It is expected that the effect of m6A on transcription is executed through nuclear localized m6A interactors. Several nuclear localized m6A regulators have been recently described and some evidence indicates that some of these might regulate transcription. For instance, two recent studies suggested a possible role for the m6A regulator, RBMX (hnRNPG) in control of nascent transcription. RBMX has previously been implicated with m6A in regulating alternative splicing, however, recent evidence link its function to the control of nascent transcription of CBX5 (HP1α) an important regulator of H3K9me3 [Citation41]. Furthermore, RBMX and Ythdc1 binding to the 5’-ends of m6A RNA were recently found to protect nascent mRNAs from premature Integrator-mediated termination, thus promoting productive transcription [Citation42]. We can therefore assume that several additional m6A binders, which possess transcription regulatory role, might be discovered soon.
3.5 m6A and the coupling with the epigenetic regulation
Ythdc1-mediated transcriptional regulation can be achieved by several distinct mechanisms, including a direct interaction with chromatin remodeling proteins. Consistent with this possibility, a recent work in mouse embryonic stem cells (mESCs) shows that co-transcriptional deposition of m6A directs KDM3B to chromatin through the m6A reader Ythdc1 (). This leads to demethylation of H3K9me2 and subsequently to activation of gene expression (). The uncovered mechanism indicates a direct cross-talk between RNA modification and chromatin state, involving a positive feedback loop that drives gene expression.
Table 1. m6A functions related to epigenetic regulation
Contrary to the overall positive impact seen previously on mRNA and eRNA transcription, an interesting report show that Ythdc1 recruits SETDB1, which is required to maintain mESC identity and retrotransposon repression by recognizing a subset of m6A-marked TE-derived transcripts (). Specifically, SETDB1-mediated H3K9me3 is dependent on Ythdc1 and m6A RNA to prevent the de-differentiation of mESCs into two-cell (2C) stage like transition, and to keep retrotransposons silenced (). Additionally, m6A regulates the level of histone methyltransferase Ezh2 of Polycomb Repressive Complex 2 (PRC2), and its reduction upon Mettl3 knockdown decreased both Ezh2 protein expression and consequent H3K27me3 levels [Citation43]. Interestingly, SETDB1 also modulates the recruitment of PRC2. In a subset of SETDB1 bound peaks, loss of SETDB1 also resulted in loss of Ezh2 binding leading to decrease in H3K27me3 by PRC2 complex [Citation44]. These results as well as other reports reveal a previously undefined key role of RNA m6A modification and the reader Ythdc1 in chromatin remodeling leading to transcriptional repression at specific retrotransposon loci, heterochromatin formation and regulating early development [Citation45–47]; .
These findings add yet another layer of function to the dynamic epitranscriptome and highlight the integration of epitranscriptomic signaling into the fine tuning of gene expression.
4. m6A is required for splicing regulation
Numerous studies have demonstrated differential exon usage upon the loss of function of m6A MTC, suggesting its role in the regulation of splicing [Citation49,Citation50]. Yet, the role of m6A in splicing somehow started on a controversial footing. A study noted that nascent RNA, although containing many unspliced introns, rarely harbored m6A in introns. Additionally, the authors highlighted that the m6A sites were distal to the splice sites and the m6A harboring exons were spliced similarly in the absence of Mettl3, suggesting that m6A on the RNAs are inconsequential for splicing [Citation51]. However, several later studies highlighted the direct role of m6A in splicing regulation. One such study found that local m6A-dependent RNA structures facilitate binding of heterogeneous ribonucleoprotein C (HNRNPC) to mRNA. Any changes in m6A level led to structural switch dependent changes in HNRNPC binding, in turn affecting the alternative splicing of target mRNAs [Citation20]. A study by Xiao and colleagues showed that Ythdc1 promotes exon inclusion in targeted mRNAs through binding of pre-mRNA splicing factor SRSF3 while blocking SRSF10 binding to mRNA. This study clearly demonstrated that m6A directly regulates mRNA splicing through recruiting and modulating pre-mRNA splicing factors [Citation25]. Furthermore, multiple studies suggest that the m6A MTC complex affects the binding of ribonucleoprotein complexes (RNPs), reviewed in [Citation52]. In another study from Xiao and colleagues, hnRNPG is shown to interact with m6A on nascent RNA and CTD of RNA Pol II to regulate alternative splicing [Citation53]. Finally, m6A modification on 3’ splice site is shown to block U2AF35 binding which resulted in inhibition of splicing. The change in the methylation pattern is triggered by the change in diet and hence the subsequent splicing inhibition [Citation54].
There is growing evidence that shows a tight coupling of RNA processing events with transcription. The center piece of this coupling is the C-terminal domain (CTD) of RNA Pol II which facilitates the interactions between the transcription machinery and RNA processing factors including, but not limited to, splicing and RNA modification factors.
As transcription plays a role in exon definition, many splicing factors in-turn are also known to regulate exon definition by precisely controlling transcriptional state. This includes core spliceosome factors as well as spliceosome associated factors, which are shown to regulate transcriptional dynamics including transition from pausing to elongation as well as processivity [Citation55–57]. With recent studies demonstrating role of m6A MTC in modulating the transcriptional dynamics by affecting pausing [Citation39], chromatin context [Citation58], and eRNAs [Citation29,Citation30], further work is expected to demonstrate how m6A dependent modulation of transcription directly affects splicing outcomes.
5. Future directions
5.1 m6A-mediated R-loops resolution or accumulation?
Recent studies highlight the role RNA: DNA hybrids, called R-loops, play in the regulation of transcription, DNA repair, or even in genomic instability. The R-loops are formed when the nascent RNA hybridizes with the DNA template and the remaining single stranded non-template DNA. These structures are mainly found in TSS regions and positively correlate with RNAPII pausing [Citation59]. Indeed, blocking transcription elongation with DRB reagent, which increases the RNA Pol II pausing, results in increased R-loop formation at the TSS. Several other studies with R-ChIP and GRO-seq experiment have shown a decrease in R-loops and RNAPII occupancy at TSS after DRB removal, consistent with the idea that DRB removal allows transition into elongation. However, the causal relationship between these two processes, R-loop formation and RNA Pol II pausing, is not clearly established. It still needs to be conclusively shown that whether R-loops establish pausing or it is a consequence of stalled polymerase at the TSS.
A recent study has shown in HeLa cells that m6A RNA participates in the termination of transcription through the formation of R-loops. The accumulation of R-loops at the transcription end site decreases in the METTL3 KD condition [Citation60]. Interestingly, it was also shown that the accumulation of R-loops increases under the METTL3 KD condition in hPSCs [Citation61]. In this study, authors identified several m6A binding proteins, such as YTHDF1 and 2 together with METTL3, in interaction with R-loops. Another finding indicates that the transcriptional regulator tonicity-responsive enhancer-binding protein (TonEBP, also called NFAT5) recognizes R-loops, recruits METTL3 to methylate RNA, which helps an RNA cleavage enzyme to resolve R-loops, thus maintaining genome integrity [Citation62].
Together, these studies suggest a context dependent relationship between m6A and R-loop formation. Thus, it would be interesting to check if m6A could, in certain cellular contexts, promote the release of RNAPII promoter pausing through the destabilization of R-loops. Furthermore, these results raise interesting questions regarding how METTL3 contacts R-loops and what mechanism supports the accumulation or resolution of R-loops?
In conclusion, the link between R-loops formation/resolution and m6A deposition needs to be clarified, especially in the context of transcription regulation at promoter.
5.2 Chromatin interaction topology
In Drosophila, m6A MTC components preferentially bind to promoters (65%). However, a large proportion of binding sites are also located in intergenic (15%) and intronic (13%) regions, where regulatory sequences such as enhancers are located. These results along with the recently discovered m6A methylation of eRNA could indicate a potential role of m6A MTC in chromatin organization and enhancer-promoter interactions. The transcriptional impact associated with knockdown of m6A MTC components may be mediated by the role of m6A in these chromatin interactions. Indeed, it has been proposed that eRNAs directly promote enhancer-promoter looping and gene activation [Citation63–65]. The ability to produce eRNA seems to occur independently of the transcription of the target gene [Citation66]. The functional role of eRNAs remains unclear but several evidence show that eRNA transcription is highly correlated with enhancer activity and induced gene expression in Drosophila [Citation67] and vertebrates [Citation65,Citation68–72]. Furthermore, in neurons Schaukowitch and colleagues showed that, upon stimulation, the transient release of NELF from promoters of Immediate Early Genes (IEG) depends on the presence of the specific eRNA from the enhancer that stimulates respective IEG [Citation73]. The role of eRNA in the release of paused RNAPII would be then interesting to follow in the context where these eRNAs contain m6A modifications, and whether this release is sensitive to the methylation state of the eRNAs. Further work will be required to validate if a link exists between these processes, identify the molecular actors involved, and understand the specificity of m6A-mediated enhancer-promoter recognition and the correlation with RNAPII pausing release.
6. Conclusions and outlooks
Recent works have highlighted the key regulatory role RNA modifications play in control of gene expression. Not surprisingly, the modification itself is tightly regulated and their mis-regulation is associated with developmental and physiological abnormalities. Due to the co-transcriptional nature of their deposition, there is extensive possibilities of the dialogue between RNA modification and other co-transcriptional processes, including splicing. Indeed, recent studies point toward the crosstalk between splicing, transcription, epigenetic modifications, and m6A MTC. The extent of direct regulation of transcription by m6A MTC is limited and seems to be context dependent. The regulation of eRNAs by m6A MTC also brings an interesting perspective to this regulation, where the transcriptional effect can be developmental and context dependent, including both the transcriptional activation and repression. But future studies are needed to further extend this knowledge and to uncover the extent of m6A MTC impact on transcriptional regulation. It can be also speculated that this crosstalk might not be exclusive to the m6A modification of mRNA, but potentially also exist for other RNA modifications. With constant development of new tools and assays to precisely map RNA modifications, including direct sequencing of RNA molecules with nanopore sequencing, the true extent of the crosstalk between RNA modifications and transcription will be uncovered.
Acknowledgments
This work was supported by the Agence Nationale de la Recherche (ANR-13-JSV2-0001) to G.J.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Yue Y, Liu J, He C. RNA N 6 -methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 2015;29(13):1343–1355.
- Frye M, Haranda T, Behm B, et al. RNA modifications modulate gene expression during development. Science. 2018;361(80):1346–1349.
- Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, et al. The dynamic N1 -methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016;530(7591):441–446.
- Linder B, Grozhik AV, Olarerin-George AO, et al. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods. 2015;12(8):767–772.
- Kan L, Grozhik AV, Vedanayagam J, et al. The m 6 A pathway facilitates sex determination in Drosophila. Nat Commun. 2017;8(1):1–16.
- Worpenberg L, Paolantoni C, Longhi S, et al. Ythdf is a N6-methyladenosine reader that modulates Fmr1 target mRNA selection and restricts axonal growth in Drosophila. EMBO J. 2021;40(4):1–20.
- Zhou J, Wan J, Shu XE, et al. N6-Methyladenosine guides mRNA alternative translation during integrated stress response. Mol Cell. 2018;69(4):636–647.e7.
- Dominissini D, Moshitch-Moshkovitz S, Salmon-Divon M, et al. Transcriptome-wide mapping of N6-methyladenosine by m 6A-seq based on immunocapturing and massively parallel sequencing. Nat Protoc. 2013;8(1):176–189.
- Meyer KD, Saletore Y, Zumbo P, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149(7):1635–1646.
- Shulman Z, Stern-Ginossar N. The RNA modification N 6-methyladenosine as a novel regulator of the immune system. Nat Immunol. 2020;21(5):501–512.
- Schwartz S. Cracking the epitranscriptome. RNA. 2016;22(2):169–174.
- Patil DP, Chen CK, Pickering BF, et al. M6 A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537(7620):369–373.
- Uranishi H, Zolotukhin AS, Lindtner S, et al. The RNA-binding motif protein 15B (RBM15B/OTT3) acts as cofactor of the nuclear export receptor NXF1. J Biol Chem. 2009;284(38):26106–26116.
- Zolotukhin AS, Uranishi H, Lindtner S, et al. Nuclear export factor RBM15 facilitates the access of DBP5 to mRNA. Nucleic Acids Res. 2009;37(21):7151–7162.
- Knuckles P, Lence T, Haussmann IU, et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m 6 A machinery component Wtap/Fl(2)d. Genes Dev. 2018;32(5–6):415–429.
- Xu C, Liu K, Tempel W, et al. Structures of human ALKBH5 demethylase reveal a unique binding mode for specific single-stranded N6-methyladenosine RNA demethylation. J Biol Chem. 2014a;289(25):17299–17311.
- Zheng G, Dahl JA, Niu Y, et al. ALKBH5 Is a Mammalian RNA demethylase that Impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49(1):18–29.
- Xu C, Wang X, Liu K, et al. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat Chem Biol. 2014b;10(11):927–929.
- Xu C, Liu K, Ahmed H, et al. Structural basis for the discriminative recognition of N6-Methyladenosine RNA by the human YT521-B homology domain family of proteins. J Biol Chem. 2015;290(41):24902–24913.
- Liu N, Dai Q, Zheng G, et al. N6 -methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518(7540):560–564.
- Zhou KI, Shi H, Lyu R, et al. Regulation of Co-transcriptional Pre-mRNA Splicing by m6A through the Low-Complexity Protein hnRNPG. Mol Cell. 2019;76(1):70–81.e9.
- Alarcón CR, Goodarzi H, Lee H, et al. HNRNPA2B1 is a mediator of m6A-Dependent Nuclear RNA processing events. Cell. 2015;162(6):1299–1308.
- Du H, Zhao Y, He J, et al.YTHDF2 destabilizes m 6 A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Commun Nat.2016;7:12626.
- Shi H, Wang X, Lu Z, et al. YTHDF3 facilitates translation and decay of N 6-methyladenosine-modified RNA. Cell Res. 2017;27(3):315–328.
- Xiao W, Adhikari S, Dahal U, et al. Nuclear m6A reader YTHDC1 regulates mRNA Splicing. Mol Cell. 2016a;61(4):507–519.
- Roundtree IA, Luo GZ, Zhang Z, et al. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. Elife. 2017;6:1–28.
- Slobodin B, Han R, Calderone V, et al. Transcription impacts the efficiency of mRNA translation via Co-transcriptional N6-adenosine Methylation. Cell. 2017;169(2):326–337.e12.
- Ji Q, Zong X, Mao Y, et al. A heat shock–responsive lncRNA Heat acts as a HSF1-directed transcriptional brake via m 6 A modification. Proc Natl Acad Sci U S A. 2021;118(25):1–11.
- Liu J, Dou X, Chen C, et al. N 6 -methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science. 2020;8(6477):580–586.
- Lee JH, Wang R, Xiong F, et al. Enhancer RNA m6A methylation facilitates transcriptional condensate formation and gene activation. Mol Cell. 2021;81(16):3368–3385.e9.
- Sabari BR, Dall’Agnese A, Boija A, et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018;379(80):361.
- Nayler O, Hartmann AM, Stamm S. The ER repeat protein YT521-B localizes to a novel subnuclear compartment. J Cell Biol. 2000;150(5):949–961.
- Brien TO, Hardlnu S, Greenleaft A, et al. Phosphorylation of RNA polymerase lie-terminal elongation. Nature. 1993;75–77.
- Hsin JP, Manley JL. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 2012;26(19):2119–2137.
- Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14(19):2452–2460.
- Lagha M, Bothma JP, Esposito E, et al. XPaused Pol II coordinates tissue morphogenesis in the drosophila embryo. Cell. 2013;153(5):976.
- Vos SM, Farnung L, Urlaub H, et al. Structure of paused transcription complex Pol II–DSIF–NELF. Nature. 2018a;560(7720):601–606.
- Vos SM, Farnung L, Boehning M, et al. Structure of activated transcription complex Pol II–DSIF–PAF–SPT6. Nature. 2018b;560(7720):607–612.
- Akhtar J, Renaud Y, Albrecht S, et al. m6A RNA methylation regulates promoter- proximal pausing of RNA polymerase II. Mol Cell. 2021;81(16):3356–3367.e6.
- Huang H, Weng H, Zhou K, et al. Histone H3 trimethylation at lysine 36 guides m6A RNA modification co-transcriptionally. Nature; 2019.
- Prieto C, Nguyen DTT, Liu Z, et al. Transcriptional control of CBX5 by the RNA-binding proteins RBMX and RBMXL1 maintains chromatin state in myeloid leukemia. Nat. Cancer. 2021;2(7):741–757.
- Xu W, He C, Kaye EG, et al. Dynamic control of chromatin-associated m6A methylation regulates nascent RNA synthesis. Mol Cell. 2022;82:1–13.
- Chen J, Zhang YC, Huang C, et al. m6A Regulates neurogenesis and neuronal development by modulating histone methyltransferase Ezh2. Genom Proteom Bioinform. 2019;17(2):154–168.
- Fei Q, Yang X, Jiang H, et al. SETDB1 modulates PRC2 activity at developmental genes independently of H3K9 trimethylation in mouse ES cells. Genome Res. 2015;25(9):1325–1335.
- Chen C, Liu W, Guo J, et al. Nuclear m6A reader YTHDC1 regulates the scaffold function of LINE1 RNA in mouse ESCs and early embryos. Protein Cell. 2021;12(6):455–474.
- Liu J, Gao M, He J, et al. The RNA m6A reader YTHDC1 silences retrotransposons and guards ES cell identity. Nature. 2021;591(7849):322–326.
- Xu W, Li J, He C, et al. METTL3 regulates heterochromatin in mouse embryonic stem cells. Nature. 2021;591(7849):317–321.
- Wang Y, Li Y, Yue M, et al. N 6-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat Neurosci. 2018;21(2):195–206.
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485(7397):201–206.
- Zhao X, Yang Y, Sun BF, et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014;24(12):1403–1419.
- Ke S, Pandya-Jones A, Saito Y, et al. m 6 A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev. 2017;31(10):990–1006.
- Lewis CJT, Pan T, Kalsotra A. RNA modifications and structures cooperate to guide RNA-protein interactions. Nat Rev Mol Cell Biol. 2017;18(3):202–210.
- Xiao W, Adhikari S, Xiao W, et al. Nuclear m 6 A Reader YTHDC1 Regulates mRNA Splicing. Mol Cell. 2016b;61:1–13.
- Mendel M, Delaney K, Pandey RR, et al. Splice site m6A methylation prevents binding of U2AF35 to inhibit RNA splicing. Cell. 2021;184(12):3125–3142.e25.
- Akhtar J, Kreim N, Marini F, et al. Promoter-proximal pausing mediated by the exon junction complex regulates splicing. Nat Commun. 2019;10(1):521.
- Ji X, Zhou Y, Pandit S, et al. SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase. Cell. 2013;153(4):855–868.
- Luco RF, Misteli T. More than a splicing code: integrating the role of RNA, chromatin and non-coding RNA in alternative splicing regulation. Curr Opin Genet Dev. 2011;21(4):366–372.
- Li Y, Xia L, Tan K, et al. N 6-Methyladenosine co-transcriptionally directs the demethylation of histone H3K9me2. Nat Genet. 2020;52(9):870–877.
- Chen L, Chen JY, Zhang X, et al. R-ChIP using inactive RNase H reveals dynamic coupling of R-loops with transcriptional pausing at gene promoters. Mol Cell. 2017;68(4):745–757.e5.
- Yang X, Liu QL, Xu W, et al. m6A promotes R-loop formation to facilitate transcription termination. Cell Res. 2019;29(12):1035–1038.
- Abakir A, Giles TC, Cristini A, et al. N 6-methyladenosine regulates the stability of RNA:DNA hybrids in human cells. Nat Genet. 2020;52(1):48–55.
- Kang HJ, Cheon NY, Park H, et al. TonEBP recognizes R-loops and initiates m6A RNA methylation for R-loop resolution. Nucleic Acids Res. 2021;49(1):269–284.
- Hsieh CL, Fei T, Chen Y, et al. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc Natl Acad Sci U S A. 2014;111(20):7319–7324.
- Li W, Notani D, Ma Q, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498(7455):516–520.
- Wang D, Garcia-Bassets I, Benner C, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474(7351):390–397.
- Vernimmen D. Uncovering enhancer functions using the α-Globin Locus. PLoS Genet. 2014;10. DOI:https://doi.org/10.1371/journal.pgen.1004668
- Mikhaylichenko O, Bondarenko V, Harnett D, et al. The degree of enhancer or promoter activity is reflected by the levels and directionality of eRNA transcription. Genes Dev. 2018;32(1):42–57.
- Hah N, Murakami S, Nagari A, et al. Enhancer transcripts mark active estrogen receptor binding sites. Genome Res. 2013;23(8):1210–1223.
- Kaikkonen MU, Spann NJ, Heinz S, et al. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol Cell. 2013;51(3):310–325.
- Kim TK, Hemberg M, Gray JM, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465(7295):182–187.
- Melo CA, Drost J, Wijchers PJ, et al. ERNAs are required for p53-Dependent enhancer activity and gene transcription. Mol Cell. 2013;49(3):524–535.
- Vernimmen D, De GM, Sloane-Stanley JA, et al. Long-range chromosomal interactions regulate the timing of the transition between poised and active gene expression. EMBO J. 2007;26(8):2041–2051.
- Schaukowitch K, Joo JY, Liu X, et al. Enhancer RNA facilitates NELF release from immediate early genes. Mol Cell. 2014;56(1):29–42.