ABSTRACT
Initially discovered by genetic screens in budding yeast, SPT5 and its partner SPT4 form a stable complex known as DSIF in metazoa, which plays pleiotropic roles in multiple steps of transcription. SPT5 is the most conserved transcription elongation factor, being found in all three domains of life; however, its structure has evolved to include new domains and associated posttranslational modifications. These gained features have expanded transcriptional functions of SPT5, likely to meet the demand for increasingly complex regulation of transcription in higher organisms. This review discusses the pleiotropic roles of SPT5 in transcription, including RNA polymerase II (Pol II) stabilization, enhancer activation, Pol II pausing and its release, elongation, and termination, with a focus on the most recent progress of SPT5 functions in regulating metazoan transcription.
Introduction
RNA polymerase II (Pol II)-mediated transcription is a highly regulated dynamic process that generally consists of three phases: initiation, elongation, and termination. Transcription initiation requires the assembly of a preinitiation complex (PIC) composed of Pol II and general transcription factors (GTFs) on core promoters. Initiation begins with the opening of double-stranded DNA and synthesis of RNA transcripts before the transcription machinery pauses promoter proximally, which has been widely accepted as a regulatory step of metazoan transcription deciding whether and, if yes, how often Pol II is released into productive elongation. Pol II speeds up gradually to several kb per minute during elongation, passing through exons and introns to influence co-transcriptional splicing. When arriving at the 3′ end of genes, messenger RNAs (mRNAs) are cleaved and polyadenylated while Pol II is evicted from the DNA template, leading to transcription termination and the formation of processed or partially processed transcripts ready to be exported to the cytoplasm.
The precise regulation of transcription depends on a panel of core factors or complexes that control each step of transcription and coordinate the transitions from one step to another. Among these factors, the evolutionarily conserved transcription regulator SPT5 (suppressor of Ty5 homolog), termed the DRB (5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole) sensitivity-inducing factor (DSIF) when in complex with SPT4 in metazoans, stands out for its critical roles in virtually all steps of transcription including enhancer activation, pausing maintenance, pause release, elongation, RNA processing, and termination. Despite the crucial pleiotropic roles of SPT5 in transcription, until recently, there was limited mechanistic understanding, especially in mammalian cells, of how SPT5 controls each transcriptional step and how it coordinates their stepwise transitions. Moreover, controversies remain concerning several aspects of SPT5 functions. For example, it is still debated, for protein-coding genes and their enhancers, whether SPT5 is simply an activator of transcription or if it can both upregulate and downregulate gene expression in a context-dependent manner, as well as whether SPT5 is simply a general or sometimes tissue-specific transcriptional regulator.
In this review, we summarize recent discoveries in SPT5 regulation, with a focus on metazoan transcription, particularly pausing, elongation, and enhancer activation. We also discuss how recent progress could address long-lasting questions and controversies regarding SPT5′s multifaceted functions and their underlying mechanisms.
The identification and functions of SPT5 and DSIF
SPT5, together with SPT4, was initially identified by the Winston group through the selection of mutants in the budding yeast Saccharomyces cerevisiae (S. cerevisiae) that suppress the inhibition caused by Ty transposon insertions in the 5’ noncoding regions of the HIS4 gene [Citation1]. Subsequent studies by Winston and colleagues linked SPT5 with transcription elongation and found that it might form complex with SPT4 [Citation2–4]. SPT5 is the only known transcription elongation factor conserved in all three domains of life (). SPT5 in bacteria (called NusG) and archaea share one structurally conserved NusG N-terminal (NGN) domain and one Kyprides, Ouzounis, Woese (KOW) domain with eukaryotes (KOW1 for eukaryotes). However, SPT5 in eukaryotes has evolved a negatively charged N-terminal region (SPT5N), several additional KOW domains, and one or two C-terminal repeat regions (CTR). In higher organisms, SPT5 is subject to extensive phosphorylation and contains at least two hotspots within the KOWx-4/5-linker (a flexible linker between KOWx-4 and KOW5 domains) and the CTR1 [Citation5–15], respectively. The phosphorylation status at these sites, along with the kinases and phosphatases catalyzing them, is crucial to the regulation of the different SPT5-dependent transcription steps (discussed below).
Figure 1. Domains and structures of SPT5/NusG in different organisms.
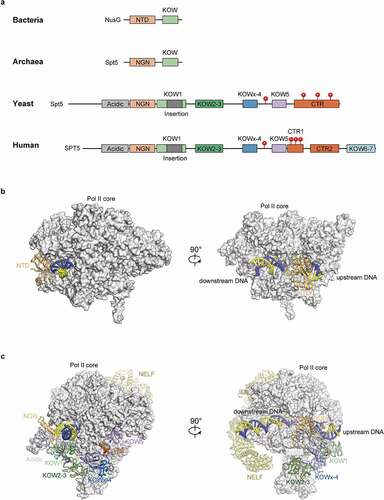
Using in vitro transcription assays in combination with biochemical fractionation of Hela cell extracts, the Handa group identified DSIF, composed of SPT5 and SPT4, as a DRB sensitivity factor, and for the first time connected SPT5 with the stabilization of promoter-proximal pausing [Citation16]. An independent study later purified DSIF through the identification of factors that mediate Mediator responsiveness in transcription, corroborating DSIF functioning in early steps of transcription in mammalian cells [Citation17]. Subsequent reports found that SPT5 associates with the actively elongating form of Pol II from 5’ to 3’ ends of genes upon transcriptional activation in Drosophila, highlighting the conserved positive role of SPT5 in transcription in higher organisms [Citation4,Citation18]. These seminal studies above have laid the foundation for future investigation of this essential, pleiotropic transcriptional regulator.
Stabilization of pol II protein by SPT5
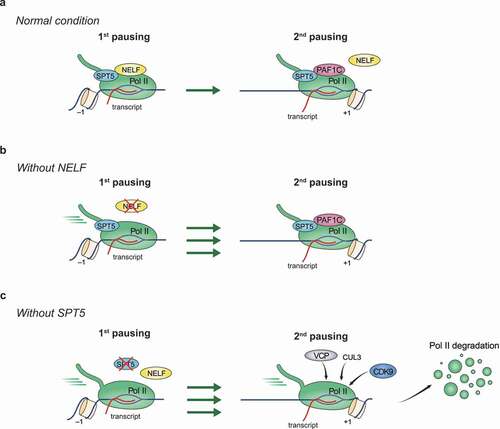
Apart from the well-established roles of SPT5 in multiple transcriptional steps, recent studies from the Shilatifard group and our lab revealed the unexpected SPT5 function in stabilizing Pol II protein [Citation19,Citation20]. In both reports, proteolysis-targeting chimera (PROTAC)-induced degradation of endogenous SPT5 in different human cells causes an immediate, substantial degradation of the largest Pol II subunit RPB1, supporting a direct regulation of Pol II stability (). Mechanistically, the Shilatifard group found that SPT5 loss induces the enrichment of the E3 ligase Cullin 3 and the unfoldase VCP/p97 on chromatin, especially at promoters, where it initiates RPB1 degradation. The degradation requires the kinase activity of P-TEFb, suggesting that the phosphorylation of Pol II itself, or of other factors, can be critical for Pol II degradation [Citation19]. SPT5 depletion mainly (but not exclusively) destabilizes paused Pol II in mammalian cells. Although SPT4 is required for optimal DSIF functions in transcription, its loss has no evident effect on Pol II stability [Citation19]. Despite the lack of typical Pol II pausing in S. cerevisiae, SPT5-dependent Pol II stabilization appears to be conserved in budding yeast, as Spt5 removal leads to Rpb1 degradation, and Rpb1 levels are rescued by Cdc48 (a yeast ortholog of VCP) removal [Citation19].
Figure 2. Functions of SPT5 and NELF in stabilizing promoter-proximal Pol II pausing.
When transcribing Pol II encounters damaged DNA, it stalls and induces transcription-coupled DNA repair (TCR) to remove DNA lesions [Citation21–24]. This process also involves the degradation of RPB1 following its ubiquitination at lysine 1268 (K1268) [Citation25,Citation26]. Upon the initiation of TCR, Cockayne syndrome group B (CSB, also known as ERCC6) (or its orthologue Rad26 in S. cerevisiae) is among the first batch of proteins to be recruited to the arrested Pol II [Citation21,Citation24,Citation27–34] directly. Structures of Pol II bound by CSB/Rad26 reveal notable steric clashes between CSB/Rad26 and DSIF, suggesting that DSIF needs to be displaced by CSB/Rad26 in TCR [Citation35,Citation36]. Reciprocally, DSIF plays a regulatory role in TCR by hindering unscheduled CSB/Rad26 loading, TCR commencement, and presumably TCR-induced Pol II degradation. Therefore, cells lacking either SPT5 or SPT4 circumvent the requirement for CSB/Rad26 to induce TCR [Citation37–41].
It is as yet unclear whether or not SPT5 (or DSIF) mediated Pol II stabilization in normal transcription and TCR share a common pathway. Based on the current data, one noticeable difference is that SPT4 is dispensable for stabilizing Pol II during normal transcription, but it is crucial for regulating CSB loading during TCR [Citation19,Citation37]. Future investigations are warranted to unveil the underlying mechanisms of SPT5-dependent Pol II stabilization in different contexts.
SPT5 function in pausing
Although a relative enrichment of engaged Pol II at promoters is observed for some genes in lower organisms such as Schizosaccharomyces pombe (S. pombe) and S. cerevisiae [Citation42,Citation43], promoter-proximal pausing is substantially more apparent and pervasive in more advanced species and has been considered as a gained regulatory transcriptional step through evolution to meet the demand for more precise and prompt control of gene expression in complex processes such as development [Citation44–48]. The paused Pol II typically dwells within a 20–120 bp window downstream of the transcription start site (TSS). SPT5, along with NELF, plays a crucial role in stabilizing paused Pol II. On the basis that RPB1 protein is degraded upon acute SPT5 depletion as discussed above, we now can anticipate two non-exclusive mechanisms whereby SPT5 stabilizes paused Pol II (): one is that SPT5 prevents the targeting of paused Pol II by proteasome-mediated degradation on chromatin, the other is that it could restrain Pol II release toward downstream regions. Unlike SPT5, NELF can only inhibit the downstream shift of paused Pol II but does not regulate RPB1 protein stability [Citation19]. DSIF and NELF directly interact with each other, and the association of NELF with Pol II is dependent on the presence of DSIF, while the DSIF-Pol II interaction does not require NELF () [Citation49–51]. Therefore, it is plausible that SPT5-dependent suppression of downstream Pol II release is mediated through NELF. Given that the phenotype of RPB1 protein degradation is more dominant than that of downstream Pol II shift following acute SPT5 depletion, the RPB1 protein degradation pathway needs to be inhibited before revealing the pervasive SPT5 role in keeping Pol II at pausing sites [Citation19,Citation20].
High-resolution Pol II profiling methods such as PRO-seq, combined with the interference of key transcription regulators, have enabled the discovery of two sequential pausing regions for one promoter () [Citation52,Citation53]. The upstream one (first pausing) is mainly located within the 20–120 bp pausing window and is generally the one we traditionally found and widely studied. The downstream one (second pausing) is mainly positioned proximally to the dyad of +1 nucleosomes, suggesting the implication of chromatin context in controlling the second pausing. In contrast to the first pausing sites that are occupied by the majority of paused Pol II population, the second pausing is more obscure or even hidden and thus depends on disruption of the first pausing to be clearly manifested genome-wide. With the help of DSIF, NELF contributes to the stabilization of the first but not the second pausing [Citation52,Citation53], with its depletion resulting in the downstream shift of Pol II to the second pausing sites around the +1 nucleosome dyad. The stabilization of Pol II at the second pausing sites is aided by the pausing and elongation regulator PAF1 complex (PAF1C) and probably chromatin remodelers [Citation54–56], whereas Pol II release from the second pausing sites requires factors such as P-TEFb and MYC [Citation52,Citation53,Citation57].
Structural comparison of NELF- and PAF1C-containing Pol II complexes reveals steric clashes between NELF and PAF1C, thus eliciting a plausible model that the shift of Pol II from the first to the second pausing sites is accompanied by a substantial change in the composition of Pol II complex, such as the replacement of NELF by PAF1C [Citation15,Citation54,Citation58]. Different from the eviction of NELF during this transition, DSIF remains with Pol II at the second pausing site (). Under the condition that Pol II degradation is blocked, rapid SPT5 depletion leads to further release of Pol II from the second pausing sites into gene bodies [Citation19], indicating the function of SPT5 in stabilizing Pol II at the second pausing site.
SPT5 function in pause release
The master regulator of pause release for most if not all paused genes is P‑TEFb, comprising CDK9 and cyclin T1 [Citation59,Citation60]. Recruitment of P‑TEFb at promoters is mediated by specific transcription factors or cofactors via different mechanisms [Citation47,Citation61–64]. Moreover, P‑TEFb activity is modulated by interacting factors or complexes. For example, incorporating into the super elongation complex (SEC) or binding with BRD4 boosts its catalytic activity while its integration into the 7SK complex limits its activity [Citation61,Citation62,Citation65–68]. Close to 100 putative substrates of P-TEFb have been identified, which are enriched for proteins involved in transcription and RNA catabolism [Citation6]. Among them, the most prominent and presumably crucial ones that primarily occupy promoters include Pol II itself, NELF, and SPT5 (). The phosphorylation of NELF elicits its dissociation from chromatin and thus the relief of the first pausing [Citation52,Citation53,Citation69], while phosphorylation of Pol II CTD at serine 2 and SPT5 may partially induce the release of Pol II to proceed into productive elongation [Citation5,Citation70].
Figure 3. Schematic of SPT5 function in regulating pause release.
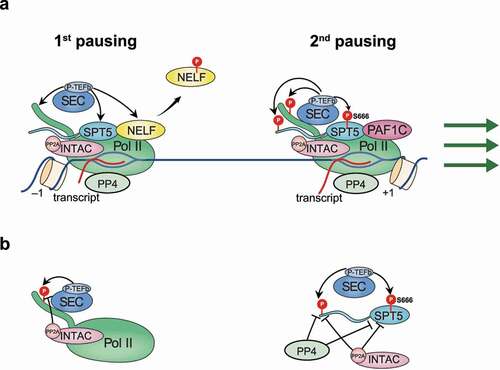
SPT5 contains two major phosphorylation hotspots, serine 666 (S666) within the KOWx-4/5-linker, and the fourth threonine residues of the CTR1 repeats (). It had previously been undetermined whether, and if yes, which hotspot is implicated in pause release regulation by SPT5. To address this issue, we recently conducted rescue experiments by inducing the expression of wild-type SPT5 or mutants mimicking unphosphorylated SPT5 at S666 and the CTR1 threonines, respectively, in cells with depletion of endogenous SPT5 [Citation20]. Our results unambiguously indicate that phosphorylation at S666 of the KOWx-4/5-linker, but not CTR1, is critical for P-TEFb mediated release of paused Pol II into elongation. The underlying mechanism could be explained by structural studies: KOWx-4 and KOW5 form an “RNA clamp” over the RNA exit tunnel of Pol II, while the positively charged KOWx-4/5-linker that connects KOWx-4 to KOW5 passes the negatively charged exiting RNA and thus potentially retains it in the exit tunnel [Citation7]. S666 phosphorylation could reduce the net positive change of the linker to disrupt the SPT5-RNA interaction, thus enabling a conformational change of Pol II to allow further synthesis/extension of RNA out of the exit tunnel during the transition to productive elongation. Notably, mutations of SPT5 at either the KOWx-4/5-linker or CTR1 exert no effect on RPB1 protein stability, indicating that SPT5-dependent stabilization of Pol II does not require phosphorylation at either hotspot [Citation20].
Targeting SPT5 by phosphatases
Targeted phosphorylation by P-TEFb and other transcription-activating kinases is counterbalanced by different classes of phosphatases throughout the entire transcription cycle [Citation71]. For most of these phosphatases, their catalytic specificity and detailed functions remain uncertain or even controversial. For example, we recently showed a lack of catalytic activity for RPAP2 [Citation72], a previously reported putative Pol II phosphatase [Citation73–75]. To date, identified phosphatases targeting SPT5 include PP1, PP4, and PP2A () [Citation11,Citation12,Citation76,Citation77]. PP4 dephosphorylates both S666 of the KOWx-4/5-linker and CTR1 of SPT5, whereas PP1 targets the SPT5 CTR1 [Citation12]. Supporting the requirement for S666 but not CTR1 phosphorylation in promoting release from pausing [Citation20], only depletion of PP4 but not PP1 induces the release of Pol II into gene bodies [Citation12]. Consistently, PP4 is predominantly located between the TSS and ~2-3 kb downstream of genes. In addition to the antagonistic regulation in phosphorylation of common substrates, P-TEFb can direct phosphorylate and thus inhibit the activity of both PP1 and PP4, adding another layer of complexity to their crosstalk ().
Figure 4. Schematic of SPT5 function in elongation-termination transition.
Similar to PP4, PP2A dephosphorylates both S666 and CTR1 of SPT5 (). Both the chromatin association and catalytic activation of PP2A rely on the incorporation of the PP2A core enzyme into the Integrator complex to assemble a new subfamily of PP2A holoenzyme, which we named INTAC (Integrator and PP2A-AC) complex [Citation76–78]. Disruption of the INTAC phosphatase module induces transcriptional activation by the release of Pol II from pausing, at least partially through the elevation of SPT5 phosphorylation at S666 [Citation76,Citation78]. Therefore, loss of INTAC phosphatase activity confers cellular resistance to P-TEFb inhibition () [Citation77]. The balance between P-TEFb and INTAC in modulating pausing and pause release could be harnessed by other transcription regulators such as PAF1C, which can directly interact with INTAC [Citation54,Citation58]. Through this interaction, PAF1 can recruit INTAC to genomic regions, including promoters, to suppress SPT5 phosphorylation, which could potentially explain the broad function of PAF1 in restraining the release of Pol II from the second pausing sites [Citation54–56]. However, it is important to highlight that SPT5 is not the only player in the antagonism between P-TEFb and INTAC, since disruption of the INTAC phosphatase module in SPT5-null cells still stimulates transcriptional activation [Citation19]. Therefore, additional common targets, such as Pol II itself, could function synergistically with SPT5 in mediating the balance of transcriptional regulation between P-TEFb and INTAC.
SPT5 function in productive elongation
In addition to stabilizing paused Pol II at promotors in metazoa, SPT5, along with SPT4, has been more well known for its conserved function in promoting elongation processivity and velocity since the discovery of DSIF () [Citation8,Citation11,Citation16,Citation59,Citation60,Citation79–84]. Supporting its multifaceted roles, SPT5 remains associated with Pol II during elongation and termination after exiting the pausing sites [Citation85,Citation86]. SPT5 function in productive transcription was already established by a host of studies in yeast, which lacks a pervasive pausing phenotype and thus is an ideal model to evaluate regulation of processivity by SPT5. In S. cerevisiae, Spt5 is required to sustain global transcription and cell survival, with its mutation or depletion leading to a genome-wide defect in Pol II progression in gene bodies [Citation2,Citation81,Citation87,Citation88]. Consistently, a vast majority of genes are downregulated at levels of both nascent and mature RNAs upon SPT5 depletion in Drosophila [Citation89].
Depletion of SPT5 in mammalian cells results in accumulation of Pol II in the early gene body region, as measured by Pol II ChIP-seq or PRO-seq [Citation20,Citation79,Citation85]. This type of alteration could be interpreted as either enhanced release of Pol II into gene bodies or impaired Pol II processivity during the productive elongation stage, or both. Nonetheless, the enhanced release of Pol II from the pause site would normally upregulate nascent RNA synthesis, while impaired Pol II processivity would dampen nascent RNA synthesis. Therefore, quantification of nascent RNA levels by techniques such as 4SU-seq or transient transcriptome sequencing (TT-seq) is crucial in distinguishing between these two scenarios. Our recent study using spike-in normalized TT-seq revealed that rapid SPT5 degradation in human cells leads to a genome-wide reduction in nascent RNA levels [Citation20]. This change is vastly different from that caused by acute loss of PAF1C in the same or different cell types, which can exhibit an increase or decrease in nascent transcription in a gene-specific manner [Citation54,Citation90]. These results support a dominant role of SPT5 in promoting Pol II processivity. Moreover, the rapid depletion system allows the separation of SPT5’s role in pausing and productive elongation, with the former to stabilize paused Pol II mainly by limiting its degradation or disassociation from chromatin as elaborated above.
The evolutionary conservation of SPT5 elongation function can be traced back to its counterparts in bacteria (called NusG) and archaea (), where SPT5/NusG primarily promotes elongation progression [Citation91–93]. The elongation-promoting function of SPT5 is in part mediated by the suppression of Pol II arrest during elongation, as SPT5 conveys resistance to nucleotide-limiting conditions prone to inducing elongation arrest [Citation16,Citation94]. As described above, SPT5/NusG from all three domains of life share one NGN domain and at least one KOW domain (corresponding to KOW1 in eukaryotes), implying the functional importance of these regions in mediating productive elongation () [Citation93,Citation95]. Structures of SPT4-SPT5 in complex with the polymerase and biochemical studies have shed light on the underlying molecular mechanisms: The NGN domain directly binds to RPB1, RPB2, and the upstream DNA duplex to form a DNA clamp that facilitates the maintenance of a closed active center cleft for elongation. KOW/KOW1 and SPT4 together conceal and further stabilize the DNA clamp [Citation15,Citation95–101]. Moreover, SPT5 CTR, especially its phosphorylated form, further stimulates elongation by recruiting other elongation factors such as PAF1C [Citation102–104].
The regulation of nucleosome dynamics and histone marks by SPT5
The elongation process implicates constant disassembly and reassembly of nucleosomes [Citation105]. As revealed by the structure of elongating Pol II with nucleosome, SPT5, with the help of Elf1/ELOF1, intervenes between Pol II and the nucleosome to promote the progression of the polymerase through nucleosomal barriers [Citation106]. This study provides a molecular basis for the reported SPT5 function in promoting elongation through nucleosomes in both eukaryotes and archaea [Citation107,Citation108]. Interestingly, despite the presence of SPT5/NusG in all three domains of life, SPT4 only exists in eukaryotes and archaea, coincident with the existence of nucleosomes or nucleosome-like structures in these two domains [Citation109]. Notably, a recent study in S. cerevisiae shows that Spt4 not only facilitates transcription through nucleosomal barriers but also regulates nucleosome positioning, although the mechanism is still unclear [Citation110]. From an evolutionary perspective, SPT5 gains a negatively charged N-terminal region (SPT5N) that interacts with the H2A-H2B dimer and the H3-H4 tetramer [Citation106,Citation111,Citation112]. This interaction was recently found to assist in the redeposition of nucleosomal histones and thus preserve chromatin during elongation [Citation112].
The influence of SPT5 on nucleosomes is also reflected by its regulation of epigenetic marks on histones. Loss of SPT5 induces a widespread decline in multiple histone modifications, including H2B ubiquitination (H2Bub), H3K4me3, H3K36me3, and H3K79me2 [Citation113,Citation114]. Apart from the influence of histone marks by the SPT5-mediated elongation control, the regulation of histone marks by SPT5 can also be mediated through its direct impact on nucleosome dynamics as discussed above or indirectly through its cooperation with enzymes or factors governing the deposition of these marks [Citation59,Citation115]. For instance, SPT5 contributes to the optimal recruitment of PAF1C, which acts as a platform for the recruitment of multiple chromatin-modifying enzymes including Rad6-Bre1, DOT1L, COMPASS, and SETD2 [Citation54,Citation55,Citation102–104,Citation116–129]. Future studies are needed to extricate how SPT5ʹs direct impact on nucleosome dynamics to influence the epigenetic landscape coordinates with its indirect regulation of epigenetic marks through enzymes or factors controlling them.
SPT5 function in termination
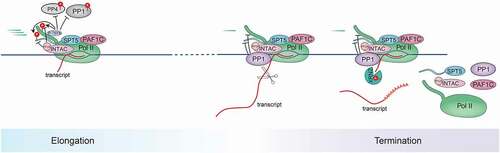
The transcription termination process includes the modulation of the polymerase elongation capacity in order to facilitate the dismantling and perhaps recycling of the transcription machinery. In contrast to the impaired productive elongation induced by SPT5 loss as described above, the depletion of SPT5 also leads to extended transcription beyond the transcription end site (TES) in both mammalian cells [Citation79] and yeast [Citation130], indicating that SPT5 is engaged in the induction of efficient termination. Using a rapid degradation system, acute loss of SPT5 leads to widespread transcriptional readthrough, corroborating its direct regulation of termination () [Citation20]. SPT5 function in termination is likely attributed to its association with, and thus recruitment of, termination factors, including the cleavage stimulation factor (CstF) [Citation20,Citation131,Citation132], although the mechanistic details remain to be explored.
Phosphorylation of SPT5 could further influence its function in termination. The Bentley group has proposed a “sitting duck torpedo” mechanism whereby the dephosphorylation of SPT5 CTR1 by PP1 decelerates transcription in the termination zone and thus converts Pol II into a viable target for termination () [Citation8]. Our recent results show that the disruption of SPT5 phosphorylation at CTR1, but not S666 within the KOWx-4/5-linker, induces more efficient termination around the TES [Citation20], confirming the critical role of CTR1 phosphorylation status in termination. One possible mechanism underlying this regulation is that SPT5 dephosphorylation decommissions other elongation factors such as PAF1C, which is also involved in the termination process and which preferentially binds phosphorylated SPT5 [Citation54,Citation102,Citation113,Citation133–135]. However, further investigation is required to determine the primary role of the dynamics of SPT5 CTR1 phosphorylation during termination in detail.
Compared with the role of SPT5 phosphorylation sites in transcription termination, the functions of the kinases and phosphatases catalyzing SPT5 phosphorylation in this process are more enigmatic, presumably because each of them has multiple additional substrates exerting differential impacts on transcriptional regulation. As elaborated above, P-TEFb is the major kinase responsible for the phosphorylation of CTR1, while the corresponding phosphatases include PP1, PP4, and PP2A. The Fisher lab showed that P-TEFb blocking leads to transcriptional readthrough at the 3’ end of genes in part due to its substrate, the exoribonuclease XRN2, the phosphorylation of which enhances its enzymatic activity and thus ensures efficient termination [Citation6]. Conversely, the Murphy lab reported a poly(A)–associated elongation checkpoint regulated by P-TEFb, with inhibition of P-TEFb inducing Pol II to prematurely terminate transcription around the TES [Citation136]. P-TEFb inhibition-induced termination defects are readily rescued by inhibition of PP2A but not inhibition of PP1, likely through PP2A targeting of the splicing factor SF3B1 [Citation137]. Disruption of PP1 leads to SPT5 CTR1 hyperphosphorylation at the TES, enhanced elongation velocity, and delayed termination [Citation8,Citation11,Citation12,Citation132,Citation138]. However, it remains to be determined how different phosphatases cooperate in neutralizing P-TEFb activity and which substrates are vital in mediating the kinase-phosphatase balance during the orchestration of the elongation-termination transition.
Enhancer regulation by SPT5
Enhancer regions can be actively transcribed to synthesize enhancer RNAs (eRNAs), a process which potentially facilitates target gene activation [Citation139,Citation140]. Enhancer transcription harbors some typical regulatory features shared by active genes, such as bidirectional transcription, Pol II pausing, pause release, and early termination, despite each step being regulated to a different degree compared with protein-coding genes [Citation20,Citation89,Citation141,Citation142]. Similar to its global importance for protein-coding gene transcription, SPT5 is globally required for the synthesis of noncoding RNAs, including eRNAs in Drosophila [Citation89]. Although SPT5 depletion in mouse B cells causes altered expression for a subset of enhancers [Citation80], the acute and near-complete degradation of SPT5 in human cells dampens most if not all transcriptional activation of active enhancers [Citation20], in line with SPT5 function in Drosophila [Citation89].
Paused Pol II is less stable at enhancers than promoters, being more susceptible to early termination [Citation89,Citation143], yet SPT5 is critical in maintaining the stability of paused Pol II at enhancers [Citation20]. Therefore, it is tempting to speculate that SPT5 mediated RPB1 protein stabilization contributes to preserving paused Pol II for its ensuing release at enhancers, as it does at promoters [Citation19,Citation20]. Nonetheless, the regulation of pause release by SPT5 phosphorylation status at enhancers is poorly investigated. We surmise that the balance between P-TEFb and INTAC in modulating pausing and pause release, partially by controlling SPT5 phosphorylation as discussed above [Citation76–78], is also in play at enhancers as promoters do, because PAF1-dependent INTAC recruitment at enhancers prevents the release of paused Pol II and eRNA accumulation [Citation54,Citation56,Citation58]. In addition to the INTAC phosphatase module, the RNA endonuclease activity in the Integrator module of INTAC could additionally promote termination to suppress enhancer transcription [Citation58,Citation143,Citation144].
Numerous studies have reported the regulatory role of eRNAs in activating target genes [Citation145–147]. For instance, eRNAs could interact with NELF to facilitate its eviction, and thus promote the release of paused Pol II from promotor-proximal regions [Citation148,Citation149], in accordance with the increased paused Pol II release upon enhancer activation [Citation56]. This suggests that there are at least two layers of gene transcriptional control by SPT5: one is through its direct association with Pol II at promoters and gene bodies, and the other is through indirectly influencing target genes by modulating activation of their corresponding enhancers.
The presence of H3K27ac and nascent transcription have been used to distinguish active enhancers from inactive ones [Citation139,Citation146,Citation150]. SPT5 depletion results in decreased H3K27ac levels on a subset of initially highly transcribed enhancers [Citation20,Citation80], possibly due to reduced production of eRNAs, which can stimulate the histone acetyltransferase activity of CBP [Citation151]. Moreover, active enhancers generally harbor accessible chromatin structures to expose specific sequence motifs that are recognized by DNA-binding transcription factors that boost transcription. Therefore, the maintenance of enhancer accessibility and transcription activity requires continuous chromatin remodeling [Citation152,Citation153]. However, less is known about how transcription regulators, including SPT5, influence chromatin accessibility and landscape. Rapid SPT5 depletion results in decreased chromatin accessibility for a group of enhancers, which exhibit a reduction in SWI/SNF occupancy [Citation20]. This can be explained by the physical association and thus recruitment to chromatin of SWI/SNF by SPT5. However, alterations in chromatin accessibility are not correlated or coupled with the transcription changes at enhancers upon SPT5 loss, suggesting that SPT5 might have independent roles in modulating enhancer chromatin and transcription [Citation20]. Future studies are warranted to interrogate details of the underlying mechanisms.
Remaining questions
How to delineate the general and gene-specific functions of SPT5? Strategies combining near-complete SPT5 depletion and spike-in normalized nascent transcriptome profiling reveal that SPT5 is crucial for the transcriptional activation of most if not all protein-coding genes in Drosophila and human cells [Citation20,Citation89]. Yet studies also indicate that a group of genes related to development, hematopoietic stem cell formation, and erythropoiesis seem more susceptible to the perturbation of SPT5 in zebrafish and Drosophila [Citation154–157]. Future studies employing more efficient SPT5 depletion and calibrated sequencing techniques in specific biological contexts would help reconcile the inconsistency and explain the differential susceptibility of SPT5 target genes.
What are the mechanisms of SPT5 mediated enhancer activation, and how does it contribute to protein-coding gene transcription? Compared to our understanding of SPT5 functions on protein-coding genes, much less attention has been paid to the elucidation of SPT5-mediated enhancer regulation. For example, it is unclear whether SPT5 function (or lack of its function) is related to the higher termination frequency and lower elongation capacity at enhancers relative to promoters. Also, we are uncertain as to what extent SPT5 contributes to the shape of the chromatin landscape at enhancers. Moreover, it would be interesting to dissect how SPT5-mediated enhancer activation cooperates with its direct roles in pausing and elongation of the corresponding genes.
How to correlate SPT5 function in transcription to developmental and disease processes? Likely because SPT5 is an essential protein implicated in various steps of transcription, studies trying to unravel the biological significance of SPT5 in specific contexts, and how each SPT5-regulated transcriptional step relates to its biological functions, are lacking. However, with our deepened mechanistic understanding of SPT5 function in transcription and improved capability to decouple its roles in different transcription steps, it is now possible to conduct more systematic investigations to interrogate SPT5 functions in varying physiological or pathological processes, ideally using more advanced model systems with transgenic approaches.
What is the role of SPT5 in RNA Polymerase I (Pol I) transcription? As a multifaceted transcriptional factor, SPT5 is also required for efficient Pol I transcription and ribosomal RNAs synthesis [Citation158]. SPT5 interacts with Pol I directly [Citation158,Citation159] and regulates Pol I activity positively and negatively [Citation160]. Other SPT5 binding factors that are involved in Pol II transcription might also regulate Pol I transcription. For example, PAF1C, which plays an important role in promoting Pol II transcription, may be recruited to ribosomal DNA through its interaction with SPT5 to stimulate Pol I elongation [Citation161,Citation162]. Given that SPT5 has multiple critical roles in the control of Pol I transcription, it would be interesting to utilize advanced approaches to dissect its direct functions at the molecular level.
Acknowledgments
We apologize to those colleagues whose work has not been cited due to space limitations. The Chen lab is currently funded by grants from the National Key R&D Program of China (2021YFA1301700), the National Natural Science Foundation of China (32070636), and the Shanghai Natural Science Foundation (20ZR1412100).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Winston F, Chaleff DT, Valent B, et al. MUTATIONS AFFECTING TY-MEDIATED EXPRESSION OF THE HIS4 GENE OF SACCHAROMYCES CEREVISIAE. Genetics. 1984;107(2):179–197.
- Hartzog GA, Wada T, Handa H, et al. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 1998;12(3):357–369.
- Swanson MS, Malone EA, Winston F. SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat. Mol Cell Biol. 1991;11(8):4286.
- Kaplan CD, Morris JR, Wu C, et al. Spt5 and Spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes Dev. 2000;14(20):2623–2634.
- Yamada T, Yamaguchi Y, Inukai N, et al. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol Cell. 2006;21(2):227–237.
- Sanso M, Levin RS, Lipp JJ, et al. P-TEFb regulation of transcription termination factor Xrn2 revealed by a chemical genetic screen for Cdk9 substrates. Genes Dev. 2016;30(1):117–131.
- Bernecky C, Plitzko JM, Cramer P. Structure of a transcribing RNA polymerase II–DSIF complex reveals a multidentate DNA–RNA clamp. Nat Struct Mol Biol. 2017;24(10):809–815.
- Cortazar MA, Sheridan RM, Erickson B, et al. Control of RNA Pol II speed by PNUTS-PP1 and Spt5 dephosphorylation facilitates termination by a “sitting duck torpedo” mechanism. Molecular Cell. 2019;76(6):896–908 e894.
- Kim JB, Sharp PA. Positive transcription elongation factor B phosphorylates hSPT5 and RNA polymerase II carboxyl-terminal domain independently of cyclin-dependent kinase-activating kinase. J Biol Chem. 2001;276(15):12317–12323.
- Ni Z, Saunders A, Fuda NJ, et al. P-TEFb is critical for the maturation of RNA polymerase II into productive elongation in vivo. Mol Cell Biol. 2008;28(3):1161–1170.
- Parua PK, Booth GT, Sansó M, et al. A Cdk9–PP1 switch regulates the elongation–termination transition of RNA polymerase II. Nature. 2018;558(7710):460–464.
- Parua PK, Kalan S, Benjamin B, et al. Distinct Cdk9-phosphatase switches act at the beginning and end of elongation by RNA polymerase II. Nat Commun. 2020;11(1):4338.
- Qiu Y, Gilmour DS. Identification of regions in the Spt5 subunit of DRB sensitivity-inducing factor (DSIF) that are involved in promoter-proximal pausing. J Biol Chem. 2017;292(13):5555–5570.
- Rimel JK, et al. Selective inhibition of CDK7 reveals high-confidence targets and new models for TFIIH function in transcription. Genes Dev. 2020;34(21–22):1452–1473.
- Vos SM, Farnung L, Boehning M, et al. Structure of activated transcription complex Pol II–DSIF–PAF–SPT6. Nature. 2018;560(7720):607–612.
- Wada T, Takagi T, Yamaguchi Y, et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12(3):343–356.
- Malik S, Barrero MJ, Jones T. Identification of a regulator of transcription elongation as an accessory factor for the human mediator coactivator. Proc Natl Acad Sci U S A. 2007;104(15):6182–6187.
- Andrulis ED, Guzman E, Doring P, et al. High-resolution localization of drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev. 2000;14:2635–2649.
- Aoi Y, et al. SPT5 stabilization of promoter-proximal RNA polymerase II. Mol Cell. 2021;81(21):4413–4424 e4415.
- Hu S, Peng L, Xu C, et al. SPT5 stabilizes RNA polymerase II, orchestrates transcription cycles, and maintains the enhancer landscape. Mol Cell. 2021;81(21):4425–4439 e4426.
- Laine J-P, Egly J-M. Initiation of DNA repair mediated by a stalled RNA polymerase IIO. EMBO J. 2006;25(2):387–397.
- Bohr VA, Smith CA, Okumoto DS, et al. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40(2):359–369.
- Mellon I, Spivak G, Hanawalt PC. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51(2):241–249.
- Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9(12):958–970.
- Nakazawa Y, Hara Y, Oka Y, et al. Ubiquitination of DNA damage-stalled RNAPII promotes transcription-coupled repair. Cell. 2020;180(6):1228–1244 e1224.
- Tufegdzic Vidakovic A, Mitter R, Kelly GP, et al. Regulation of the RNAPII pool is integral to the DNA damage response. Cell. 2020;180(6):1245–1261 e1221.
- Troelstra C, van Gool A, de Wit J, et al. ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne’s syndrome and preferential repair of active genes. Cell. 1992;71(6):939–953.
- van Gool AJ, Verhage R, Swagemakers SM, et al. RAD26, the functional S. cerevisiae homolog of the Cockayne syndrome B gene ERCC6. EMBO J. 1994;13(22):5361–5369.
- van Gool AJ, Citterio E, Rademakers S, et al. The Cockayne syndrome B protein, involved in transcription-coupled DNA repair, resides in an RNA polymerase II-containing complex. EMBO J. 1997;16(19):5955–5965.
- Tantin D, Kansal A, Carey M. Recruitment of the putative transcription-repair coupling factor CSB/ERCC6 to RNA polymerase II elongation complexes. Mol Cell Biol. 1997;17(12):6803–6814.
- Selby CP, Sancar A. Human transcription-repair coupling factor CSB/ERCC6 is a DNA-stimulated ATPase but is not a helicase and does not disrupt the ternary transcription complex of stalled RNA polymerase II. J Biol Chem. 1997;272(3):1885–1890.
- Sarker AH, Tsutakawa SE, Kostek S, et al. Recognition of RNA polymerase II and transcription bubbles by XPG, CSB, and TFIIH: insights for transcription-coupled repair and Cockayne Syndrome. Mol Cell. 2005;20(2):187–198.
- van der Weegen Y, Golan-Berman H, Mevissen TET, et al. The cooperative action of CSB, CSA, and UVSSA target TFIIH to DNA damage-stalled RNA polymerase II. Nat Commun. 2020;11(1):2104.
- Epanchintsev A, Costanzo F, Rauschendorf M-A, et al. Cockayne’s syndrome A and B proteins regulate transcription arrest after genotoxic stress by promoting ATF3 degradation. Mol Cell. 2017;68(6):1054–1066 e1056.
- Xu J, Lahiri I, Wang W, et al. Structural basis for the initiation of eukaryotic transcription-coupled DNA repair. Nature. 2017;551(7682):653–657.
- Kokic G, Wagner FR, Chernev A, et al. Structural basis of human transcription–DNA repair coupling. Nature. 2021;598(7880):368–372.
- Jansen LE, et al. Spt4 modulates Rad26 requirement in transcription-coupled nucleotide excision repair. EMBO J. 2000;19(23):6498–6507.
- Ding B, LeJeune D, Li S. The C-terminal repeat domain of Spt5 plays an important role in suppression of Rad26-independent transcription coupled repair. J Biol Chem. 2010;285(8):5317–5326.
- Li W, Giles C, Li S. Insights into how Spt5 functions in transcription elongation and repressing transcription coupled DNA repair. Nucleic Acids Res. 2014;42(11):7069–7083.
- Selvam K, Ding B, Sharma R, et al. Evidence that moderate eviction of Spt5 and promotion of error-free transcriptional bypass by rad26 facilitates transcription coupled nucleotide excision repair. J Mol Biol. 2019;431(7):1322–1338.
- Duan M, Selvam K, Wyrick JJ, et al. Genome-wide role of Rad26 in promoting transcription-coupled nucleotide excision repair in yeast chromatin. Proc Natl Acad Sci U S A. 2020;117(31):18608–18616.
- Booth GT, Wang IX, Cheung VG, et al. Divergence of a conserved elongation factor and transcription regulation in budding and fission yeast. Genome Res. 2016;26(6):799–811.
- Cheon Y, Han S, Kim T, et al. The chromatin remodeler Ino80 mediates RNAPII pausing site determination. Genome Biol. 2021;22(1):294.
- Core L, Adelman K. Promoter-proximal pausing of RNA polymerase II: a nexus of gene regulation. Genes Dev. 2019;33(15–16):960–982.
- Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet. 2012;13(10):720–731.
- Kwak H, Lis JT. Control of transcriptional elongation. Annu Rev Genet. 2013;47(1):483–508.
- Jonkers I, Lis JT. Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol. 2015;16(3):167–177.
- Kwak H, Fuda NJ, Core LJ, et al. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science. 2013;339(6122):950–953.
- Missra A, Gilmour DS. Interactions between DSIF (DRB sensitivity inducing factor), NELF (negative elongation factor), and the drosophila RNA polymerase II transcription elongation complex. Proc Natl Acad Sci U S A. 2010;107(25):11301–11306.
- Yamaguchi Y, Inukai N, Narita T, et al. Evidence that negative elongation factor represses transcription elongation through binding to a DRB sensitivity-inducing factor/RNA polymerase II complex and RNA. Mol Cell Biol. 2002;22(9):2918–2927.
- Yamamoto J, Hagiwara Y, Chiba K, et al. DSIF and NELF interact with integrator to specify the correct post-transcriptional fate of snRNA genes. Nat Commun. 2014;5(1):4263.
- Aoi Y, Smith ER, Shah AP, et al. NELF regulates a promoter-proximal step distinct from RNA Pol II pause-release. Mol Cell. 2020;78(2):261–274 e265.
- Chiu AC, Suzuki HI, Wu X, et al. Transcriptional pause sites delineate stable nucleosome-associated premature polyadenylation suppressed by U1 snRNP. Mol Cell. 2018;69(4):648–663 e647.
- Wang Z, Song A, Xu H, et al. Coordinated regulation of RNA polymerase II pausing and elongation progression by PAF1. Sci Adv. 2022;8(13):eabm5504.
- Chen FX, Woodfin A, Gardini A, et al. PAF1, a molecular regulator of promoter-proximal pausing by RNA polymerase II. Cell. 2015;162(5):1003–1015.
- Chen FX, Xie P, Collings CK, et al. PAF1 regulation of promoter-proximal pause release via enhancer activation. Science. 2017;357(6357):1294–1298.
- Jaenicke LA, von Eyss B, Carstensen A, et al. Ubiquitin-dependent turnover of MYC antagonizes MYC/PAF1C complex accumulation to drive transcriptional elongation. Mol Cell. 2016;61(1):54–67.
- Liu X, Guo Z, Han J, et al. The PAF1 complex promotes 3′ processing of pervasive transcripts. Cell Rep. 2022;38(11):110519.
- Decker T-M. Mechanisms of transcription elongation factor DSIF (Spt4–Spt5). J Mol Biol. 2021;433(14):166657.
- Dollinger R, Gilmour DS. Regulation of promoter proximal pausing of RNA polymerase II in metazoans. J Mol Biol. 2021;433(14):166897.
- Zhou Q, Li T, Price DH. RNA polymerase II elongation control. Annu Rev Biochem. 2012;81(1):119–143.
- Yik JHN, Chen R, Nishimura R, et al. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol Cell. 2003;12(4):971–982.
- Yang Z, Yik JHN, Chen R, et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19(4):535–545.
- Chen R, Liu M, Li H, et al. PP2B and PP1α cooperatively disrupt 7SK snRNP to release P-TEFb for transcription in response to Ca 2+ signaling. Genes Dev. 2008;22(10):1356–1368.
- Lu X, Zhu X, Li Y, et al. Multiple P-TEFbs cooperatively regulate the release of promoter-proximally paused RNA polymerase II. Nucleic Acids Res. 2016;44(14):6853–6867.
- Takahashi H, Parmely T, Sato S, et al. Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell. 2011;146(1):92–104.
- Luo Z, Lin C, Shilatifard A. The super elongation complex (SEC) family in transcriptional control. Nat Rev Mol Cell Biol. 2012;13(9):543–547.
- Xue Y, Yang Z, Chen R, et al. A capping-independent function of MePCE in stabilizing 7SK snRNA and facilitating the assembly of 7SK snRNP. Nucleic Acids Res. 2010;38(2):360–369.
- Fujinaga K, Irwin D, Huang Y, et al. Dynamics of human immunodeficiency virus transcription: p-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol Cell Biol. 2004;24(2):787–795.
- Bowman EA, Kelly WG. RNA polymerase II transcription elongation and Pol II CTD Ser2 phosphorylation: a tail of two kinases. Nucleus. 2014;5(3):224–236.
- Cossa G, Parua PK, Eilers M, et al. Protein phosphatases in the RNAPII transcription cycle: erasers, sculptors, gatekeepers, and potential drug targets. Genes Dev. 2021;35(9–10):658–676.
- Wang X, Qi Y, Wang Z, et al. RPAP2 regulates a transcription initiation checkpoint by inhibiting assembly of pre-initiation complex. Cell Rep. 2022;39(4):110732.
- Mosley AL, Pattenden SG, Carey M, et al. Rtr1 is a CTD phosphatase that regulates RNA polymerase II during the transition from serine 5 to serine 2 phosphorylation. Mol Cell. 2009;34(2):168–178.
- Xiang K, Manley JL, Tong L. The yeast regulator of transcription protein Rtr1 lacks an active site and phosphatase activity. Nat Commun. 2012;3(1):946.
- Egloff S, Zaborowska J, Laitem C, et al. Ser7 phosphorylation of the CTD recruits the RPAP2 Ser5 phosphatase to snRNA genes. Mol Cell. 2012;45(1):111–122.
- Huang K-L, Jee D, Stein CB, et al. Integrator recruits protein phosphatase 2A to prevent pause release and facilitate transcription termination. Mol Cell. 2020;80(2):345–358 e349.
- Vervoort SJ, Welsh SA, Devlin JR, et al. The PP2A-integrator-CDK9 axis fine-tunes transcription and can be targeted therapeutically in cancer. Cell. 2021;184(12):3143–3162 e3132.
- Zheng H, Qi Y, Hu S, et al. Identification of integrator-PP2A complex (INTAC), an RNA polymerase II phosphatase. Science. 2020;370(6520)
- Fitz J, Neumann T, Pavri R. Regulation of RNA polymerase II processivity by Spt5 is restricted to a narrow window during elongation. EMBO J. 2018;37(8)
- Fitz J, et al. Spt5-mediated enhancer transcription directly couples enhancer activation with physical promoter interaction. Nat Genet. 2020;52(5):505–515.
- Shetty A, et al. Spt5 plays vital roles in the control of sense and antisense transcription elongation. Mol Cell. 2017;66(1):77–88 e75.
- Xu J, Chong J, Wang D. Opposite roles of transcription elongation factors Spt4/5 and Elf1 in RNA polymerase II transcription through B-form versus non-B DNA structures. Nucleic Acids Res. 2021;49(9):4944–4953.
- Rosen GA, Baek I, Friedman LJ, et al. Dynamics of RNA polymerase II and elongation factor Spt4/5 recruitment during activator-dependent transcription. Proc Natl Acad Sci U S A. 2020;117(51):32348–32357.
- Bourgeois CF, Kim YK, Churcher MJ, et al. Spt5 cooperates with human immunodeficiency virus type 1 Tat by preventing premature RNA release at terminator sequences. Mol Cell Biol. 2002;22(4):1079–1093.
- Rahl PB, Lin CY, Seila AC, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141(3):432–445.
- Pavri R, Gazumyan A, Jankovic M, et al. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 2010;143(1):122–133.
- Swanson MS, Malone EA, Winston F. SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat. Mol Cell Biol. 1991;11(8):3009–3019.
- Swanson MS, Winston F. SPT4, SPT5 and SPT6 interactions: effects on transcription and viability in Saccharomyces cerevisiae. Genetics. 1992;132(2):325–336.
- Henriques T, et al. Widespread transcriptional pausing and elongation control at enhancers. Genes Dev. 2018;32(1):26–41.
- Zumer K, Maier KC, Farnung L, et al. Two distinct mechanisms of RNA polymerase II elongation stimulation in vivo. Mol Cell. 2021;81(15):3096–3109 e3098.
- Burova E, Hung SC, Sagitov V, et al. Escherichia coli NusG protein stimulates transcription elongation rates in vivo and in vitro. J Bacteriol. 1995;177(5):1388–1392.
- Kang JY, Mooney RA, Nedialkov Y, et al. Structural basis for transcript elongation control by NusG family universal regulators. Cell. 2018;173(7):1650–1662 e1614.
- Werner F. A nexus for gene expression—molecular mechanisms of Spt5 and NusG in the three domains of life. J Mol Biol. 2012;417(1–2):13–27.
- Zhu W, Wada T, Okabe S, et al. DSIF contributes to transcriptional activation by DNA-binding activators by preventing pausing during transcription elongation. Nucleic Acids Res. 2007;35(12):4064–4075.
- Hirtreiter A, Damsma GE, Cheung ACM, et al. Spt4/5 stimulates transcription elongation through the RNA polymerase clamp coiled-coil motif. Nucleic Acids Res. 2010;38(12):4040–4051.
- Klein BJ, Bose D, Baker KJ, et al. RNA polymerase and transcription elongation factor Spt4/5 complex structure. Proc Natl Acad Sci U S A. 2011;108(2):546–550.
- Meyer PA, Li S, Zhang M, et al. Structures and functions of the multiple KOW domains of transcription elongation factor Spt5. Mol Cell Biol. 2015;35(19):3354–3369.
- Martinez-Rucobo FW, Sainsbury S, Cheung AC, et al. Architecture of the RNA polymerase-Spt4/5 complex and basis of universal transcription processivity. EMBO J. 2011;30(7):1302–1310.
- Ehara H, Yokoyama T, Shigematsu H, et al. Structure of the complete elongation complex of RNA polymerase II with basal factors. Science. 2017;357(6354):921–924.
- Crickard JB, Fu J, Reese JC. Biochemical analysis of yeast suppressor of Ty 4/5 (Spt4/5) reveals the importance of nucleic acid interactions in the prevention of RNA polymerase II arrest. J Biol Chem. 2016;291(19):9853–9870.
- Yakhnin AV, Murakami KS, Babitzke P. NusG is a sequence-specific RNA polymerase pause factor that binds to the non-template DNA within the paused transcription bubble. J Biol Chem. 2016;291(10):5299–5308.
- Wier AD, Mayekar MK, Heroux A, et al. Structural basis for Spt5-mediated recruitment of the Paf1 complex to chromatin. Proc Natl Acad Sci U S A. 2013;110(43):17290–17295.
- Chen JJ, Mbogning J, Hancock MA, et al. Spt5 phosphorylation and the Rtf1 Plus3 domain promote Rtf1 function through distinct mechanisms. Mol Cell Biol. 2020;40(15)
- Mayekar MK, Gardner RG, Arndt KM. The recruitment of the Saccharomyces cerevisiae Paf1 complex to active genes requires a domain of Rtf1 that directly interacts with the Spt4-Spt5 complex. Mol Cell Biol. 2013;33(16):3259–3273.
- Kujirai T, Kurumizaka H. Transcription through the nucleosome. Curr Opin Struct Biol. 2020;61:42–49.
- Ehara H, Kujirai T, Fujino Y, et al. Structural insight into nucleosome transcription by RNA polymerase II with elongation factors. Science. 2019;363(6428):744–747.
- Crickard JB, Lee J, Lee T-H, et al. The elongation factor Spt4/5 regulates RNA polymerase II transcription through the nucleosome. Nucleic Acids Res. 2017;45(11):6362–6374.
- Sanders TJ, Lammers M, Marshall CJ, et al. TFS and Spt4/5 accelerate transcription through archaeal histone-based chromatin. Mol Microbiol. 2019;111(3):784–797.
- Mattiroli F, Bhattacharyya S, Dyer PN, et al. Structure of histone-based chromatin in Archaea. Science. 2017;357(6351):609–612.
- Uzun Ü, Brown T, Fischl H, et al. Spt4 facilitates the movement of RNA polymerase II through the +2 nucleosomal barrier. Cell Rep. 2021;36(13):109755.
- Farnung L, Ochmann M, Engeholm M, et al. Structural basis of nucleosome transcription mediated by Chd1 and FACT. Nat Struct Mol Biol. 2021;28(4):382–387.
- Evrin C, Serra‐Cardona A, Duan S, et al. Spt5 histone binding activity preserves chromatin during transcription by RNA polymerase II. EMBO J. 2022;41(5):e109783.
- Zhou K, Kuo WHW, Fillingham J, et al. Control of transcriptional elongation and cotranscriptional histone modification by the yeast BUR kinase substrate Spt5. Proc Natl Acad Sci U S A. 2009;106(17):6956–6961.
- Chen Y, Yamaguchi Y, Tsugeno Y, et al. DSIF, the Paf1 complex, and Tat-SF1 have nonredundant, cooperative roles in RNA polymerase II elongation. Genes Dev. 2009;23(23):2765–2777.
- Hartzog GA, Fu J. The Spt4–Spt5 complex: a multi-faceted regulator of transcription elongation. Biochim Biophys Acta. 2013;1829(1):105–115.
- Qiu H, Hu C, Gaur NA, et al. Pol II CTD kinases Bur1 and Kin28 promote Spt5 CTR-independent recruitment of Paf1 complex. EMBO J. 2012;31(16):3494–3505.
- Squazzo SL, Costa PJ, Lindstrom DL, et al. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 2002;21(7):1764–1774.
- Zhu B, Mandal SS, Pham A-D, et al. The human PAF complex coordinates transcription with events downstream of RNA synthesis. Genes Dev. 2005;19(14):1668–1673.
- Krogan NJ, Dover J, Wood A, et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell. 2003;11(3):721–729.
- Ng HH, Robert F, Young RA, et al. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11(3):709–719.
- Wood A, Schneider J, Dover J, et al. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J Biol Chem. 2003;278(37):34739–34742.
- Wood A, Schneider J, Dover J, et al. The Bur1/Bur2 complex is required for histone H2B monoubiquitination by Rad6/Bre1 and histone methylation by COMPASS. Mol Cell. 2005;20(4):589–599.
- Ng HH, Dole S, Struhl K. The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J Biol Chem. 2003;278(36):33625–33628.
- Tenney K, Gerber M, Ilvarsonn A, et al. Drosophila Rtf1 functions in histone methylation, gene expression, and Notch signaling. Proc Natl Acad Sci U S A. 2006;103(32):11970–11974.
- Xiao T, Kao C-F, Krogan NJ, et al. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol Cell Biol. 2005;25(2):637–651.
- Van Oss SB, Shirra MK, Bataille AR, et al. The histone modification domain of Paf1 complex subunit Rtf1 directly stimulates H2B ubiquitylation through an interaction with Rad6. Mol Cell. 2016;64(4):815–825.
- Tomson BN, Arndt KM. The many roles of the conserved eukaryotic Paf1 complex in regulating transcription, histone modifications, and disease states. Biochim Biophys Acta. 2013;1829(1):116–126.
- Liu Y, Warfield L, Zhang C, et al. Phosphorylation of the transcription elongation factor Spt5 by yeast Bur1 kinase stimulates recruitment of the PAF complex. Mol Cell Biol. 2009;29(17):4852–4863.
- Chen F, Liu B, Guo L, et al. Biochemical insights into Paf1 complex–induced stimulation of Rad6/Bre1-mediated H2B monoubiquitination. Proc Natl Acad Sci U S A. 2021;118(33)
- Baejen C, Andreani J, Torkler P, et al. Genome-wide analysis of RNA polymerase II termination at protein-coding genes. Mol Cell. 2017;66(1):38–49 e36.
- Mayer A, Schreieck A, Lidschreiber M, et al. The Spt5 C-terminal region recruits yeast 3′ RNA cleavage factor I. Mol Cell Biol. 2012;32(7):1321–1331.
- Kecman T, Kuś K, Heo D-H, et al. Elongation/termination factor exchange mediated by PP1 phosphatase orchestrates transcription termination. Cell Rep. 2018;25(1):259–269 e255.
- Nordick K, Hoffman MG, Betz JL, et al. Direct interactions between the Paf1 complex and a cleavage and polyadenylation factor are revealed by dissociation of Paf1 from RNA polymerase II. Eukaryot Cell. 2008;7(7):1158–1167.
- Rozenblatt-Rosen O, Nagaike T, Francis JM, et al. The tumor suppressor Cdc73 functionally associates with CPSF and CstF 3′ mRNA processing factors. Proc Natl Acad Sci U S A. 2009;106(3):755–760.
- Chen FX, Smith ER, Shilatifard A. Born to run: control of transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol. 2018;19(7):464–478.
- Laitem C, Zaborowska J, Isa NF, et al. CDK9 inhibitors define elongation checkpoints at both ends of RNA polymerase II–transcribed genes. Nat Struct Mol Biol. 2015;22(5):396–403.
- Tellier M, Zaborowska J, Neve J, et al. CDK9 and PP2A regulate RNA polymerase II transcription termination and coupled RNA maturation. bioRxiv. 2021. 2006.2021.449289 (2022).
- Eaton JD, Francis L, Davidson L, et al. A unified allosteric/torpedo mechanism for transcriptional termination on human protein-coding genes. Genes Dev. 2020;34(1–2):132–145.
- Field A, Adelman K. Evaluating enhancer function and transcription. Annu Rev Biochem. 2020;89(1):213–234.
- Tippens ND, Vihervaara A, Lis JT. Enhancer transcription: what, where, when, and why? Genes Dev. 2018;32(1):1–3.
- Core LJ, et al. Defining the status of RNA polymerase at promoters. Cell Rep. 2012;2(4):1025–1035.
- Andersson R, Sandelin A, Danko CG. A unified architecture of transcriptional regulatory elements. Trends Genet. 2015;31(8):426–433.
- Lai F, Gardini A, Zhang A, et al. Integrator mediates the biogenesis of enhancer RNAs. Nature. 2015;525(7569):399–403.
- Lykke-Andersen S, Žumer K, Molska EŠ, et al. Integrator is a genome-wide attenuator of non-productive transcription. Mol Cell. 2021;81(3):514–529 e516.
- Rahnamoun H, Orozco P, Lauberth SM. The role of enhancer RNAs in epigenetic regulation of gene expression. Transcription. 2020;11(1):19–25.
- Rickels R, Shilatifard A. Enhancer logic and mechanics in development and disease. Trends Cell Biol. 2018;28(8):608–630.
- Arnold PR, Wells AD, Li XC. Diversity and emerging roles of enhancer RNA in regulation of gene expression and cell fate. Front Cell Dev Biol. 2020;7:377.
- Schaukowitch K, Joo J-Y, Liu X, et al. Enhancer RNA facilitates NELF release from immediate early genes. Mol Cell. 2014;56(1):29–42.
- Gorbovytska V, Kim S-K, Kuybu F, et al. Enhancer RNAs stimulate Pol II pause release by harnessing multivalent interactions to NELF. Nat Commun. 2022;13(1):2429.
- Creyghton MP, Cheng AW, Welstead GG, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107(50):21931–21936.
- Bose DA, Donahue G, Reinberg D, et al. RNA binding to CBP stimulates histone acetylation and transcription. Cell. 2017;168(1–2):135–149 e122.
- Iurlaro M, Stadler MB, Masoni F, et al. Mammalian SWI/SNF continuously restores local accessibility to chromatin. Nat Genet. 2021;53(3):279–287.
- Schick S, Grosche S, Kohl KE, et al. Acute BAF perturbation causes immediate changes in chromatin accessibility. Nat Genet. 2021;53(3):269–278.
- Guo S, Yamaguchi Y, Schilbach S, et al. A regulator of transcriptional elongation controls vertebrate neuronal development. Nature. 2000;408(6810):366–369.
- Jennings BH, Shah S, Yamaguchi Y, et al. Locus-specific requirements for Spt5 in transcriptional activation and repression in drosophila. Curr Biol. 2004;14(18):1680–1684.
- Krishnan K, Salomonis N, Guo S. Identification of Spt5 target genes in zebrafish development reveals its dual activity in vivo. PLoS One. 2008;3(11):e3621.
- Bai X, Trowbridge JJ, Riley E, et al. TiF1-gamma plays an essential role in murine hematopoiesis and regulates transcriptional elongation of erythroid genes. Dev Biol. 2013;373(2):422–430.
- Schneider DA, French SL, Osheim YN, et al. RNA polymerase II elongation factors Spt4p and Spt5p play roles in transcription elongation by RNA polymerase I and rRNA processing. Proc Natl Acad Sci U S A. 2006;103(34):12707–12712.
- Viktorovskaya OV, Appling FD, Schneider DA. Yeast transcription elongation factor Spt5 associates with RNA polymerase I and RNA polymerase II directly. J Biol Chem. 2011;286(21):18825–18833.
- Anderson SJ, Sikes ML, Zhang Y, et al. The transcription elongation factor Spt5 influences transcription by RNA polymerase I positively and negatively. J Biol Chem. 2011;286(21):18816–18824.
- Zhang Y, Sikes ML, Beyer AL, et al. The Paf1 complex is required for efficient transcription elongation by RNA polymerase I. Proc Natl Acad Sci U S A. 2009;106(7):2153–2158.
- Zhang Y, T. Smith AD, Renfrow MB, et al. The RNA polymerase-associated factor 1 complex (Paf1C) directly increases the elongation rate of RNA polymerase I and is required for efficient regulation of rRNA synthesis. J Biol Chem. 2010;285(19):14152–14159.
- Zuber PK, et al. Structure and nucleic acid binding properties of KOW domains 4 and 6–7 of human transcription elongation factor DSIF. Sci Rep. 2018;8(1):11660.
