abstract
Occupying 17% of human genome, the mobile long interspersed element 1 (LINE-1 or L1) continues to modulate the landscape of our genome by inserting into new loci and, as a result, causing sporadic diseases. It is not surprising that human cells have evolved a battery of mechanisms to control and limit the activity of LINE-1. Our recent study unravels such a mechanism that is imposed by the stress granule pathway. This mechanism functions by sequestering the LINE-1 RNA-protein complex within the cytoplasmic stress granules and thus inhibiting the nuclear import of LINE-1 RNA and its subsequent reverse transcription and integration into cellular DNA. Conditions that promote stress granule formation, such as expression of the SAMHD1 protein, further reduce LINE-1 retrotransposition.
Approximately 45% of human genome has been derived from transposable elements.Citation1 These include DNA transposons, long terminal repeat (LTR) retrotransposons (also called endogenous retroviruses), and non-LTR retrotransposons. Long interspersed element 1 (LINE-1) belongs to non-LTR retrotransposons and comprises ∼17% of human genome.Citation1 Compared to the other transposons that have mostly become inactive, approximately 100 copies of LINE-1 are still active.Citation2 Retrotransposition of these LINE-1s is associated with nearly 100 human diseases.Citation3 LINE-1 encodes two proteins called ORF1 and ORF2. ORF1 is an RNA-binding protein and associates with LINE-1 RNA.Citation4-7 ORF2 is an enzyme that has endonuclease and reverse transcriptase activities.Citation8,9 ORF1, ORF2 and LINE-1 RNA together form an RNP complex that needs to enter the nucleus where LINE-1 RNA is reverse transcribed and integrated into cellular DNA.Citation10-12
Humans have survived LINE-1 invasion and amplification over millions of years thanks to the evolution of a battery of mechanisms that control LINE-1 activity. Some of these mechanisms begin to be unraveled as a result of intensive research in the past couple of decades. One such mechanism is suppression of LINE-1 transcription by methylating LINE-1 DNA.Citation13-15 In support of this mechanism, knockdown or knockout genes that are involved in DNA methylation leads to increase in the activities of LINE-1 and other transposons.Citation13 In the course of embryonic development there are a couple of waves of DNA demythlyation. DNA demethylation inevitably activates LINE-1 RNA expression.Citation16 To control retrotransposition of LINE-1 and other transposable elements, primordial germ cells (PGCs) are equipped with the piRNA machinery to inactivate LINE-1 so as to protect the integrity of genome DNA in germ cells.Citation17,18
Recent studies have revealed that cells have a rich layer of mechanisms that check LINE-1 activity at the post-transcription stage. Many of these mechanisms involve cellular factors that have been shown to restrict viral infections. One example is the APOBEC family of proteins that are cytidine deaminase and inactivate viral or LINE-1 DNA by introducing lethal mutations.Citation19-23 An RNA helicase MOV10 inhibits retrotransposition of LINE-1 by associating with LINE-1 RNP and diminishing LINE-1 RNA level.Citation24-26 A recent study by Goodier et al tested a panel of viral restriction factors and showed that many of them, including BST-2, ISG20, MAVS, Mx2 and ZAP, strongly reduce LINE-1 activity.Citation27 The anti-LINE-1 activity of ZAP was also reported by Moran group.Citation28 It appears that cells have evolved mechanisms that can restrict both infective viruses and endogenous retroelements. In support of this scenario, results from our group and Yu lab have demonstrated that a viral restriction factor called SAMHD1 restricts LINE-1 retrotransposition.Citation29,30
As a deoxynucleotide triphosphate (dNTP) triphosphohydrolase, SAMHD1 inhibits HIV-1 infection in non-cycling cells by reducing dNTP level and thereby abrogating viral reverse transcription.Citation31,32 In contrast, in dividing cells, SAMHD1 is phosphorylated at amino acid T592 by cyclin A2/CDK1 and, as a result, loses its antiviral function.Citation33,34 The anti-LINE-1 activity of SAMHD1 was quickly tested by Zhao et al. Similar to other viral restriction factors, Zhao et al showed that SAMHD1 suppresses retrotransposition of LINE-1 by reducing the expression of ORF2 and thus impairing reverse transcription of LINE-1 RNA ().Citation30 Zhu et al. also found that dGTP-triggered tetramer formation of SAMHD1 is important for dNTP depletion and SAMHD1-mediated inhibition of LINE-1 transposition.Citation35 Results from our group confirmed the restriction of LINE-1 by SAMHD1 and also suggested an alternative mechanism of action.Citation29
Figure 1. Restriction of LINE-1 by SAMHD1 and stress granules. LINE-1 ORF1p and ORF2p associate with LINE-1 RNA and together form RNP complexes. LINE-1 RNP complexes enter the nucleus where LINE-1 RNA is reverse transcribed into DNA by a target-primed mechanism. Formation of stress granules is stimulated by eIF2α phosphorylation that is catalyzed by different kinases that are activated in response to different types of stresses. The LINE-1 restriction factors including G3BP1, TIA1, ZAP, PABP, APOBEC and MOV10 are co-localized with LINE-1 RNP in stress granules or P-bodies. Stress granules appear to serve as the cytoplasmic structure where LINE-1 RNP is sequestered in association with anti-LINE-1 factors that together achieve the maximal control of LINE-1. SAMHD1 enhances stress granules formation via modulating the phosphorylation of eIF2α by mechanisms to be elucidated.
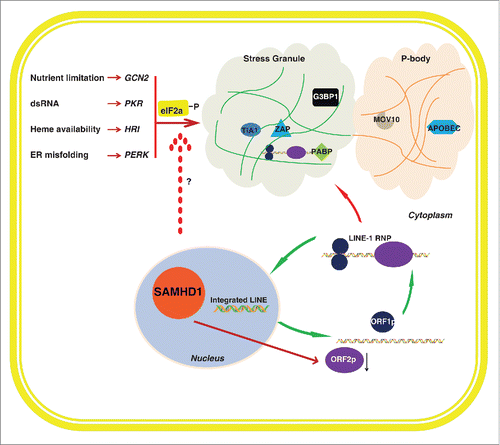
The first key observation of our study is that SAMHD1 expression enhances the localization of LINE-1 RNP into cytoplasmic stress granules. In most cases, the stress-induced phosphorylation of the translation initiation factor eIF2α induces stress granule assembly by preventing or delaying translational initiation. A family of structurally related eIF2α kinases, each activated by a different type of stress, phosphorylates the regulatory serine of eIF2α. Of these kinases, PKR is activated by double-stranded RNA, PERK is activated by endoplasmic reticulum stress, HRI is activated by oxidative stress, and GCN2 is activated by nutrient stress ().Citation36 One major function of stress granules is to temporarily store translation inert RNA molecules. These RNA molecules are either released for later translation or delivered to P-bodies for degradation.Citation37 Goodier et al have observed the localization of LINE-1 RNP to stress granules, albeit that the effect on LINE-1 activity was not fully investigated.Citation38,39 We have performed immunofluorescence experiments and utilized the automated Image Stream System to demonstrate that SAMHD1 expression causes sequestration of LINE-1 ORF1p in large cytoplasmic granule structures that are positively stained with stress granule markers G3BP1 and TIA1. This colocalization of LINE-1 components with stress granules was further confirmed by the results of co-immunoprecipitation and RT-PCR assays showing the association of stress granule marker protein G3BP1 with LINE-1 ORF1p and LINE-1 RNA.
The second key finding we made is that abrogating stress granule formation by knocking down G3BP1 or TIA1 increases LINE-1 retrotransposition whereas inducing stress granule formation with arsenite treatment suppresses LINE-1 activity. These data demonstrate the intrinsic role of stress granule pathway in controlling LINE-1 activity. Since stress granules sequester RNA molecules away from being translated, localizing to stress granules is expected to suppress the translation of LINE-1 RNA. This mechanism of inhibition at least partially explains the decrease in LINE-1 ORF2 when SAMHD1 is overexpressed and drives sequestration of LNE-1 RNA in stress granules.Citation29,30 We also note that SAMHD1 specifically reduces the level of ORF2p but not ORF1p.Citation30 This is because the translation initiation of ORF2p is different from that of ORF1p. Since the inter-ORF spacer is dispensable for ORF2p translation, the translation of ORF2p most likely relies on the re-activation of ribosome that finishes the translation of ORF1p but has not dissociated from LINE-1 RNA.Citation40 The detailed mechanism by which SAMHD1 impairs ORF2p production awaits further investigation. A second consequence of localizing to stress granules is the blockade of LINE-1 RNP from entering the nucleus where LINE-1 RNA is reverse transcribed and integrated into cellular DNA.
The third key observation of our study is that knocking down G3BP1 or TIA1 abrogates the ability of SAMHD1 to inhibit LINE-1, which strongly suggests that a functional stress granule pathway is indispensable for SAMHD1 to exert its anti-LINE-1 effect.
The role of stress granule pathway in controlling LINE-1 activity has been suggested in several other studies. Goodier et al first reported the localization of LINE-1 RNP components with stress granule markers.Citation38,39 The LINE-1 restriction factors including G3BP1,Citation29 TIA1,Citation29 ZAP,Citation27 PABP,Citation39,40 APOBCCitation19-23 and MOV10Citation24-26 co-localize with LINE-1 RNP in stress granules or P-bodies (). It is likely that stress granules serve as the cytoplasmic loci or structure where LINE-1 RNA is sequestered in association with anti-LINE-1 factors that together achieve the maximal control of LINE-1 activity (). Not surprisingly, cells have applied this mechanism of action to control viral infections.Citation41-45 In turn, viruses have evolved strategies to counter this restriction by abrogating the stress granule pathway such as cleaving the key stress granule factors G3BP1.Citation46 Stress granule pathway may thus represent an ancient form of innate immunity that cells have evolved to battle not only exogenous viruses but also endogenous retroelements.
Several questions remain unanswered. First, how is LINE-1 RNP recruited into stress granules? We have performed co-immunoprecipitation and RT-PCR experiments and found that stress granule marker protein G3BP1 associates with LINE-1 ORF1 in an RNA-dependent manner, yet we have not identified the genetic determinants that lead to the localization of LINE-1 RNP into stress granules. Several possible scenarios may exist. First, a specific sequence in LINE-1 RNA may be bound by a stress granule resident protein. Second, ORF1 or ORF2 itself may specifically interact with a stress granule component. Both mechanisms can lead to localization of LINE-1 RNP to stress granules.
Second, how does SAMHD1 enhance stress granule formation? We have observed that SAMHD1 expression increases eIF2α phosphorylation and disrupts the interaction of eIF4A and eIF4G. Both events suppress translation and are bound to stimulate stress granule formation. Given that SAMHD1 is a nuclear protein, it is unclear how it exerts its effect on translation events that take place in the cytoplasm. Further studies are warranted to identify the underlying mechanism. In addition to translation arrest, DNA damage has also been reported to stimulate stress granule formation.Citation47 The involvement of SAMHD1 in DNA damage response has been supported by its localization to the sites of DNA damage and its interaction with factors of the DNA damage repair pathway.Citation48 In addition, knockdown of SAMHD1 activates DNA damaging signaling and SAMHD1 has a role in maintaining genome stability.Citation49 These observations suggest a second mechanism that SAMHD1 may modulate stress granule formation via its involvement in DNA damage response.
It also remains to be studied whether other anti-LINE-1 factors including MOV10 and ZAP depend on the stress granule pathway to suppress LINE-1. For example, would depletion of stress granule marker proteins G3BP1 or TIA1 abrogate the anti-LINE-1 activities of MOV10 and ZAP? Are there more cellular proteins that associate with LINE-1 RNP and impair LINE-1 retrotransposition? Given that most of the studies have been performed in transformed cell lines, it would be worth investigating at which stage of embryonic development and in which cell types each of these LINE-1 restriction mechanisms functions.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This study was supported by funds from the Ministry of Science and Technology of China (2012CB911103, 2011CB5048002, 2012ZX10001006-003 and 2013ZX10001005-002), from the Nature Science Foundation of China (81371808, 81301439 and 81528012) and Canadian Institutes of Health Research (CCI-132561). The funding agencies had no role in the study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
References
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature 2001; 409:860-921; PMID:11237011; http://dx.doi.org/10.1038/35057062
- Beck CR, Garcia-Perez JL, Badge RM, Moran JV. LINE-1 elements in structural variation and disease. Annu Rev Genomics Hum Genet 2014; 12:187-215; http://dx.doi.org/10.1146/annurev-genom-082509-141802
- Hancks DC, Kazazian HH, Jr. Active human retrotransposons: variation and disease. Curr Opin Genet Dev 2012; 22:191-203; PMID:22406018; http://dx.doi.org/10.1016/j.gde.2012.02.006
- Martin SL. Ribonucleoprotein particles with LINE-1 RNA in mouse embryonal carcinoma cells. Mol Cell Biol 1991; 11:4804-7; PMID:1715025; http://dx.doi.org/10.1128/MCB.11.9.4804
- Holmes SE, Singer MF, Swergold GD. Studies on p40, the leucine zipper motif-containing protein encoded by the first open reading frame of an active human LINE-1 transposable element. J Biol Chem 1992; 267:19765-8; PMID:1328181
- Martin SL, Branciforte D, Keller D, Bain DL. Trimeric structure for an essential protein in L1 retrotransposition. Proc Natl Acad Sci U S A 2003; 100:13815-20; PMID:14615577; http://dx.doi.org/10.1073/pnas.2336221100
- Hohjoh H, Singer MF. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J 1996; 15:630-9; PMID:8599946
- Mathias SL, Scott AF, Kazazian HH, Jr., Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science 1991; 254:1808-10; PMID:1722352; http://dx.doi.org/10.1126/science.1722352
- Feng Q, Moran JV, Kazazian HH, Jr., Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 1996; 87:905-16; PMID:8945517; http://dx.doi.org/10.1016/S0092-8674(00)81997-2
- Kulpa DA, Moran JV. Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nat Struct Mol Biol 2006; 13:655-60; PMID:16783376; http://dx.doi.org/10.1038/nsmb1107
- Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Boeke JD, Moran JV. Human L1 retrotransposition: cis preference versus trans complementation. Mol Cell Biol 2001; 21:1429-39; PMID:11158327; http://dx.doi.org/10.1128/MCB.21.4.1429-1439.2001
- Kulpa DA, Moran JV. Ribonucleoprotein particle formation is necessary but not sufficient for LINE-1 retrotransposition. Hum Mol Genet 2005; 14:3237-48; PMID:16183655; http://dx.doi.org/10.1093/hmg/ddi354
- Kato Y, Kaneda M, Hata K, Kumaki K, Hisano M, Kohara Y, Okano M, Li E, Nozaki M, Sasaki H. Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum Mol Genet 2007; 16:2272-80; PMID:17616512; http://dx.doi.org/10.1093/hmg/ddm179
- Bourc'his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 2004; 431:96-9; PMID:15318244; http://dx.doi.org/10.1038/nature02886
- Walsh CP, Chaillet JR, Bestor TH. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet 1998; 20:116-7; PMID:9771701; http://dx.doi.org/10.1038/2413
- Woodcock DM, Lawler CB, Linsenmeyer ME, Doherty JP, Warren WD. Asymmetric methylation in the hypermethylated CpG promoter region of the human L1 retrotransposon. J Biol Chem 1997; 272:7810-6; PMID:9065445; http://dx.doi.org/10.1074/jbc.272.12.7810
- Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell 2007; 12:503-14; PMID:17395546; http://dx.doi.org/10.1016/j.devcel.2007.03.001
- Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 2007; 318:761-4; PMID:17975059; http://dx.doi.org/10.1126/science.1146484
- Niewiadomska AM, Tian C, Tan L, Wang T, Sarkis PT, Yu XF. Differential inhibition of long interspersed element 1 by APOBEC3 does not correlate with high-molecular-mass-complex formation or P-body association. J Virol 2007; 81:9577-83; PMID:17582006; http://dx.doi.org/10.1128/JVI.02800-06
- Wissing S, Montano M, Garcia-Perez JL, Moran JV, Greene WC. Endogenous APOBEC3B restricts LINE-1 retrotransposition in transformed cells and human embryonic stem cells. J Biol Chem 2011; 286:36427-37; PMID:21878639; http://dx.doi.org/10.1074/jbc.M111.251058
- Richardson SR, Narvaiza I, Planegger RA, Weitzman MD, Moran JV. APOBEC3A deaminates transiently exposed single-strand DNA during LINE-1 retrotransposition. Elife 2014; 3:e02008; PMID:24843014
- Chen H, Lilley CE, Yu Q, Lee DV, Chou J, Narvaiza I, Landau NR, Weitzman MD. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr Biol 2006; 16:480-5; PMID:16527742; http://dx.doi.org/10.1016/j.cub.2006.01.031
- Bogerd HP, Wiegand HL, Hulme AE, Garcia-Perez JL, O'Shea KS, Moran JV, Cullen BR. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc Natl Acad Sci U S A 2006; 103:8780-5; PMID:16728505; http://dx.doi.org/10.1073/pnas.0603313103
- Arjan-Odedra S, Swanson CM, Sherer NM, Wolinsky SM, Malim MH. Endogenous MOV10 inhibits the retrotransposition of endogenous retroelements but not the replication of exogenous retroviruses. Retrovirology 2012; 9:53; PMID:22727223; http://dx.doi.org/10.1186/1742-4690-9-53
- Goodier JL, Cheung LE, Kazazian HH, Jr. MOV10 RNA helicase is a potent inhibitor of retrotransposition in cells. PLoS Genet 2012; 8:e1002941; PMID:23093941; http://dx.doi.org/10.1371/journal.pgen.1002941
- Li X, Zhang J, Jia R, Cheng V, Xu X, Qiao W, Guo F, Liang C, Cen S. The MOV10 helicase inhibits LINE-1 mobility. J Biol Chem 2013; 288:21148-60; PMID:23754279; http://dx.doi.org/10.1074/jbc.M113.465856
- Goodier JL, Pereira GC, Cheung LE, Rose RJ, Kazazian HH, Jr. The Broad-Spectrum Antiviral Protein ZAP Restricts Human Retrotransposition. PLoS Genet 2015; 11:e1005252; PMID:26001115; http://dx.doi.org/10.1371/journal.pgen.1005252
- Moldovan JB, Moran JV. The Zinc-Finger Antiviral Protein ZAP Inhibits LINE and Alu Retrotransposition. PLoS Genet 2015; 11:e1005121; PMID:25951186; http://dx.doi.org/10.1371/journal.pgen.1005121
- Hu S, Li J, Xu F, Mei S, Le Duff Y, Yin L, Pang X, Cen S, Jin Q, Liang C, et al. SAMHD1 Inhibits LINE-1 Retrotransposition by Promoting Stress Granule Formation. PLoS Genet 2015; 11:e1005367; PMID:26134849; http://dx.doi.org/10.1371/journal.pgen.1005367
- Zhao K, Du J, Han X, Goodier JL, Li P, Zhou X, Wei W, Evans SL, Li L, Zhang W, et al. Modulation of LINE-1 and Alu/SVA retrotransposition by Aicardi-Goutieres syndrome-related SAMHD1. Cell Rep 2013; 4:1108-15; PMID:24035396; http://dx.doi.org/10.1016/j.celrep.2013.08.019
- Goldstone DC, Ennis-Adeniran V, Hedden JJ, Groom HC, Rice GI, Christodoulou E, Walker PA, Kelly G, Haire LF, Yap MW, et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 2011; 480:379-82; PMID:22056990; http://dx.doi.org/10.1038/nature10623
- Powell RD, Holland PJ, Hollis T, Perrino FW. Aicardi-Goutieres syndrome gene and HIV-1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase. J Biol Chem 2011; 286:43596-600; PMID:22069334; http://dx.doi.org/10.1074/jbc.C111.317628
- White TE, Brandariz-Nunez A, Valle-Casuso JC, Amie S, Nguyen LA, Kim B, Tuzova M, Diaz-Griffero F. The retroviral restriction ability of SAMHD1, but not its deoxynucleotide triphosphohydrolase activity, is regulated by phosphorylation. Cell Host Microbe 2013; 13:441-51; PMID:23601106; http://dx.doi.org/10.1016/j.chom.2013.03.005
- Cribier A, Descours B, Valadao AL, Laguette N, Benkirane M. Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell Rep 2013; 3:1036-43; PMID:23602554; http://dx.doi.org/10.1016/j.celrep.2013.03.017
- Zhu C, Gao W, Zhao K, Qin X, Zhang Y, Peng X, Zhang L, Dong Y, Zhang W, Li P, et al. Structural insight into dGTP-dependent activation of tetrameric SAMHD1 deoxynucleoside triphosphate triphosphohydrolase. Nat Commun 2013; 4:2722; PMID:24217394
- Anderson P, Kedersha N. Stress granules. Curr Biol 2009; 19:R397-8; PMID:19467203; http://dx.doi.org/10.1016/j.cub.2009.03.013
- Balagopal V, Parker R. Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Curr Opin Cell Biol 2009; 21:403-8; PMID:19394210; http://dx.doi.org/10.1016/j.ceb.2009.03.005
- Goodier JL, Mandal PK, Zhang L, Kazazian HH, Jr. Discrete subcellular partitioning of human retrotransposon RNAs despite a common mechanism of genome insertion. Hum Mol Genet 2010; 19:1712-25; PMID:20147320; http://dx.doi.org/10.1093/hmg/ddq048
- Goodier JL, Zhang L, Vetter MR, Kazazian HH, Jr. LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol Cell Biol 2007; 27:6469-83; PMID:17562864; http://dx.doi.org/10.1128/MCB.00332-07
- Alisch RS, Garcia-Perez JL, Muotri AR, Gage FH, Moran JV. Unconventional translation of mammalian LINE-1 retrotransposons. Genes Dev 2006; 20:210-24; PMID:16418485; http://dx.doi.org/10.1101/gad.1380406
- Ariumi Y, Kuroki M, Kushima Y, Osugi K, Hijikata M, Maki M, Ikeda M, Kato N. Hepatitis C virus hijacks P-body and stress granule components around lipid droplets. J Virol 2011; 85:6882-92; PMID:21543503; http://dx.doi.org/10.1128/JVI.02418-10
- Reineke LC, Lloyd RE. Diversion of stress granules and P-bodies during viral infection. Virology 2013; 436:255-67; PMID:23290869; http://dx.doi.org/10.1016/j.virol.2012.11.017
- Cristea IM, Rozjabek H, Molloy KR, Karki S, White LL, Rice CM, Rout MP, Chait BT, MacDonald MR. Host factors associated with the Sindbis virus RNA-dependent RNA polymerase: role for G3BP1 and G3BP2 in virus replication. J Virol 2010; 84:6720-32; PMID:20392851; http://dx.doi.org/10.1128/JVI.01983-09
- Pager CT, Schutz S, Abraham TM, Luo G, Sarnow P. Modulation of hepatitis C virus RNA abundance and virus release by dispersion of processing bodies and enrichment of stress granules. Virology 2013; 435:472-84; PMID:23141719; http://dx.doi.org/10.1016/j.virol.2012.10.027
- Reineke LC, Lloyd RE. The stress granule protein G3BP1 recruits protein kinase R to promote multiple innate immune antiviral responses. J Virol 2015; 89:2575-89; PMID:25520508; http://dx.doi.org/10.1128/JVI.02791-14
- Ng CS, Jogi M, Yoo JS, Onomoto K, Koike S, Iwasaki T, Yoneyama M, Kato H, Fujita T. Encephalomyocarditis virus disrupts stress granules, the critical platform for triggering antiviral innate immune responses. J Virol 2013; 87:9511-22; PMID:23785203; http://dx.doi.org/10.1128/JVI.03248-12
- Verkaik NS, Persengiev S. Induction of Stress Granule Assembly is Essential for the Orchestration of DNA Damage Response. Nature Precedings 2008; 1591:1–23
- Clifford R, Louis T, Robbe P, Ackroyd S, Burns A, Timbs AT, Wright Colopy G, Dreau H, Sigaux F, Judde JG, et al. SAMHD1 is mutated recurrently in chronic lymphocytic leukemia and is involved in response to DNA damage. Blood 2014; 123:1021-31; PMID:24335234; http://dx.doi.org/10.1182/blood-2013-04-490847
- Kretschmer S, Wolf C, Konig N, Staroske W, Guck J, Hausler M, Luksch H, Nguyen LA, Kim B, Alexopoulou D, et al. SAMHD1 prevents autoimmunity by maintaining genome stability. Ann Rheum Dis 2015; 74:e17; PMID:24445253; http://dx.doi.org/10.1136/annrheumdis-2013-204845
