ABSTRACT
Ty1 is a long terminal repeat (LTR) retrotransposon belonging to the Ty1/copia family and is present in up to 32 full-length copies in Saccharomyces. Like retroviruses, Ty1 contains GAG and POL genes, LTRs, and replicates via an RNA intermediate within a virus-like particle (VLP). Although Ty1 retrotransposition is not infectious, uncontrolled replication can lead to detrimental effects on the host genome, including insertional mutagenesis and chromosomal rearrangements. Ty1 copy number control (CNC) limits replication and is mediated through a self-encoded protein called p22. p22 is translated from a subgenomic Ty1 RNA and encodes an amino-truncated version of the Gag protein. We highlight a recent study identifying Ty1 Gag, which comprises the VLP capsid and provides nucleic acid chaperone functions, as a direct target of p22-mediated inhibition. CNC-resistant (CNCR) mutations map within predicted helical domains of Gag, including those in the Ty1/copia pfam domain Retrotran_gag_2 (formerly UBN2) and a central region we refer to as the CNCR domain. CNCR Gag forms VLPs that exclude p22, thus restoring Ty1 replication. We discuss possible mechanisms for p22 inclusion in Ty1 VLPs and compare Ty1 CNC with retroviral restriction factors targeting capsid (CA).
The Ty1 element of Saccharomyces is a well-studied member of the widely dispersed Ty1/copia class of long-terminal repeat (LTR) retrotransposons.Citation1 An active Ty1 element is about 5.9 kb in length and consists of 2 overlapping open reading frames, GAG and POL, which are bracketed by LTRs. The Ty1 life cycle resembles retroviral replication, as the element produces a near full-length mRNA that serves as both a template for translation and reverse transcription (). Ty1 protein products Gag and Gag-Pol are produced in a ˜20:1 ratio, respectively, due to a +1 ribosomal frameshifting event that is required to express POL.Citation2 Gag is the capsid protein that forms the virus-like particle (VLP) and provides nucleic acid chaperone (NAC) functions. Gag-Pol also assembles into the VLP, where it is cleaved into mature Gag, protease, integrase, and reverse transcriptase. Unlike retroviruses, Ty1 retrotransposition is not infectious and VLPs remain intracellular. Instead, packaged Ty1 mRNA is reverse transcribed within a mature VLP and the cDNA is integrated back into the host genome from which it originated. A comparison between Ty1 replication and the stages of retroviral infection is shown in . Early stages of retroviral infection occur post-entry into a new host cell and include capsid uncoating, reverse transcription, and integration. During the late stages of retroviral infection, viral RNA and proteins are expressed from the genome-integrated retroviral sequence called a provirus and assembled into viral particles in the cytoplasm or at the plasma membrane prior to budding from the cell.
Figure 1. Comparison of Ty1 versus retrovirus replication and restriction. (A) Ty1 replication begins with transcription of the element in the host genome. Ty1 mRNA is exported from the nucleus and is translated to form Ty1 proteins, Gag and Gag-Pol. Ty1 mRNA and proteins form cytoplasmic foci called retrosomes, in which Gag multimerization and VLP assembly occur. These stages are similar to the late stages of retroviral infection and are most sensitive to restriction by p22. After assembly, Ty1 VLPs undergo maturation via Ty1 protease. Packaged Ty1 mRNA is reverse transcribed to form Ty1 cDNA. Ty1 cDNA and at least Ty1 integrase form the preintegration complex (PIC). The Ty1 PIC is imported into the nucleus via a nuclear localization signal within integrase and is integrated into the host genome. (B) Late stage replication begins when an integrated provirus is transcribed and viral RNA is translated into viral proteins. Immature virus particles can assemble in the cytoplasm or at the plasma membrane before budding out of the cell. The sheep restriction factor enJS56A1 acts in the late stage, where it blocks effective trafficking of JSRV viral particles to the plasma membrane. Virus particle maturation occurs extracellularly and early stage infection begins with binding and entry of the mature virion into the host cell. The capsid core is released into the cytoplasm and undergoes uncoating and reverse transcription. The PIC is formed prior to nuclear entry and integration. Several known restriction factors block early stage retroviral replication by binding to the incoming capsid core. TRIM5α inhibits uncoating prior to reverse transcription, while Mx2/MxB and Fv1 inhibit a step post reverse transcription but prior to integration.
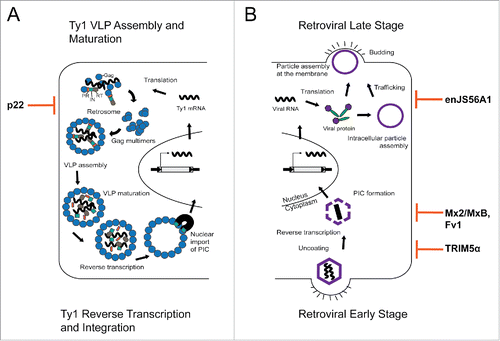
The replicative nature of the Ty1 replication cycle can cause problems for its yeast host, because integration of Ty1 elements in or near host genes can modulate their expression or cause genomic instability.Citation3-6 Rampant Ty1 retrotransposition is limited by an intrinsic system called copy number control (CNC), which is defined by a reduction in Ty1 mobility when Ty1 element copy number increases.Citation7,8 Recently, we discovered that Ty1 retrotransposition is restricted by a 22-kilodaltons protein (p22) encoded by internally initiated Ty1i transcripts.Citation9 Ty1i RNA expression is initiated ˜800 bp downstream of the full length Ty1 mRNA transcription start site and both full length Ty1 mRNA and Ty1i RNA likely utilize the same termination signals. Because the first AUG within Ty1i RNA is in the same reading frame as GAG, the protein sequence of p22 is identical to the C-terminal half of Gag and spans a portion of the Retrotran_gag_2 domain (PF14223; previously named UBN2) and includes the NAC region ().Citation10 We have shown that p22 binds Gag and disrupts the concentration of Ty1 mRNA and Gag protein in cytoplasmic foci termed retrosomes or T-bodies, which are the sites of VLP assembly (). p22 is present within higher order Gag complexes, including isolated VLPs.Citation9,11 Once associated with the VLPs, p22 is processed to p18 using the same Ty1 protease cleavage site found in full-length Gag (). Although p18 can bind Ty1 RNA and exhibits NAC activity to some extent, a derivative of p18 lacking NAC activity can still restrict Ty1 retrotransposition, likely due to its ability to inhibit the nucleic acid chaperone function of the Gag C-terminal region.Citation10 The presence of p22 in VLPs leads to aberrant VLPs that are defective for reverse transcription.Citation8,9
Figure 2. CNCR mutations map within specific domains of Ty1 Gag. Ty1 Gag sequence was analyzed for secondary structure prediction and domain identification. Two Pfam domains, TYA (PF01021, residues 17–114) and Retrotran_gag_2 (PF14223, residues 245–356), are present in Ty1 Gag. Gray boxes represent predicted helical regions. CNCR mutations clustered between residues 170–220, which we define as the CNCR domain, and within Retrotran_gag_2. Other features include the Ty1 p22 protein and the NAC region, the protease cleavage sites in Ty1 Gag and p22 proteins (scissors), and W184, which is a conserved residue found in Ty1/copia and is required for Ty1 retrotransposition.Citation11,15
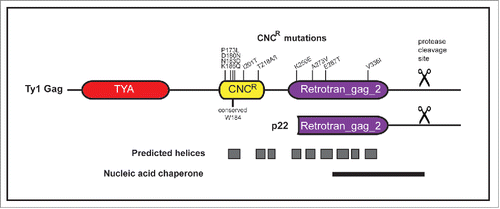
To understand the molecular mechanism of CNC, we searched for a target of p22 involved in Ty1 inhibition. Our approach included a forward genetic screen using randomly mutagenized Ty1 elements to select for resistance to CNC.Citation11 CNC-resistance (CNCR) mutations map within GAG (), suggesting Ty1 Gag is the target for p22 action. However, inferring a mechanism of resistance is complicated given that Ty1 Gag is not well characterized at a functional or structural level. Most of our knowledge of Gag structure comes from the study of VLPs isolated from yeast overexpressing Ty1.Citation12 Our efforts to purify and crystallize Gag and Gag segments have failed, likely due to the fact that recombinant Gag proteins require high salt concentrations to remain soluble (Nishida Y., unpublished data). Therefore, we took a bioinformatics approach to enhance our understanding of Ty1 Gag structural domains.
The majority of the N-terminal region of Gag, which contains the TYA pfam domain, is predicted to lack secondary structure (). Epitope mapping suggests that the N-terminal region of Gag faces the outside of the VLPsCitation13 and low resolution cryo-electron microscopy (38 Å) supports the view that multimerization of Gag trimeric clusters underlies VLP structure.Citation14 Current advanced cryo-electron tomography techniques will help elucidate the arrangement of Gag subunits, which is required for understanding both VLP assembly and restriction by p22 via Gag-binding. In comparison, the central and more C-terminal regions of Gag are predicted to contain 9 helical stretches, reminiscent of the high helical content of retroviral capsid (CA) proteins (). Strikingly, nearly all the CNCR mutations map within these predicted helical domains and cluster into 2 regions. The first cluster of mutations coincides within the CNCR domain, a central portion of Gag containing the first helix and a conserved tryptophan residue (W184A) present in diverse Ty1/copia Gag proteins.Citation15 The second cluster of mutations maps within the Retrotran_gag_2 domain of Ty1 Gag, which is a domain conserved in Ty1/copia type Gag proteins across Eukarya. Locating the Retrotran_gag_2 domain in Ty1 Gag required more sensitive profile-based search methods, but the discovery of this domain is intriguing. The Retrotran_gag_2 helices have been implicated in Ty1 VLP assembly,Citation16 therefore, this region likely plays a vital role in forming the contacts between Gag proteins within the VLP for many eukaryotic retrotransposons. This is in contrast to the TYA domain present in Ty1 Gag, whose presence seems to be limited to Saccharomyces. The region of Gag responsible for its NAC activity partially overlaps with the Retrotran_gag_2 domain ().Citation17 Very few CNCR mutations mapped within the NAC region, but it is interesting to consider that CNCR mutations in the NAC region might restore Ty1 Gag/mRNA interactions, such as RNA packaging.Citation10,18 This would be in contrast to characterized resistance mutations within the CNCR domain, which do not rescue Ty1 mRNA levels in VLPs. Citation11 In addition, CNCR mutations only partially rescue Ty1 retrotransposition frequency, highlighting the idea that p22 may disrupt multiple Gag-directed activities.
A striking phenotype observed with CNCR Ty1 elements is the decreased levels of mature restriction factor p18.Citation11 Although p18 levels are low, steady state levels of unprocessed restriction factor (p22) are similar between strains expressing wild type and CNCR elements, making a reduction of restriction factor synthesis less likely. The loss of p18 may result from reduced cleavage of p22, which could occur through 2 post-translational scenarios. Either p22 no longer assembles with CNCR VLPs, or p22 is not recognized by Ty1 protease inside CNCR VLPs. To distinguish between these 2 possibilities, we compared the sedimentation profiles of p22/p18 and Gag by sucrose gradient analysis. In the presence of wild type Ty1 elements, p18 co-sediments with the higher molecular weight Gag complexes that likely contain VLPs. Remarkably, p22/p18 is not detected in the CNCR VLP fractions. Therefore, p22 may not gain access to Ty1 protease since it is not associated with assembled CNCR VLPs. It is logical to expect higher levels of immature p22 in CNCR strains if cleavage is inhibited. Perhaps the comparable levels of p22 in WT and CNCR strains indicates that VLP incorporation can protect p22 and/or p18 from cytoplasmic degradation, but this needs to be experimentally confirmed.
Although p22/p18 is excluded from CNCR VLPs, this is not due to extreme perturbation of binding between p22/p18 and CNCR Gag.Citation11 Using a tagged version of p22, we detect equivalent levels of wild type and CNCR Gag in coimmunoprecipitated material. To understand how p22 binds CNCR Gag, but is not coassembled or packaged within CNCR VLPs will require additional studies. Perhaps p22 preferentially binds to specific Gag multimerization states during assembly. There is precedence for this in the case of Gag-binding retroviral restriction factors. For example, early stage restriction factors such as Fv1, which is derived from an ancient retroviral GAG gene, Mx2/MxB, and TRIM5α bind the incoming capsid core ().Citation19,20 Interaction between these restriction factors and capsid in vitro requires capsid in a polymerized state, which more closely resembles an intact core.Citation19,21,22 Also, a Gag-like sequence called enJS56A1 was co-opted to restrict infection by the oncogenic retrovirus JSRV in sheep.Citation23 enJS56A1 acts in the late stages of retroviral infection, after integration into the host genome when viral production is initiated (). This endogenous retroviral locus produces a Gag protein that has a trans-dominant effect on JSRV replication. enJS56A1 Gag is thought to interact with JSRV Gag, disrupting its normal trafficking through the cell and resulting in its degradation. Because we know that p22 disrupts retrosome formation and limits VLP production, we favor the idea that p22 is more potent in the stages just prior to VLP assembly, which is equivalent to late stage restriction factors (). Furthermore, p22 probably interacts with Gag in multiple ways. If p22 binds monomeric Gag, then it could disrupt Gag trimer formation, assuming trimer assembly occurs as predicted. If p22 binds trimeric Gag, then it could be inhibiting the formation of higher multimerization states of Gag. It is also possible that p22 only interacts with fully assembled VLPs. These possibilities should to be carefully addressed to understand how p22/p18 interferes with Ty1 replication.
Conclusion
Our study of Ty1 CNC by the self-encoded restriction factor p22 revealed mutations in Gag that increase retrotransposition levels. We have examined Ty1 Gag sequence using bioinformatics approaches and characterized resistance mutations in the CNCR and Retrotran_gag_2 domains of Ty1 Gag. Considering that the Gag Retrotran_gag_2 domain is conserved throughout the Ty1/copia superfamily, further study of this region will enhance our understanding of retrotransposon Gag structure and function. In addition, analyzing how p22 interacts with Ty1 Gag is key to revealing the structural and functional basis of CNC in Saccharomyces as well as other restriction mechanisms that involve Gag-binding.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Natarajan Kannan for help with the bioinformatic analyses.
Funding
This work was supported by the National Institutes of Health, Grant: GM095622 (DJG), and the National Science Foundation Graduate Fellowship: 1011RH25213 (JMT).
References
- Curcio MJ, Lutz S, Lesage P. The Ty1 LTR-Retrotransposon of Budding Yeast, Saccharomyces cerevisiae. Microbiol Spectr 2015; 3:MDNA3-0053-2014; PMID:26104690; http://dx.doi.org/10.1128/microbiolspec.MDNA3-0053-2014
- Farabaugh PJ. Post-transcriptional regulation of transposition by Ty retrotransposons of Saccharomyces cerevisiae. J Biol Chem 1995; 270:10361-4; PMID:7737964; http://dx.doi.org/10.1074/jbc.270.18.10361
- Maxwell PH, Burhans WC, Curcio MJ. Retrotransposition is associated with genome instability during chronological aging. Proc Natl Acad Sci U S A 2011; 108:20376-81; PMID:22021441; http://dx.doi.org/10.1073/pnas.1100271108
- Patterson MN, Scannapieco AE, Au PH, Dorsey S, Royer CA, Maxwell PH. Preferential retrotransposition in aging yeast mother cells is correlated with increased genome instability. DNA Repair (Amst) 2015; 34:18-27; PMID:26298836; http://dx.doi.org/10.1016/j.dnarep.2015.07.004
- Chan JE, Kolodner RD. A genetic and structural study of genome rearrangements mediated by high copy repeat Ty1 elements. PLoS Genet 2011; 7:e1002089; PMID:21637792; http://dx.doi.org/10.1371/journal.pgen.1002089
- Garfinkel DJ. Genome evolution mediated by Ty elements in Saccharomyces. Cytogenet Genome Res 2005; 110:63-9; PMID:16093659; http://dx.doi.org/10.1159/000084939
- Garfinkel DJ, Nyswaner K, Wang J, Cho JY. Post-transcriptional cosuppression of Ty1 retrotransposition. Genetics 2003; 165:83-99; PMID:14504219; http://dx.doi.org/10.1534/genetics.107.082602
- Matsuda E, Garfinkel DJ. Posttranslational interference of Ty1 retrotransposition by antisense RNAs. Proc Natl Acad Sci U S A 2009; 106:15657-62; PMID:19721006; http://dx.doi.org/10.1073/pnas.0908305106
- Saha A, Mitchell JA, Nishida Y, Hildreth JE, Ariberre JA, Gilbert WV, Garfinkel DJ. A trans-dominant form of Gag restricts Ty1 retrotransposition and mediates copy number control. J Virol 2015; 89(7):3922-38; PMID:25609815; http://dx.doi.org/10.1128/JVI.03060-14
- Nishida Y, Pachulska-Wieczorek K, Blaszczyk L, Saha A, Gumna J, Garfinkel DJ, Purzycka KJ. Ty1 retrovirus-like element Gag contains overlapping restriction factor and nucleic acid chaperone functions. Nucleic Acids Res 2015; 43:7414-31; PMID:26160887; http://dx.doi.org/10.1093/nar/gkv695
- Tucker JM, Larango ME, Wachsmuth LP, Kannan N, Garfinkel DJ. The Ty1 Retrotransposon Restriction Factor p22 Targets Gag. PLoS Genet 2015; 11:e1005571; PMID:26451601; http://dx.doi.org/10.1371/journal.pgen.1005571
- Roth J-F. The Ty virus-like particles. Yeast 2000; 16:785-95; PMID:10861903; http://dx.doi.org/10.1002/1097-0061(20000630)16:9%3c785::AID-YEA550%3e3.0.CO;2-L
- Brookman JL, Stott AJ, Cheeseman PJ, Burns NR, Adams SE, Kingsman AJ, Gull K. An immunological analysis of Ty1 virus-like particle structure. Virology 1995; 207:59-67; PMID:7532885; http://dx.doi.org/10.1006/viro.1995.1051
- Al-Khayat HA, Bhella D, Kenney JM, Roth JF, Kingsman AJ, Martin-Rendon E, Saibil HR. Yeast Ty retrotransposons assemble into virus-like particles whose T-numbers depend on the C-terminal length of the capsid protein. J Mol Biol 1999; 292:65-73; PMID:10493857; http://dx.doi.org/10.1006/jmbi.1999.3055
- Peterson-Burch BD, Voytas DF. Genes of the Pseudoviridae (Ty1/copia retrotransposons). Mol Biol Evol 2002; 19:1832-45; PMID:12411593; http://dx.doi.org/10.1093/oxfordjournals.molbev.a004008
- Martin-Rendon E, Marfany G, Wilson S, Ferguson DJ, Kingsman SM, Kingsman AJ. Structural determinants within the subunit protein of Ty1 virus-like particles. Mol Microbiol 1996; 22:667-79; PMID:8951814; http://dx.doi.org/10.1046/j.1365-2958.1996.d01-1716.x
- Cristofari G, Ficheux D, Darlix JL. The GAG-like protein of the yeast Ty1 retrotransposon contains a nucleic acid chaperone domain analogous to retroviral nucleocapsid proteins. J Biol Chem 2000; 275:19210-7; PMID:10766747; http://dx.doi.org/10.1074/jbc.M001371200
- Purzycka KJ, Legiewicz M, Matsuda E, Eizentstat LD, Lusvarghi S, Saha A, Le Grice SF, Garfinkel DJ. Exploring Ty1 retrotransposon RNA structure within virus-like particles. Nucleic Acids Res 2013; 41:463-73; PMID:23093595; http://dx.doi.org/10.1093/nar/gks983
- Fricke T, White TE, Schulte B, de Souza Aranha Vieira DA, Dharan A, Campbell EM, Brandariz-Nunez A, Diaz-Griffero F. MxB binds to the HIV-1 core and prevents the uncoating process of HIV-1. Retrovirology 2014; 11:68; PMID:25123063; http://dx.doi.org/10.1186/s12977-014-0068-x
- Sanz-Ramos M, Stoye JP. Capsid-binding retrovirus restriction factors: discovery, restriction specificity and implications for the development of novel therapeutics. J Gen Virol 2013; 94:2587-98; PMID:24026671; http://dx.doi.org/10.1099/vir.0.058180-0
- Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H, Diaz-Griffero F, Anderson DJ, Sundquist WI, Sodroski J. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci U S A 2006; 103:5514-9; PMID:16540544; http://dx.doi.org/10.1073/pnas.0509996103
- Hilditch L, Matadeen R, Goldstone DC, Rosenthal PB, Taylor IA, Stoye JP. Ordered assembly of murine leukemia virus capsid protein on lipid nanotubes directs specific binding by the restriction factor, Fv1. Proc Natl Acad Sci U S A 2011; 108:5771-6; PMID:21436027; http://dx.doi.org/10.1073/pnas.1100118108
- Murcia PR, Arnaud F, Palmarini M. The transdominant endogenous retrovirus enJS56A1 associates with and blocks intracellular trafficking of Jaagsiekte sheep retrovirus Gag. Journal of virology 2007; 81:1762-72; PMID:17135320; http://dx.doi.org/10.1128/JVI.01859-06
