ABSTRACT
The horizontal gene transfer facilitated by mobile genetic elements impacts almost all areas of bacterial evolution, including the accretion and dissemination of antimicrobial-resistance genes in the human and animal pathogen Staphylococcus aureus. Genome surveys of staphylococcal plasmids have revealed an unexpected paucity of conjugation and mobilization loci, perhaps suggesting that conjugation plays only a minor role in the evolution of this genus. In this letter we present the DNA sequences of historically documented staphylococcal conjugative plasmids and highlight that at least 3 distinct and widely distributed families of conjugative plasmids currently contribute to the dissemination of antimicrobial resistance in Staphylococcus. We also review the recently documented “relaxase-in trans” mechanism of conjugative mobilization facilitated by conjugative plasmids pWBG749 and pSK41, and discuss how this may facilitate the horizontal transmission of around 90% of plasmids that were previously considered non-mobilizable. Finally, we enumerate unique sequenced S. aureus plasmids with a potential mechanism of mobilization and predict that at least 80% of all non-conjugative S. aureus plasmids are mobilizable by at least one mechanism. We suggest that a greater research focus on the molecular biology of conjugation is essential if we are to recognize gene-transfer mechanisms from our increasingly in silico analyses.
Text
Conjugation and mobilization
Conjugation is a highly evolved and efficient mechanism facilitating DNA transfer in bacteria, therefore the characterization of DNA sequences, genes and proteins involved in conjugation is crucial if we are to accurately appraise the potential for gene transfer from bioinformatics analyses. Several bacterial genome surveys have revealed an unexpected paucity of recognized conjugation loci in many bacterial genera.Citation1,2 In staphylococci, only an estimated 5% of plasmids carry conjugation-gene clusters required for autonomous conjugative transfer. In contrast, the evidence for horizontal transfer of staphylococcal plasmids between distinct lineages is abundant, leading some to propose that bacteriophage transduction may account for much of this transfer.Citation3-5 However, several new mechanisms of conjugative mobilization have recently been elucidated, which we believe may resolve the paradoxical underrepresentation of conjugative plasmids in staphylococci.Citation6-8
Autonomously-transferring conjugative plasmids carry both mating-pore genes and genes for DNA processing, single-stranded DNA (ssDNA) replication and recruitment of ssDNA to the mating pore. DNA is recruited to the mating pore by the relaxase protein, which binds, cleaves and covalently attaches to a recognition sequence called the origin-of-transfer (oriT), forming (often with accessory proteins) a nucleoprotein complex referred to as the relaxasome.Citation9 The relaxasome is recruited to the mating-pore through interactions with a mating-pore component called the VirD4 coupling protein, after which it is transferred to recipient cells through a type-IV secretion system (). Relaxases can additionally be involved in rolling-circle-like plasmid replication in the donor bacterium and recircularization and replication of plasmid DNA in the recipient. Plasmid conjugation systems can therefore be considered an evolutionary amalgamation of type IV protein secretion systems that have evolved to transfer protein-tethered DNA, with rolling-circle replicases (or recombinases)Citation10,11 that have evolved an association with the type IV secretion system through relaxase-coupling-protein interactions.Citation12
Figure 1. Mechanisms of conjugative mobilization in Staphylococci. The conjugative plasmid encodes all genes required for formation of the mating pore, as well as the coupling protein, DNA relaxase and an oriT. Mobilizable plasmids can exploit the conjugative-plasmid mating pore by either: (A) encoding a mimic sequence of the conjugative-plasmid oriT; (B) encoding a distinct relaxase (Mob) compatible with the conjugative plasmids coupling protein and its own cognate oriT or; (C) by carrying a replicative relaxase (Rep) compatible with the conjugative-plasmid coupling protein.
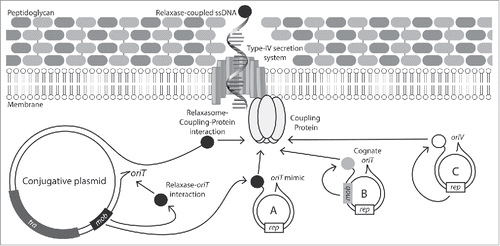
Mobilizable plasmids are those which carry DNA-transfer genes required for formation of all or part of the relaxasome, but lack genes required for mating pore formation. Mobilizable plasmids have an ability to exploit conjugative plasmids for horizontal dissemination, but are non-mobile in cells that lack mobile elements carrying compatible mating-pore genes. The vast majority of documented mobilizable plasmids exploit conjugative element mating-pores by encoding their own relaxase (Mob) that acts on the plasmid's cognate oriT ().Citation1
Three distinct families of staphylococcal conjugative plasmids
Staphylococcal isolates are frequent hosts to diverse antimicrobial-resistance plasmids, but until recently only one family of staphylococcal conjugative plasmids had been characterized at a molecular level. Closely related members of the pSK41/pGO1 plasmid family were first documented by several groups in the early 1980s as the basis of emergent gentamicin resistance.Citation13-16 In addition to aminoglycoside resistance, these plasmids have been found to variously confer resistance to penicillins, trimethoprim, bleomycin, tetracycline, antiseptics and disinfectants, mupirocin, and macrolides, lincosamides and streptogramin B. The resistance genes responsible for these phenotypes are usually encoded by small plasmids cointegrated between copies of IS257/IS431 within the pSK41/pGO1 plasmid.Citation17 Notably, plasmids of this type have subsequently been associated with linezolid and high-level vancomycin resistance.Citation18,19 pSK41/pGO1 family plasmids share a near-identical syntenic backbone that encodes resolution, partitioning and replication functions, and a cluster of approximately 15 transfer genes for proteins implicated in conjugation, including a predicted coupling protein and homologues of type-IV secretion system componentsCitation16,20 A conserved relaxase gene, nes, is located elsewhere within the plasmid backbone, adjacent to the oriT site at which its product nicks the plasmid DNA to initiate conjugative transfer.Citation21,22
The pSK41/pGO1 family conjugative plasmids mobilise some smaller plasmids (3.5–14.5 kb) when co-resident in the same cell. Mobilizable staphylococcal plasmids such as pSK639 and pC221 contain a DNA segment encoding the genes mobCAB.Citation23-25 The MobC protein binds to an adjacent cognate oriT sequence that is nicked by the MobA relaxase to initiate transfer.Citation26 Numerous other small plasmids (3.5–14.5 kb) instead encode a distinct locus implicated as another relaxase-in cis mobilization system. The pre gene and site, RSA, were originally characterized as a site-specific recombination function carried by staphylococcal plasmids.Citation27 However, this system has been implicated in mobilization of the S. aureus plasmid pUB110 in BacillusCitation28 and a homologous locus carried by the streptococcal plasmid pMV158 was subsequently shown to comprise a gene, mobM, encoding a relaxase that acts on a corresponding oriT site to facilitate mobilization of that plasmid.Citation29,30
More recently a second distinct family of staphylococcal conjugative plasmids was characterized. pWBG749 was found in a strain from a remote indigenous Australian community in 1995.Citation3,7,31 pWBG749-family conjugative plasmids carrying penicillin, aminoglycoside and vancomycin-resistance genes have since been identified.Citation8,32,33 The predicted proteins encoded by the pWBG749 conjugation-gene cluster are distinct from those of pSK41 and include a distinct putative relaxase SmpP and a distinct oriT sequence. Interestingly, the majority of pWBG749-like plasmids identified in sequence databases do not appear to carry antimicrobial-resistance loci, perhaps explaining why they have been largely overlooked.Citation8
In the 1980s at least 3 families of distinct staphylococcal conjugative plasmids were identified in the Grubb laboratory from S. aureus isolated from Australia, Africa and Asia, and at least 2 of these plasmid families were clearly distinct from pSK41-family plasmids based on their restriction and incompatibility profiles. Conjugative plasmid pWBG637, was originally isolated in 1985 from a Nigerian hospital S. aureus isolate and subsequently identified in several other hospital-associated strains with distinct chromosomal lineages.Citation34-36 Like pWBG749, pWBG637 does not carry resistance genes.Citation37,38 pWBG637 is capable of conjugation to other S. aureus, S. epidermidis and Enterococcus faecalis strains and is able to mobilise the conjugative transfer of several coresident antimicrobial-resistance plasmids.Citation34,39 In work presented here, we sequenced pWBG637 to further clarify its relatedness to other conjugative plasmids. BLASTN alignments of the pWBG637 sequence (KX086582) revealed it shared 99% nucleotide identity with pWBG749 over 91% of its 38 kb length. Therefore, while members of the pWBG749-family of conjugative plasmids have only recently been sequenced, they have been documented as a distinct family of staphylococcal conjugative plasmids capable of mobilising other antimicrobial-resistance plasmids since the 1980s.
A third distinct family of “Diffusible pigment” conjugative plasmids, which we have designated the pWBG4 family, were first identified as a third unique type of conjugative plasmid in 1985 by Townsend et al. The first of these, pWBG14,Citation40 confers aminoglycoside, macrolide, lincosamide and spectinomycin resistance and was identified in a strain originally isolated from Royal Perth Hospital in 1968. Conjugative transfer of related plasmids pWBG4 and pWBG25 was subsequently demonstrated.Citation41,42 A pWBG4-family plasmid conferring trimethoprim resistance, pWBG707, was identified in a Malaysian isolate in 1992.Citation43 In work presented here, we sequenced pWBG4 (KX149096) and pWBG707 (KX149097). Bioinformatic analyses of these sequences revealed pWBG4 and pWBG707 shared a 19-KB gene cluster of 23 co-oriented open-reading frames encoding predicted products related to VirD2, VirB2 and proteins of various other gram-positive conjugative elements () (also noted by Shore et al.).Citation44 Interestingly, pWBG4 was found to be almost identical to pSK73 (GQ915269.1), a recently-sequenced plasmid carried by a strain originally isolated in Melbourne in 1966.
Figure 2. pWBG4, a third family of staphylococcal conjugative plasmids. The internal circle represents the gene map of pWBG4, showing the positions and predicted products of the putative pWBG4 conjugation cluster detA-detV (white arrows with black outlines) and other open-reading frames. The outer circles represent ungapped circular BLASTN alignments of pWBG4-family plasmids, created using BRIG software.Citation63
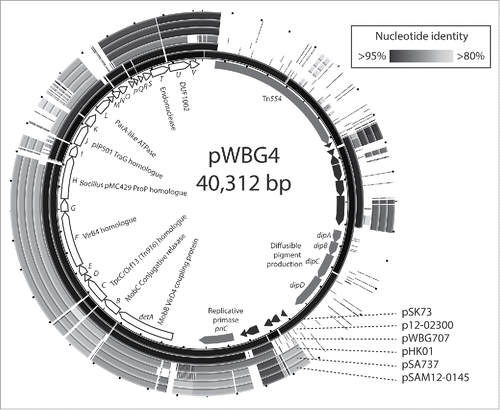
The putative pWBG4 conjugation gene cluster, which we here name detA-detV, is clearly distinct from that of pSK41 and pWBG749. Analysis of DetB using CONJscan,Citation45 placed DetB in the MobC-family of conjugative relaxases. Furthermore, DetA encoded a putative VirD4/MobB coupling protein, thus the detA-detB region shared the same Mob-gene arrangement as mobBC on the mobilizable plasmids ClodDF13 (of Escherichia coli CloDF13) and pUA140 (of Streptococcus mutans) and the conjugative Yersinia enterocolitica plasmid p29930.Citation46-48 The detA-detV region was additionally identified on several contemporary staphylococcal plasmids associated with the dissemination of cfr-gene-encoded linezolid resistance in both human and animal-isolated staphylococci. The pWBG4-family plasmid pSA737, was found in the first American example of cfr-mediated linezolid resistance in a human S. aureus isolateCitation49 and was subsequently identified in 19 isolates from 2 Ohio hospitals.Citation50 pWBG4-family cfr-carrying plasmids have now been identified in China (pHK01),Citation51 Germany (p12-02300),Citation18 and Ireland (pSAM12-0145).Citation44 Despite the wide distribution of pWBG4-like plasmids, the clinical importance of linezolid and the conspicuousness of the detA-detV conjugation gene cluster, only one of these studies reported laboratory conjugation experiments confirming that the cfr-carrying pWBG4-like plasmid was conjugative.Citation44
In summary, it is clear that there are at least 3 distinct families of conjugative plasmids (based on their distinct conjugation-gene clusters) in staphylococci. Members of each of the pSK41, pWBG749 and pWBG4 families of conjugative plasmids can be identified in extant and historical staphylococcal lineages and each is currently contributing to the horizontal spread of resistance mechanisms against last-resort antimicrobials such as vancomycin and linezolid.
Relaxase-in trans mobilization
There is also a relative paucity of Mob-gene loci on non-conjugative plasmids in Staphylococcus aureus.Citation3,4 Non-conjugative staphylococcal plasmids range from small (<5 kb) rolling-circle plasmids that often only carry genes for replication and a single antimicrobial resistance, to larger (up to ∼65 kb) plasmids which carry multiple resistances. A genome survey of staphylococcal plasmids isolated since the 1940s revealed that most staphylococci carry at least one plasmid > 20kb. But, the 3 most common large plasmid families, pMW2, pIB485 and pUSA300HOUMR, which represent 43 % of all plasmids >20 kb identified, lack conjugation or mobilization genes.Citation3
A surprising observation from conjugation experiments with pWBG749, is that it is able to mobilise several large multiresistance plasmids that lack Mob-gene loci, including a member of the pIB485 family. In our recent Nucleic Acids Research article, we demonstrate that the mechanism by which pWBG749 mobilises non-conjugative plasmids is distinct from that of most previously described systems.Citation8 Plasmids mobilised by pWBG749 carry sequences that mimic the oriT of pWBG749, indicating that the putative relaxase of pWBG749, SmpP, recognizes these oriT mimics and recruits the mobilizable plasmid DNA to the pWBG749-produced mating pore. Each pWBG749-family oriT mimic, minimally contains a 126-bp region encompassing three overlapping inverted-repeat sequences (IR1-3), at least one copy of an accessory repeat (AR) and a defined relaxase core region ().Citation8 The relaxase-in trans mechanism is not technically novel, but likely because it has rarely been recognized in nature, like pWBG749, it has been overlooked. What is most astounding from our analyses is the extensive distribution of oriT sequences on staphylococcal plasmids. Over 50% of non-identical sequenced plasmids carry 1–3 copies of a pWBG749-like oriT mimic ().Citation8
Figure 3. The diversity of oriT mimics on large and small resistance plasmids in staphylococci. (A) The plasmid map of the rolling-circle plasmid pNewbould305, illustrates the presence of 3 potential mobilization mechanisms. The repB gene of pNewbould305 shares 56% amino-acid identity over 96% of its length with the pBS42/pUB110 RepB protein, which enables mobilization by ICEBs1-family elements. Downstream of the repB gene is an oriT mimic sequence of the pWBG749-family, subfamily OT45 and a pSK41-like oriT mimic sequence. (B) The atypically large staphylococcal plasmid pWBG762, carries 4 oriT mimics. Three are of the pWBG749 family and one is of the pSK41 family. (C) Alignment of the pWBG749-family oriT mimic sequences carried by pNewbould305 and pWBG762 below the pWBG749 oriT region, illustrating IR2 sequence divergence. Conserved nucleotides are shaded. The AR1-AR3 repeats of the full oriT required for mobilization by pWBG749Citation8 have been truncated in this figure for clarity (D) Alignment of the pSK41-like oriT mimic sequences from pNewbould305 and pWBG762, below the pSK41 oriT region, showing divergence of the IR sequence; the Nes relaxase nick site is denoted by a vertical arrowhead.
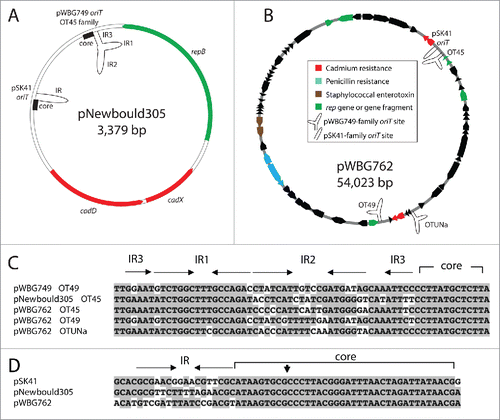
Five variants of the pWBG749-family oriT sequence have been identified, named OT49, OT45, OTUNa, OTSep and OT408. Each variant differs in a predicted inverted-repeat sequence 2 (IR2) (). Putative pWBG749-family conjugative plasmids, individually carrying each of the five distinct oriT variants, are represented in DNA sequence databases.Citation8 The divergence of the oriT sequences suggests that each pWBG749-family is likely specific for a single oriT variant. Consistent with this hypothesis, pWBG749, which carries an OT49 oriT, is not able to mobilise pMW2, which carries an OT45 sequence, nor can it mobilise an OT45 oriT sequence cloned from the conjugative plasmid pWBG745. A putative ribbon-helix-helix DNA binding protein, SmpO, encoded by all pWBG749-family plasmids, appears to dictate the specificity of mobilization for these oriT sequences. Cloning of the pWBG745-encoded SmpO sequence along with the OT45 oriT, enables OT45-oriT mobilization by pWBG749. This suggests that the pWBG749 relaxase SmpP, is able to interact with both OT49 and OT45 oriT variants, but the presence of the oriT-specific SmpO protein is required for efficient mobilization. The reasons for the divergence of conjugative-plasmid oriTs and oriT mimic sequences is not clear, but it could signal an underlying frequency-dependent selection against the most common oriT variants, perhaps due to a reduction in conjugative-plasmid fitness resulting from mobilizable-plasmid exploitation.
Coincident with the discovery of the relaxase-in trans mechanism of mobilization based on the carriage of pWBG749-like oriT mimics, sequences resembling the pSK41 oriT were similarly detected on a wide range of non-conjugative staphylococcal plasmids, recently reported in our Journal of Bacteriology article.Citation52 These pSK41-like oriT mimics possess a core sequence identical to the conjugative oriT sequence, which includes the Nes nick site. However, they differ in an adjacent inverted repeat that forms a DNA hairpin important for interactions with the Nes relaxase (). Nonetheless, purified pSK41 Nes binds and processes 2 different oriT mimics in vitro, albeit at lower efficiency than its cognate oriT sequence. Furthermore, pSK41 mobilises a recombinant plasmid carrying a copy of the pSK41 oriT, suggesting that pSK41-like conjugative plasmids are also capable of performing the relaxase-in trans mechanism of mobilization. However, pSK41 did not mobilise recombinant plasmids containing either of the 2 oriT mimics that had been shown to be substrates in vitro. It would seem likely that nucleotide differences in the inverted repeat flanking the pSK41-like oriT mimics confer specificity on the mobilization process, paralleling the variant specificity described above for pWBG749 relaxase-in trans mobilization. The involvement of accessory proteins, analogous to pWBG749 SmpO, in pSK41 relaxasome formation is currently under investigation. Four pSK41 mimic sequence types were identified on 83 non-conjugative staphylococcal plasmids, but notably none were identical to the oriT sequence found on all sequenced pSK41/pGO1 family conjugative plasmids.Citation52 Taken together, these observations raise the provocative possibility that many characterized plasmids carry oriT mimics specific for divergent pSK41-like conjugative plasmids that are yet to be detected.
What proportion of non-conjugative staphylococcal plasmids are potentially mobilizable?
The discovery of the relaxase-in trans mechanism of mobilization greatly increases the number of non-conjugative plasmids in S. aureus that can be considered to be potentially mobilizable, as illustrated by our previous analysis of a database of 280 non-identical S. aureus plasmids collected from NCBI.Citation8,52 Only 18 (6 %) of these plasmids carry a relaxase gene resembling that of either pWBG749 (SmpP; n = 4) or pSK41 (Nes; n = 14). Examination here of the remaining 262 non-conjugative plasmids (Table S1) for the mobilization relaxase genes mobA and pre, and the recently identified oriT mimic sequences (pWBG749-family and pSK41-family) revealed a remarkable stratification in their distribution with respect to plasmid size. mobA or pre genes were found almost exclusively on plasmids in the 3.5–14.5 kb size range. 74% (n = 49) of plasmids in this size-range encode one of these genes (but never both). No smaller plasmids carried these genes, and for plasmids over 14.5 kb (n = 135), only 2% (n = 3) contained mobA or pre (likely pseudogenes were not included); these likely represent small plasmid cointegrates present in larger chimeric plasmids. Notably, 89% (n = 120) of plasmids over 14.5 kb, which includes many multiresistance and pathogenicity plasmids, contained at least one pWBG749-family (87%) or pSK41-family (38 %) oriT mimic sequence. Equally strikingly, only a single plasmid in the 3.5–14.5 kb size range contained an oriT mimic, and that plasmid didn't have mobA or pre. Thus, mimics appear to be remarkably under-represented in 3.5–14.5 kb plasmids, which are typically mobilizable by virtue of either a mobA or pre system with a cognate oriT. In contrast, mimics were carried by 39 % of plasmids less than 3.5 kb (n = 65), but none were found on plasmids less than 2.5 kb (n = 33).
Of 262 non-conjugative S. aureus plasmids (Table S1), 56 % (n = 146) were found to carry at least one oriT mimic, with pWBG749-family mimics present on 53 % (n = 138) and pSK41 mimics evident in 23 % (n = 61); many plasmids possessed both mimic types (20 %; n = 53). Some plasmids, such as pWBG762, carried 3 copies of pWBG749-family mimics as well as a single pSK41-family mimic. Perhaps most significantly, whereas very few large (>14.5 kb) multiresistance/pathogenicity plasmids were previously thought to be potentially mobilizable (i.e., encode mobA or pre), the detection of oriT mimics on 89% of these plasmids suggest nearly all these plasmids are potentially mobilizable by pWBG749 or pSK41-family plasmids through a relaxase-in trans mechanism. The prevalence and conservation of oriT mimics not only implies far greater potential for mobilization than previously anticipated, but also carries important implications for our understanding of the evolution of staphylococcal plasmids and their hosts. Specifically, plasmids carrying these mimics effectively bear the evolutionary fingerprints of conjugative plasmids and implicate conjugative mobilization as major evolutionary driver in staphylococci.
Overall, 74% of non-conjugative S. aureus plasmids may have the capacity to be mobilised by either a relaxase-in trans (56 %) or the classical relaxase-in cis (19%) mechanism. The estimate for potential mobilization may be higher still if we consider the recently discovered “replicative relaxase” mechanisms of mobilization. The Grossman laboratory recently described the mobilization of rolling-circle-replication plasmids pC194, pBS42 and pHP13, which lack genes encoding classical Mob proteins, by the ICEBs1 family of integrative and conjugative elements. This mobilization mechanism is dependent on the plasmid-encoded rolling-circle replication protein, suggesting that these Rep proteins interact directly with the VirD4 coupling protein of ICEBs1-family conjugation systems, recruiting ssDNA to the mating pore in a Mob-independent manner ().Citation6 ICEBs1 carries a conjugation gene cluster related to that of Tn916, a family of integrative and conjugative elements that are frequently carried by staphylococci.Citation53 BLASTP searches of non-conjugative plasmids revealed that 39% (n = 103) carried a Rep gene closely resembling that of pC194/pBS42/pHP13 (E = 10−30) (Table S1). If these plasmids are indeed mobilizable through a replicative relaxase mechanism, this would raise the proportion of non-conjugative S. aureus plasmids with potential for mobilization to 80% (n = 210). We predict that this proportion may approach 100% as further types of conjugative plasmids and new mechanisms of plasmid mobilization are discovered in coming years.
Conclusions
While only around 5–6% of S. aureus plasmids are conjugative, it appears that the majority of non-conjugative plasmids, including most large multiresistance plasmids, are potentially mobilizable. While the prevalence of conjugative plasmids in isolated S. aureus is low, the acquisition of Mob and oriT sequences by most non-conjugative plasmids evidences that conjugative mobilization is a frequent enough event that most S. aureus plasmids have evolved to take advantage of it. The presence of multiple oriT sites on numerous plasmids, suggests that previous and frequent exposure to variants of both pWBG749 and pSK41-family plasmids has facilitated the transfer of the most widely distributed large multiresistance plasmids such as pMW2, pIB485 and pUSA300HOUMR. The most disturbing consequence of this mobilization, from the perspective of antimicrobial-resistance, is that while horizontal transfer of pWBG749 or pSK41 is apparently rare, when it occurs, it may facilitate the transfer of any mobilizable plasmid present in the same cell. If documented vancomycin-resistance gene carrying derivatives of pSK41Citation22 and pWBG749Citation32 were to become endemic in S. aureus populations for example, we might expect gene transfer rates for all compatible mobilizable plasmids to increase as well, along with their resistance and virulence-gene cargo. Given the high prevalence of oriT and Mob-carrying plasmids in S. aureus, it is perhaps fortuitous for our healthcare systems that the prevalence of conjugative plasmids in S. aureus is only 5–6%. However, this low apparent incidence may reflect a sequence data sample heavily biased toward disease-causing organisms. Clinical strains are isolated from specialized niches so might be quite distinct from the broader population, or yet to be defined reservoirs, from which they originate, where mechanisms of horizontal gene transfer might be more significant.
There are very few examples of the relaxase-in trans mechanism documented for other mobile genetic elements. Citation54-57 Considering its simplicity, it is intriguing why the relaxase-in trans mechanism of plasmid mobilization has not been widely detected in other species. We suspect that in general, mobilization has been favored in S. aureus due to selection against large plasmid size and/or carriage of conjugation genes. S. aureus hosts relatively small plasmids compared to other genera, suggesting constraints on plasmid size may be particularly strict.Citation3,4 Conjugation-gene clusters are much larger than mobilization loci. The carriage of mobilization loci by most S. aureus plasmids may represent an evolutionary trade-off that enables smaller plasmid size while maintaining a potential for transfer in the presence of a conjugative plasmid.Citation58 Carriage of an oriT mimic likely has even a smaller impact on plasmid size than Mob-gene carriage and the accumulation of multiple oriT mimics likely increases the opportunity for transfer. An interesting observation from the analysis of plasmid sizes and putative mobilization mechanisms, is that only 3% of plasmids over 14.5 kb carried a Mob or Pre gene, but 89% of the same plasmids carry an oriT. This could suggest that for large plasmids, there may be some deleterious effect of Mob-gene carriage. Indeed, plasmids such as pWBG762 and pMW2, carry pre pseudogenes, suggesting that they have been captured incidentally through plasmid cointegration events and subsequently mutated. Further molecular investigations are required to fully understand the nature of the selective forces shaping conjugative mobilization and reveal if the relaxase-in trans mechanism is indeed a phenomenon largely unique to staphylococci, or if it has simply been overlooked in other species.
Bacterial whole-genome sequencing has exploded in recent years and is even replacing our routine typing methods in pathogen and antimicrobial-resistance surveillance. With these increased data come the promise of an enlightened view of bacterial genome evolution and an increased understanding of the events that lead to the emergence of highly pathogenic or resistant strains. An understanding of horizontal-gene-transfer events and the mobile elements that facilitate them, is central to our understanding of bacterial evolution. We and others envisage that in the near future we will have the capacity to track horizontal spread of resistance and virulence elements throughout bacterial populations, which in turn will reveal opportunities in which we can control or prevent gene transfer from occuring.Citation2 However, how well we extract meaningful information from the multitude of sequenced genomes depends entirely on how well we understand the genes, proteins and molecular mechanisms underlying these processes. It is clear from our analyses that even for a clinically important and routinely sequenced pathogen like S. aureus, until recently, we have been oblivious to a DNA transfer mechanism that has impacted the evolution of the majority of its characterized plasmids. Several novel and diverse conjugation and mobilization systems have recently been recognized in bacteriaCitation6,8,11,52 so we would be foolish to assume that there are not additional gene-transfer mechanisms operating that are yet to be recognized.Citation6,58
Materials and methods
Plasmids pWBG637 and pWBG707 were conjugated into S. aureus WBG4515Citation59 and then the whole genomes of the resulting strains were sequenced using Illumina MiSeq with the MiSeq Reagent Kit v2 (250 × 2). Reads were assembled using SPAdes v. 3.6.0 and contigs were extended using SSPACE Standard v. 3.0.Citation60,61 Plasmid pWBG4 DNA was extracted and digested with combinations of BamHI, EcoRI, ClaI and HindIII and cloned into pBluescript SK+. Each clone was sequenced using primer walking and Sanger sequencing, producing 413 sequences covering pWBG4 in both directions. A clone carrying a 5.7 kb ClaI fragment containing dipA-dipC conferred diffusible pigment production in E. coli (). Plasmid annotations were initially carried out using Prokka (1.12-β), prior to manual annotation.Citation62 Detection of Mob-gene loci was carried out as previously describedCitation8 and the non-conjugative S. aureus plasmid list is provided in Table S1.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
2015MGE0032R-s02.xls
Download MS Excel (107.5 KB)Acknowledgments
The Authors and the School of Biomedical Sciences, Curtin University, dedicate this letter to the memory of David E. Townsend, for his substantial contribution to our understanding of conjugative plasmids in staphylococcal bacteria. Q.N. and K.J.P. credit D. Townsend for his supervision of the shotgun-cloning and Sanger-sequencing experiments leading to the sequence of pWBG4.
Funding
J.P.R. thanks the Faculty of Health Sciences and the School of Biomedical Sciences, Curtin University for funding contributions. K.Y.E. and R.J.M. thank the Australian government for their postgraduate scholarships. Research on pSK41 transfer was supported by National Health and Medical Research Council (Australia) Project Grant APP1081412 to N.F. and S.M.K.
References
- Smillie C, Garcillan-Barcia MP, Francia MV, Rocha EP, de la Cruz F. Mobility of plasmids. Microbiol Mol Biol Rev 2010; 74:434-52; PMID:20805406; http://dx.doi.org/10.1128/MMBR.00020-10
- Garcillan-Barcia MP, Alvarado A, de la Cruz F. Identification of bacterial plasmids based on mobility and plasmid population biology. FEMS Microbiol Rev 2011; 35:936-56; PMID:21711366; http://dx.doi.org/10.1111/j.1574-6976.2011.00291.x
- Shearer JE, Wireman J, Hostetler J, Forberger H, Borman J, Gill J, Sanchez S, Mankin A, Lamarre J, Lindsay JA, et al. Major families of multiresistant plasmids from geographically and epidemiologically diverse staphylococci. G3 (Bethesda) 2011; 1:581-91; PMID:22384369; http://dx.doi.org/full_text
- McCarthy AJ, Lindsay JA. The distribution of plasmids that carry virulence and resistance genes in Staphylococcus aureus is lineage associated. BMC Microbiol 2012; 12:104; PMID:22691167; http://dx.doi.org/10.1186/1471-2180-12-104
- Chua KY, Seemann T, Harrison PF, Monagle S, Korman TM, Johnson PD, Coombs GW, Howden BO, Davies JK, Howden BP, et al. The dominant Australian community-acquired methicillin-resistant Staphylococcus aureus clone ST93-IV [2B] is highly virulent and genetically distinct. PLoS One 2011; 6:e25887; PMID:21991381; http://dx.doi.org/10.1371/journal.pone.0025887
- Lee CA, Thomas J, Grossman AD. The Bacillus subtilis conjugative transposon ICEBs1 mobilizes plasmids lacking dedicated mobilization functions. J Bacteriol 2012; 194:3165-72; PMID:22505685; http://dx.doi.org/10.1128/JB.00301-12
- O'Brien FG, Ramsay JP, Monecke S, Coombs GW, Robinson OJ, Htet Z, Alshaikh FA, Grubb WB. Staphylococcus aureus plasmids without mobilization genes are mobilized by a novel conjugative plasmid from community isolates. J Antimicrob Chemother 2015; 70:649-52; PMID:25411186; http://dx.doi.org/10.1093/jac/dku454
- O'Brien FG, Yui Eto K, Murphy RJ, Fairhurst HM, Coombs GW, Grubb WB, Ramsay JP. Origin-of-transfer sequences facilitate mobilization of non-conjugative antimicrobial-resistance plasmids in Staphylococcus aureus. Nucleic Acids Res 2015; 43:7971-83; PMID:26243776; http://dx.doi.org/10.1093/nar/gkv755
- Goessweiner-Mohr N, Arends K, Keller W, Grohmann E. Conjugation in gram-positive bacteria. Microbiol Spectr 2014; 2:PLAS-0004-2013
- Rocco JM, Churchward G. The integrase of the conjugative transposon Tn916 directs strand- and sequence-specific cleavage of the origin of conjugal transfer, oriT, by the endonuclease Orf20. J Bacteriol 2006; 188:2207-13; PMID:16513750; http://dx.doi.org/10.1128/JB.188.6.2207-2213.2006
- Wisniewski JA, Traore DA, Bannam TL, Lyras D, Whisstock JC, Rood JI. TcpM: a novel relaxase that mediates transfer of large conjugative plasmids from Clostridium perfringens. Mol Microbiol 2016; 99:884-96; PMID:26560080; http://dx.doi.org/10.1111/mmi.13270
- Llosa M, Gomis-Ruth FX, Coll M, de la Cruz Fd F. Bacterial conjugation: a two-step mechanism for DNA transport. Mol Microbiol 2002; 45:1-8; PMID:12100543; http://dx.doi.org/10.1046/j.1365-2958.2002.03014.x
- Archer GL, Johnston JL. Self-transmissible plasmids in staphylococci that encode resistance to aminoglycosides. Antimicrob Agents Chemother 1983; 24:70-7; PMID:6625557; http://dx.doi.org/10.1128/AAC.24.1.70
- Townsend DE, Bolton S, Ashdown N, Grubb WB. Transfer of plasmid-borne aminoglycoside-resistance determinants in staphylococci. J Med Microbiol 1985; 20:169-85; PMID:2931527; http://dx.doi.org/10.1099/00222615-20-2-169
- Goering RV, Ruff EA. Comparative analysis of conjugative plasmids mediating gentamicin resistance in Staphylococcus aureus. Antimicrob Agents Chemother 1983; 24:450-2; PMID:6639004; http://dx.doi.org/10.1128/AAC.24.3.450
- Liu MA, Kwong SM, Jensen SO, Brzoska AJ, Firth N. Biology of the staphylococcal conjugative multiresistance plasmid pSK41. Plasmid 2013; 70:42-51; PMID:23415796; http://dx.doi.org/10.1016/j.plasmid.2013.02.001
- Firth N, Skurray R. Genetics: Accessory Elements and Genetic Exchange. In: Fischetti VA, Novick RP, Ferretti JJ, Portnoy DA, Rood JI, eds. Gram-Positive Pathogens. Washington DC: ASM Press, 2006:413-26
- Bender J, Strommenger B, Steglich M, Zimmermann O, Fenner I, Lensing C, Dagwadordsch U, Kekulé AS, Werner G, Layer F. Linezolid resistance in clinical isolates of Staphylococcus epidermidis from German hospitals and characterization of two cfr-carrying plasmids. J Antimicrob Chemother 2015; 70:1630-8; PMID:25740949
- Weigel LM, Clewell DB, Gill SR, Clark NC, McDougal LK, Flannagan SE, Kolonay JF, Shetty J, Killgore GE, Tenover FC. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science 2003; 302:1569-71; PMID:14645850; http://dx.doi.org/10.1126/science.1090956
- Grohmann E, Muth G, Espinosa M. Conjugative plasmid transfer in gram-positive bacteria. Microbiol Mol Biol Rev 2003; 67:277-301, table of contents; PMID:12794193; http://dx.doi.org/10.1128/MMBR.67.2.277-301.2003
- Climo MW, Sharma VK, Archer GL. Identification and characterization of the origin of conjugative transfer (oriT) and a gene (nes) encoding a single-stranded endonuclease on the staphylococcal plasmid pGO1. J Bacteriol 1996; 178:4975-83; PMID:8759863
- Edwards JS, Betts L, Frazier ML, Pollet RM, Kwong SM, Walton WG, Ballentine WK 3rd, Huang JJ, Habibi S, Del Campo M, et al. Molecular basis of antibiotic multiresistance transfer in Staphylococcus aureus. Proc Natl Acad Sci U S A 2013; 110:2804-9; PMID:23359708; http://dx.doi.org/10.1073/pnas.1219701110
- Projan SJ, Archer GL. Mobilization of the relaxable Staphylococcus aureus plasmid pC221 by the conjugative plasmid pGO1 involves three pC221 loci. J Bacteriol 1989; 171:1841-5; PMID:2703461
- Smith MC, Thomas CD. An accessory protein is required for relaxosome formation by small staphylococcal plasmids. J Bacteriol 2004; 186:3363-73; PMID:15150221; http://dx.doi.org/10.1128/JB.186.11.3363-3373.2004
- Apisiridej S, Leelaporn A, Scaramuzzi CD, Skurray RA, Firth N. Molecular analysis of a mobilizable theta-mode trimethoprim resistance plasmid from coagulase-negative staphylococci. Plasmid 1997; 38:13-24; PMID:9281492; http://dx.doi.org/10.1006/plas.1997.1292
- Caryl JA, Thomas CD. Investigating the basis of substrate recognition in the pC221 relaxosome. Mol Microbiol 2006; 60:1302-18; PMID:16689804; http://dx.doi.org/10.1111/j.1365-2958.2006.05188.x
- Gennaro ML, Kornblum J, Novick RP. A site-specific recombination function in Staphylococcus aureus plasmids. J Bacteriol 1987; 169:2601-10; PMID:3584064
- Selinger LB, McGregor NF, Khachatourians GG, Hynes MF. Mobilization of closely related plasmids pUB110 and pBC16 by Bacillus plasmid pXO503 requires trans-acting open reading frame beta. J Bacteriol 1990; 172:3290-7; PMID:2345147
- Grohmann E, Guzman LM, Espinosa M. Mobilization of the streptococcal plasmid pMV158: interactions of MobM protein with its cognate oriT DNA region. Mol Gen Genet 1999; 261:707-15; PMID:10394908; http://dx.doi.org/10.1007/s004380050014
- Lorenzo-Diaz F, Fernandez-Lopez C, Garcillan-Barcia MP, Espinosa M. Bringing them together: plasmid pMV158 rolling circle replication and conjugation under an evolutionary perspective. Plasmid 2014; 74:15-31; PMID:24942190; http://dx.doi.org/10.1016/j.plasmid.2014.05.004
- O'Brien FG, Coombs GW, Pearman JW, Gracey M, Moss F, Christiansen KJ, Grubb WB. Population dynamics of methicillin-susceptible and -resistant Staphylococcus aureus in remote communities. J Antimicrob Chemother 2009; 64:684-93; PMID:19713400; http://dx.doi.org/10.1093/jac/dkp285
- Rossi F, Diaz L, Wollam A, Panesso D, Zhou Y, Rincon S, Narechania A, Xing G, Di Gioia TS, Doi A, et al. Transferable vancomycin resistance in a community-associated MRSA lineage. N Engl J Med 2014; 370:1524-31; PMID:24738669; http://dx.doi.org/10.1056/NEJMoa1303359
- Panesso D, Planet PJ, Diaz L, Hugonnet JE, Tran TT, Narechania A, Munita JM, Rincon S, Carvajal LP, Reyes J, et al. Methicillin-Susceptible, Vancomycin-Resistant Staphylococcus aureus, Brazil. Emerg Infect Dis 2015; 21:1844-8; PMID:26402569; http://dx.doi.org/10.3201/eid2110.141914
- Udo E, Townsend DE, Grubb WB. A conjugative staphylococcal plasmid with no resistance phenotype. FEMS Microbiol Lett 1987; 40:279-83; http://dx.doi.org/10.1111/j.1574-6968.1987.tb02039.x
- Udo EE, Grubb WB. Molecular and phage typing of Staphylococcus aureus harbouring cryptic conjugative plasmids. Eur J Epidemiol 1996; 12:637-41; PMID:8982625; http://dx.doi.org/10.1007/BF00499464
- Udo EE, Grubb WB. Excision of a conjugative plasmid from the staphylococcal chromosome. J Med Microbiol 1990; 33:227-34; PMID:2175358; http://dx.doi.org/10.1099/00222615-33-4-227
- Udo EE, Grubb WB. A new class of conjugative plasmid in Staphylococcus aureus. J Med Microbiol 1990; 31:207-12; PMID:2156076; http://dx.doi.org/10.1099/00222615-31-3-207
- Udo EE, Grubb WB. A new incompatibility group plasmid in Staphylococcus aureus. FEMS Microbiol Lett 1991; 62:33-6; PMID:2032622; http://dx.doi.org/10.1111/j.1574-6968.1991.tb04412.x
- Udo EE, Grubb WB. Conjugal transfer of plasmid pWBG637 from Staphylococcus aureus to Staphylococcus epidermidis and Streptococcus faecalis. FEMS Microbiol Lett 1990; 60:183-7; PMID:2126516; http://dx.doi.org/10.1111/j.1574-6968.1990.tb03886.x
- Townsend DE, Ashdown N, Annear DI, Grubb WB. A conjugative plasmid encoding production of a diffusible pigment and resistance to aminoglycosides and macrolides in Staphylococcus aureus. Aust J Exp Biol Med Sci 1985; 63 (Pt 5):573-86; PMID:4091763; http://dx.doi.org/10.1038/icb.1985.61
- Townsend DE, Bolton S, Ashdown N, Annear DI, Grubb WB. Conjugative, staphylococcal plasmids carrying hitch-hiking transposons similar to Tn554: intra- and interspecies dissemination of erythromycin resistance. Aust J Exp Biol Med Sci 1986; 64 (Pt 4):367-79; PMID:3024610; http://dx.doi.org/10.1038/icb.1986.39
- Townsend DE, Grubb WB, Annear DI. A plasmid for diffusible pigment production in Staphylococcus aureus. Aust J Exp Biol Med Sci 1985; 63(Pt 4):463-72; PMID:4084142; http://dx.doi.org/10.1038/icb.1985.51
- Udo EE, Wei MQ, Grubb WB. Conjugative trimethoprim resistance in Staphylococcus aureus. FEMS Microbiol Lett 1992; 76:243-8; PMID:1427013; http://dx.doi.org/10.1111/j.1574-6968.1992.tb05470.x
- Shore AC, Lazaris A, Kinnevey PM, Brennan OM, Brennan GI, O'Connell B, Feßler AT, Schwarz S, Coleman DC. First report of cfr-carrying plasmids in the pandemic sequence type 22 methicillin-resistant Staphylococcus aureus staphylococcal cassette chromosome mec type IV clone. Antimicrob Agents Chemother 2016; 60:3007-15; PMID:26953212; http://dx.doi.org/10.1128/AAC.02949-15
- Neron B, Menager H, Maufrais C, Joly N, Maupetit J, Letort S, Carrere S, Tuffery P, Letondal C. Mobyle: a new full web bioinformatics framework. Bioinformatics 2009; 25:3005-11; PMID:19689959; http://dx.doi.org/10.1093/bioinformatics/btp493
- Kraushaar B, Appel B, Lanka E, Strauch E. Entry exclusion and oriT of a conjugative system encoded by the cryptic plasmid p29930 of Yersinia enterocolitica. Plasmid 2010; 64:79-84; PMID:20470820; http://dx.doi.org/10.1016/j.plasmid.2010.05.001
- Nunez B, De La Cruz F. Two atypical mobilization proteins are involved in plasmid CloDF13 relaxation. Mol Microbiol 2001; 39:1088-99; PMID:11251827; http://dx.doi.org/10.1046/j.1365-2958.2001.02308.x
- Zou X, Caufield PW, Li Y, Qi F. Complete nucleotide sequence and characterization of pUA140, a cryptic plasmid from Streptococcus mutans. Plasmid 2001; 46:77-85; PMID:11591133; http://dx.doi.org/10.1006/plas.2001.1539
- Mendes RE, Deshpande LM, Castanheira M, DiPersio J, Saubolle MA, Jones RN. First report of cfr-mediated resistance to linezolid in human staphylococcal clinical isolates recovered in the United States. Antimicrob Agents Chemother 2008; 52:2244-6; PMID:18391032; http://dx.doi.org/10.1128/AAC.00231-08
- Mendes RE, Deshpande LM, Bonilla HF, Schwarz S, Huband MD, Jones RN, Quinn JP. Dissemination of a pSCFS3-like cfr-carrying plasmid in Staphylococcus aureus and Staphylococcus epidermidis clinical isolates recovered from hospitals in Ohio. Antimicrob Agents Chemother 2013; 57:2923-8; PMID:23571552; http://dx.doi.org/10.1128/AAC.00071-13
- Chen H, Wu W, Ni M, Liu Y, Zhang J, Xia F, He W, Wang Q, Wang Z, Cao B, et al. Linezolid-resistant clinical isolates of enterococci and Staphylococcus cohnii from a multicentre study in China: molecular epidemiology and resistance mechanisms. Int J Antimicrob Agents 2013; 42:317-21; PMID:23880167; http://dx.doi.org/10.1016/j.ijantimicag.2013.06.008
- Pollet RM, Ingle JD, Hymes JP, Eakes TC, Eto KY, Kwong SM, Ramsay JP, Firth N, Redinbo MR. Processing of nonconjugative resistance plasmids by conjugation nicking enzyme of staphylococci. J Bacteriol 2016; 198:888-97; PMID:26728193; http://dx.doi.org/10.1128/JB.00832-15
- Roberts AP, Mullany P. Tn916-like genetic elements: a diverse group of modular mobile elements conferring antibiotic resistance. FEMS Microbiol Rev 2011; 35:856-71; PMID:21658082; http://dx.doi.org/10.1111/j.1574-6976.2011.00283.x
- Daccord A, Ceccarelli D, Burrus V. Integrating conjugative elements of the SXT/R391 family trigger the excision and drive the mobilization of a new class of Vibrio genomic islands. Mol Microbiol 2010; 78:576-88; PMID:20807202; http://dx.doi.org/10.1111/j.1365-2958.2010.07364.x
- Jaworski DD, Clewell DB. A functional origin of transfer (oriT) on the conjugative transposon Tn916. J Bacteriol 1995; 177:6644-51
- Meyer R. The R1162 mob proteins can promote conjugative transfer from cryptic origins in the bacterial chromosome. J Bacteriol 2009; 191:1574-80; PMID:19074386; http://dx.doi.org/10.1128/JB.01137-08
- Strahinic I, Kojic M, Tolinacki M, Fira D, Topisirovic L. Molecular characterization of plasmids pS7a and pS7b from Lactococcus lactis subsp. lactis bv. diacetylactis S50 as a base for the construction of mobilizable cloning vectors. J Appl Microbiol 2009; 106:78-88; PMID:19040703; http://dx.doi.org/10.1111/j.1365-2672.2008.03977.x
- Fernandez-Lopez C, Bravo A, Ruiz-Cruz S, Solano-Collado V, Garsin DA, Lorenzo-Diaz F, Espinosa M. Mobilizable rolling-circle replicating plasmids from gram-positive bacteria: a low-cost conjugative transfer. Microbiol Spectr 2014; 2; PMID:26104375
- Townsend DE, Grubb WB, Ashdown N. Gentamicin resistance in methicillin-resistant Staphylococcus aureus. Pathology 1983; 15:169-74; PMID:6310471; http://dx.doi.org/10.3109/00313028309084707
- Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 2011; 27:578-9; PMID:21149342; http://dx.doi.org/10.1093/bioinformatics/btq683
- Nurk S, Bankevich A, Antipov D, Gurevich AA, Korobeynikov A, Lapidus A, Prjibelski AD, Pyshkin A, Sirotkin A, Sirotkin Y, et al. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol 2013; 20:714-37; PMID:24093227; http://dx.doi.org/10.1089/cmb.2013.0084
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 2014; 30:2068-9; PMID:24642063; http://dx.doi.org/10.1093/bioinformatics/btu153
- Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 2011; 12:402; PMID:21824423; http://dx.doi.org/10.1186/1471-2164-12-402
