Abstract
The production of new adipocytes requires the differentiation of adipocyte precursor (AP) cells residing within the adipose tissue stromal-vascular compartment. The objective was to obtain an immortalized primary adipogenic cell line derived from FACS isolated committed APs using the conditional expression of SV40 T antigen. Adipocyte precursors were isolated from white adipose tissue (WAT) using FACS to remove non-adipogenic cell populations from mice expressing a conditionally regulated SV40 T antigen. APs were maintained by continuous culture and induced to undergo adipogenic differentiation. Adipogenesis, determined by Oil Red O staining, was assessed with each passage and compared to wildtype controls. Adipogenic capability was rapidly lost with increased passage number in committed APs with concurrent reduction in cell proliferation and expression of essential late adipogenic genes, including Pparγ and C/ebpα. Thus, FACS purified committed APs have limited capability to undergo expansion and subsequent adipogenic differentiation in vitro even if they are immortalized with the SV40 T antigen.
Introduction
Obesity results from an increase in white adipose tissue (WAT) mass through an increase in the number and size of mature adipocytes, which comprise the majority of WAT mass.Citation1 Adipocytes function as a specialized cell type to store excess nutrients in the form of triglyceride to maintain energy homeostasis. Additionally, adipocytes secrete a number of cytokines (termed adipokines) such as leptin and adiponectin that impact whole body physiology, including energy intake, lipid and glucose homeostasis, insulin sensitivity, and inflammation.Citation2-5
As mature adipocytes are post-mitotic, the production of new adipocytes results from the differentiation of adipocyte precursors (APs). Adipogenesis has been extensively studied using cell culture models, including immortalized cell lines derived from mouse embryonic fibroblasts (MEF), such as 3T3L1, and primary adipose tissue derived preadipocyte cultures. Studies using these systems have yielded many insights into adipocyte biology,Citation6 including the epigenetic and transcriptional regulation of adipogenesis (for a review see refs. 4 and 7). However, the relationship of the MEF-derived cell lines to the adipocyte lineage in vivo is unknown and primary preadipocytes, including immortalized preadipocytes, are most often derived from the passaging of the entire WAT stromal vascular fraction (SVF). As the WAT SVF is composed of a heterogeneous population of cells that includes blood lineage cells, vasculature, immune cells, and APs,Citation8-10 the relationship of these preadipocytes to the in vivo adipocyte lineage is also not clear.
Cells within the adipocyte lineage can be defined by their cell surface expression allowing for purification by fluorescent activated cell sorting (FACS).Citation8,9 Recently, we proposed an adipocyte lineage model in which adipocyte progenitors, characterized by the cell surface expression of mesenchymal stem cell markers (CD45-:CD31-:CD34+:CD29+:ScaI+:CD24+), give rise to committed preadipocytes (CD45-:CD31-:CD34+:CD29+:ScaI+:CD24-) that differentiate into lipid-filled mature adipocytes in vivo.Citation8,9 Furthermore, adipocyte progenitors are capable of forming a functional fat depot in vivo. Citation8,9 Freshly FACS isolated preadipocytes express late markers of adipogenesis, including the master regulator of adipogenesis peroxisome proliferator-activated receptor gamma isoform 2 (Pparγ) and CCAAT-enhancer-binding protein α (C/ebpα).Citation4,8,9 Thus, freshly isolated APs are ‘primed’ to directly respond to adipogenic signals. Primary cultures of isolated AP populations can readily differentiate in vitro with a minimal adipogenic induction cocktail.Citation8,9,11 While FACS-isolated APs provide many advantages for the study of adipogenesis in cell culture, the requirement to freshly isolate APs limits their use for larger scale studies that require a sustainable source of cells with low variability, such as small molecule screening.
Immortalization of purified APs may yield an enhanced adipocyte-lineage committed adipogenic cell line model for studying adipogenesis and adipocyte function. To accomplish this goal, we utilized FACS to specifically isolate highly adipogenic APs from WAT Citation8,9 derived from the H-2kb-ts-A58 Immortomouse. These mice express a conditionally regulated, thermolabile simian virus 40 (SV40) large T-antigen (T-Ag). SV40 T-Ag functions as an oncoprotein capable of transforming a variety of cell lines and inducing tumor formation in animal models.Citation12-16 Cells derived from H-2kb-ts-A58 mice express active SV40 T-Ag when cultured at 33°C in the presence of interferon gamma (INFγ).Citation17,18 The removal of INFγ and an increase in temperature to 37°C prevents SV40 T-Ag expression.Citation17 The conditional expression of SV40 is necessary as SV40 inhibits adipogenesis.Citation19 Additionally, as SV40 is expressed in all isolated cells, there is no requirement for viral transduction or selection, which could alter the function of the resulting cell lines.
Results
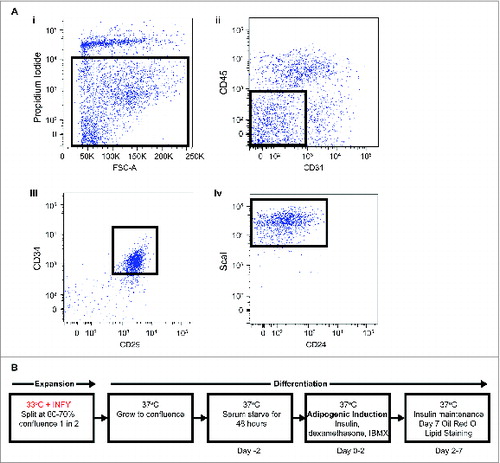
Adipocyte precursors were isolated using FACS, as previously describedCitation9,20 (), from inguinal subcutaneous WAT of H-2kb-ts-A58 transgenic mice (referred to as Immorto) and wildtype controls. Immortalization was induced through the conditional expression of the SV40 T-Ag at the permissible temperature of 33°C in the presence of interferon gamma (INFγ; ). Adipocyte precursors were maintained by continuous culture in the presence of INFγ at 33°C. Prior to differentiation, APs isolated from H-2kb-ts-A58 mice were returned to a mortal state when INFγ was removed and cells were placed at the non-permissible 37°C. Adipocyte precursors isolated from wildtype mice were included to control for the conditional expression of SV40 T-Ag during expansion. Adipocyte precursors were then grown to confluence, held for 48 hours and induced to undergo standard adipogenic differentiation with the phosphodiesterase inhibitor isobutylmethylxanthine (IBMX), the glucocorticoid dexamethonsone and insulin (MDI; Citation21,22; ).
Figure 1. (A) Identification and immortalization of APs. FACs isolation of primary APs (CD45−:CD31−:CD34+:CD29+:Scal+) (i). Selected population indicates single cells negative for propidium iodide for live cells, (ii). Selected population indicates CD45−:CD31−, (iii). Selected population indicates CD45−:CD31−:CD29+:CD34+, (iv). Selected population indicates CD45−:CD31−:CD29+:CD34+:ScaI+. (B) Immortalization procedure for APs.
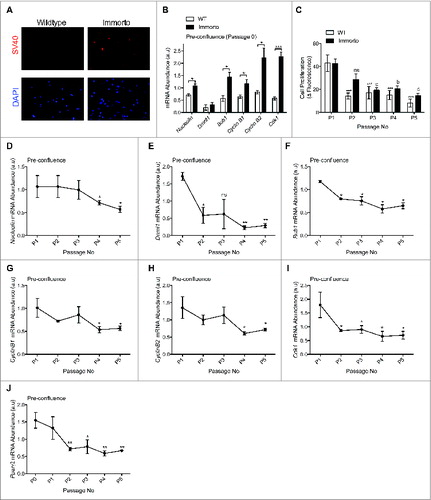
INFγ induced SV40 T-Ag expression () resulted in increased mRNA expression of SV40 T-Ag targets involved in cell cycle regulation Citation15,16,23 as compared to wildtype APs at passage zero (P0; ), indicating transgene activation. Immorto APs retained increased proliferation for 2 passages but then demonstrated reduced proliferation as compared to P0 and wildtype APs (), which exhibited reduced proliferation after only one passage in culture (). Expansion of primary APs from H-2kb-ts-A58 mice led to reduced mRNA expression of SV40 T-Ag targets involved in the cell cycle at pre-confluence (). Additionally, the expression of Pparγ2, a pro-adipogenic gene expressed in freshly FACs isolated APs,Citation8 was significantly decreased after 2 passages (), suggesting that passaging of APs in culture may perturb their commitment to the adipocyte lineage.
Figure 2. (A) Immunofluorescence of SV40 and nuclear DAPI in pre-confluent passage zero in wildtype and H-2kb-ts-A58 (immorto) derived APs. (B) Quantitative RT-PCR analysis of Nucleolin (Ncl; ID 17975), Dnmt1 (DNA methyltransferase 1; ID 13433), Bub1 (budding uninhibited by benzimidazoles 1 homolog; ID 12235), Cyclin B1 (Ccnb1; ID 268697), Cyclin B2 (Ccnb2; ID 12442) and Cdk1 (Cyclin-dependent kinase 1; ID 12534) in pre-confluent wildtype and immorto APs (n=3) at passage zero. Data are represented as mean ± SEM. T-Test for each individual gene analyzed, *P < 0.05, ***P < 0.001. (C). Adipocyte precursor proliferation after 6 hours for wildtype (WT) and immorto APs at 50-60% confluency. n = 12 per passage number. Data are represented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 for Two-way ANOVA compared to P0. a = P < 0.05, b = P < 0.01, c = P < 0.001 for Two-way ANOVA compared to Immorto P0. (D–J) Quantitative RT-PCR analysis of (D). Nucleolin (Ncl; ID 17975), (E). Dnmt1 (DNA methyltransferase 1; ID 13433), (F). Bub1 (budding uninhibited by benzimidazoles 1 homolog; ID 12235), (F). Cyclin B1 (Ccnb1; ID 268697), (H). Cyclin B2 (Ccnb2; ID 12442), (I). Cdk1 (Cyclin-dependent kinase 1; ID 12534) and (J). Pparγ2 (ID 19016) for the indicated passage numbers. n=3 per for pre-confluent wildtype (WT) and immorto APs. Data are represented as mean ± SEM. *P < 0.05 and **P < 0.01 for Two-way ANOVA compared to P1.
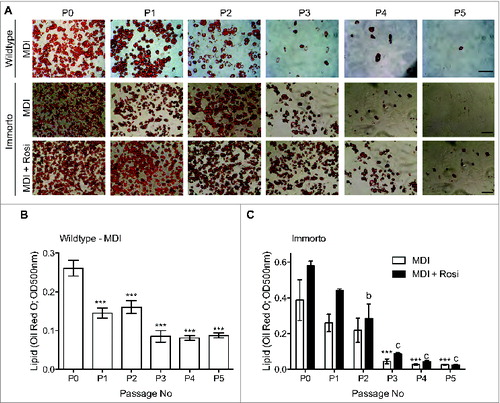
We next assessed whether Immorto and wildtype APs maintained the ability to differentiate into adipocytes in response to the standard MDI adipogenic differentiation cocktail. APs from both sources robustly differentiated through 2 passages, as assessed by lipid Oil Red O staining (), suggesting that the expression of SV40 T-Ag was sufficiently reduced upon the switch to the restrictive temperature to permit adipogenesis. However, as the passage number increased, the capability to undergo adipogenic differentiation significantly decreased, and minimal adipogenesis was observed after 3 passages (). Inclusion of rosiglitazone, a thiazolidinedione (TZD) and PPARγ agonist, with the MDI adipogenic cocktail augmented adipogenic differentiation until passage 4 (). However, minimal adipocyte differentiation, assessed by lipid Oil Red O staining, was observed after passage 5, even in the presence of rosiglitazone ().
Figure 3. (A) Passage zero (P0) through pass 5 (P5) lipid Oil Red O staining for wildtype and immorto APs at day 7 of adipogenic differentiation for MDI (top and middle for wildtype and immorto, respectively) and immorto APs with MDI and rosiglitazone (bottom). Scale bars represents 100 μm. (B) Quantification of Oil Red O for wildtype APs at day 7 of MDI adipogenic differentiation (n = 3). Data are represented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 for Two-way ANOVA compared to P0. (C) Quantification of Oil Red O for immorto APs at day 7 of adipogenic differentiation (n = 3) with MDI and MDI with rosiglitazone. Data are represented as mean ± SEM. ***P < 0.001 for Two-way ANOVA compared to MDI P0. b = P < 0.01 and c = P < 0.001 for Two-way ANOVA compared to MDI with rosiglitazone P0.
Gene expression analysis was conducted at confluence, prior to differentiation (time zero), as freshly isolatedCitation8 and non-passaged APs are enriched for late stage pro-adipogenic genes, such as Pparγ and C/ebpα, hence conferring a ‘primed’ state to directly respond to adipogenic signals. Additionally, human adipose tissue derived adult stem cells (ADASCs) isolated from the SVF exhibited a downregulation of adipogenic transcripts, including PPARγ after 4 passages in culture, as compared to freshly isolated ADASCs.Citation24 Therefore, assessing the expression of essential pro-adipogenic genes prior to differentiation represents a critical time point to indicate if APs can response to adipogenic signals.
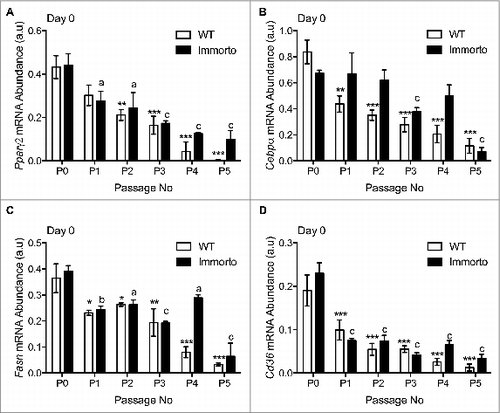
The mRNA expression of Pparγ decreased with each passage in both Immorto and wildtype APs at time zero (), consistent with reduced expression in Immorto APs at pre-confluence (). C/ebpα also exhibited reduced mRNA expression (), although immorto APs retained C/ebpα expression levels similar to P0 for 2 additional passages compared to only one passage in wildtype APs (). Key metabolic regulators of lipid biosynthesis, such as fatty acid synthase (FASN) and the fatty acid transporter cluster of differentiation 36 (CD36), are required for the production and transport of fatty acid precursors required for triglyceride synthesis, respectively. These genes displayed reduced mRNA expression after the first passage and have greatly diminished mRNA expression after 3 passages as compared to P0 ().
Figure 4. Quantitative RT-PCR analysis of (A) Pparγ2, (B) C/ebpα, (C) Fasn and (D) Cd36. n = 3 per passage number for wildtype and immorto APs at day zero before adipogenic differentiation. Data are represented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 for Two-way ANOVA compared to wildtype P0. a = P < 0.05, b = P < 0.01, c = P < 0.001 for Two-way ANOVA compared to Immorto P0.
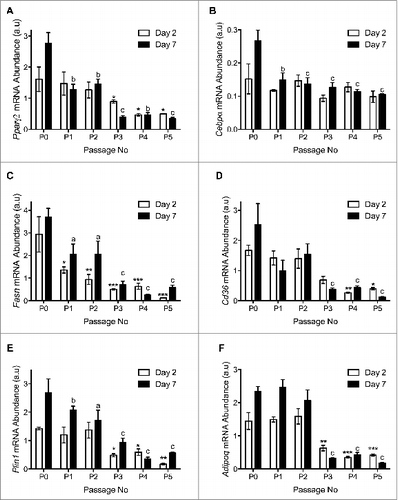
The pro-adipogenic genes Pparγ, C/ebpα, Cd36 and Fasn exhibited reduced mRNA expression throughout the differentiation time course after passage 3 as compared to non-passaged (P0) APs at 48-hours and 7 days post-adipogenic induction (). This coincided with decreased expression of the lipid droplet associated protein perilipin (Plin1) and the adipokine adiponectin (Adipoq; ), consistent with reduced lipid accumulation after passage 3 ().
Figure 5. Quantitative RT-PCR analysis of (A) Pparγ2, (B) C/ebpα, (C) Fasn and (D) Cd36. (E) Plin1 and (F) Adipoq. n=3 per passage number for wildtype and immorto APs at day 2 and day 7 after adipogenic differentiation. Data are represented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 for Two-way ANOVA compared to P0 Immorto APs for day 2. a = P < 0.05, b = P < 0.01, c = P < 0.001 for Two-way ANOVA compared to P0 Immorto APs for day 7.
Discussion
We sought to create a superior reagent for the study of adipogenesis and adipocyte function in vitro by generating an immortalized adipogenic cell line from freshly isolated, bona fide APs.Citation8,9 Toward this goal, conditional immortalization of APs was achieved using the H-2kb-ts-A58 Immortomouse, which allows for the restricted expression of SV40 T-Ag from all cells. This approach permits SV40 expression to be ablated prior to adipogenic induction and circumvents the requirement for viral transduction or selection.
Here we show that the H-2kb-ts-A58 transgene enhances proliferation only for one additional passage as compared to wildtype APs. After two passages, immorto-APs display reduced proliferation and expression of essential SV40 dependent cell cycle regulators. While freshly isolated, cultured, and non-passaged APs undergo high levels of adipogenesis in vitro,Citation8,9 APs derived from wild type or H-2kb-ts-A58 mice fail to retain the ability to robustly differentiate with a standard adipogenic induction cocktail (MDI) upon extended expansion in culture. The loss of expression of critical adipogenic genes upon passaging, including the master transcriptional regulator of adipogenesis Pparγ2 and Cebpα, and several key lipogenic genes, such as Cd36 and Fasn, correlates with the loss of adipogenic capacity of these cells.
The downregulation of proadipogenic genes, whose expression is enriched in freshly FACS isolated APs and non-passaged APs, may alter the primed state of APs to directly respond to adipogenic signals.Citation8 This downregulation may account for the substantially reduced adipogenesis observed after the third passage. Moreover, in agreement with these studies Boquest et al (2005) compared gene expression profiles from freshly isolated and cultured ADASCs and demonstrated the downregulation of transcripts associated with cell cycle, stemness, and adipocyte differentiation, including PPARγ after 4 passages.Citation24
Mature lipid-filled adipocytes are postmitotic, and therefore APs are required to exit the cell cycle during terminal differentiation. The isolation of committed APs late in the adipocyte lineage,Citation8,10 which are primed to directly respond to adipogenic signals, may have limited potential for proliferation and subsequent differentiation when removed from the in vivo microenvironment for culture in vitro. Taken together, these data indicate that passaging of APs results in a loss of the genetic repertoire required to fully support adipogenesis in vitro, regardless of the presence of SV40 during passaging.
Although SV40 T-Ag expression can overcome replicative senescence,Citation25 use of SV40 T-Ag alone may have limited capability as cells reach a second non-replicative phase termed ‘crisis’.Citation17,26 Procedures for the immortalization and subsequent differentiation of WAT-SVF derived preadipocytes have been previously described with SV40 T-Ag and have also encountered similar replicative senescence within 7 passages.Citation26,27 Moreover, genomic modification and telomere shortening induced by SV40 T-Ag immortalization have been proposed as being responsible for growth arrest and crisis.Citation28,29 The addition of telomerase activity through the coexpression of h-TERT and E7 oncoprotein from the human papillomavirus type 16 (HPV-E7) has been demonstrated to expand the proliferative and subsequent adipogenic capability in human SVF derived preadipocytes.Citation30 However, this involves further transgene insertion and selection.
The loss of adipogenic capacity upon passaging of cells is not unprecedented, as commonly used adipogenic cell lines, such as 3T3L1, also experience a decline in differentiation efficiency with increased passage number. 3T3L1 cells require the addition of TZD class of PPARγ agonists after passage 6 to achieve robust adipogenic differentiation.Citation31 Previously, approximately 25 passages of immortalized WAT SVF-derived preadipocytes were obtained from H-2kb-ts-A58 mice.Citation32 These cells retained the ability to undergo adipogenesis with MDI in combination with rosiglitazone.Citation32 Immortalized human preadipocytes also require the addition of PPARγ agonists to stimulate lipid accumulation.Citation26,30,33
Here we show that while inclusion of rosiglitazone in the standard MDI adipogenic cocktail is sufficient to enhance adipogenesis in AP-derived cells for a greater number of passages over MDI alone, the rosiglitazone induced APs still displayed reduced differentiation compared to passage zero and passage one-APs. Additionally, rosiglitazone did not significantly induce adipogenesis over MDI alone by passage 5. The augmentation of adipogenesis by the addition of rosiglitazone in the early passages can be explained by the retention of expression of Pparγ at these time points. The lack of response to rosiglitazone after further passaging indicates that the loss of Pparγ is of sufficient severity as to result in the inability to activate lipogenesis with PPARγ activation.
The decreased ability of isolated APs to retain adipogenic capacity through passaging compared to WAT-SVF-derived preadipocytesCitation32 suggests either that there are supporting cells or factors that are necessary during passaging that allow APs to retain their adipogenic capacity, or that the cells that become adipogenic after passaging of these cultures are not derived from the characterized adipocyte lineage.
Human ADASCs offer an abundant and accessible source of adult stem cells for regenerative medicine.Citation34 Based on the loss of proliferation and differentiation potential with culture,Citation24 future studies of ADASCs should investigate how to enhance the proliferative potential of cultured ADASCs to provide a sustainable source of purified cells with low variability. The recent advancement of precise genome engineering such as CRISPR/Cas9Citation35,36 will allow for sustained transcriptional manipulations necessary to maintain both proliferative and differentiation potential in immortalized physiologically relevant primary cell types. Additionally, in the search for unique cell surface markers of white, brown and beige adipocytes it was recently shown that immortalization with SV40 T-Ag or prolonged culture modified the normal gene expression observed in vivo and hence the specialized characteristics of white or brown adipocytes can be lost in vitro.Citation37 These studies demonstrate the need to use primary non-cultured adipocytes to accurately preserve adipose tissue specific gene profiles. Although the immortalization approach described here did not result in a viable immortalized cell line capable of adipogenesis, this study should guide future immortalization procedures including precise genome engineering in primary APs to mirror the transcriptional profiles of freshly isolated APs.
In closing, SV40 TAg immortalized APs have limited potential for in vitro culture, likely due to a loss of a committed late adipogenic gene expression profile. These findings indicate that SV40 immortalization will not provide an AP-derived cell line that is sufficient for large scale in vitro experiments, such as drug screening involving small molecules or siRNAs.
While other methods of immortalization may provide a greater ability to expand the cells without compromising proliferation or adipogenic capacity, techniques that require additional selection steps for transgene insertion may result in a cell line with significantly altered characteristics from freshly-isolated APs. A further approach may be to identify factors and/or supportive cell types that could be included in the cultures to maintain the adipogenic capacity of APs.
Materials and Methods
Mice
The Institutional Animal Care and Use Committee (IACUC) at Yale University approved all animal studies. All animals were kept in temperature and humidity-controlled rooms on a 12-hour:12-hour light: dark cycle, with lights on from 7:00 a.m. to 7:00 p.m. Immortomice H-2kb-ts-A58 were used as previously described Citation13,38 and genotyped according to Pvull et al (2005).Citation39
Fluorescence-activated cell sorting (FACS) of APs
Subcutaneous inguinal adipose tissue was excised from H-2kb-ts-A58 and wildtype mice and digested in collagenase as previously described.Citation20 Briefly, floating adipocytes were removed by centrifugation at 300 × g for 3 minutes and the stromal vascular fraction (SVF) was sequentially filtered through 100-, 70-, and 40–micron nylon filters (BD Biosciences). The isolated SVF was washed and resuspended in Hank's balanced salt solution (HBSS) with 3% bovine serum albumin (BSA) and stained with the following antibodies CD45 APC-eFluor 780 (eBioscience; 47-0451-80), CD31 PE-Cy7 (eBioscience, 25-0311-82), CD29 Alexa Fluor 700 (BioLegend, 102218), CD24 PerCPCy5.5 (eBioscience; 47-0242-82) and Sca-1 Pacific Blue (BD Biosciences, 560653).
Samples were sorted on a BD FACSAria cell sorter (BD Biosciences) equipped with BD FACSDiva Software. For sorting, cells were initially selected by size, on the basis of forward scatter (FSC) and side scatter (SSC), followed by exclusion of dead cells on the basis of uptake of propidium iodide. Then live cells were gated on both SSC and FSC singlets, ensuring that the staining of individual cells was analyzed. Next, the cells were separated on the basis of CD45−/CD31−, CD34+/CD29+ and Scal+ and collected into 3% BSA coated 1.5ml tubes.
Expansion and adipogenic differentiation of primary adipocyte precursors
Freshly isolated cells were placed in sterile cell culture flasks 25 cm2 (approximately 350,000 cells) or 75 cm2 (1 million cells) in DMEM (ATCC) supplemented with 10% FBS (GIBCO; Life Technologies), insulin-sodium selenite-transferrin (ITS; Sigma-Aldrich), T3 (Sigma-Aldrich), and interferon gamma (Sigma-Aldrich). Cells were allowed to grow to 60-70% confluence and were either split into 2 new flasks (1 in 2) or were frozen into aliquots using freezing media (DMEM supplemented with 10% FBS, GIBCO; Life Technologies and 10% DMSO, Sigma-Aldrich).
Adipocyte precursors were recovered from frozen stocks and plated onto carboxyl-coated 24 well plates (BD Biosciences) in DMEM supplemented with 10% FBS (GIBCO; Life Technologies), insulin-sodium selenite-transferrin (ITS), T3, and interferon gamma (10 units per ml). The cells were maintained in a 5% CO2 atmosphere at 33°C. Cells were allowed to reach 70% confluence, at which point the media was changed to DMEM supplemented with 10% FBS (GIBCO; Life Technologies) and penicillin-strep antibiotic (Life Technologies) without interferon gamma. Adipocyte precursors were transferred to 37°C, and were then held at confluence for 2 days without changing the media prior to exposure to the adipogenic differentiation cocktail in fresh DMEM media containing 1 μg/ml insulin, 0.25 μg/ml dexamethasone, and 30 μg/ml 3-isobutyl-1-methylxanthine IBMX (Sigma-Aldrich) referred to as MDI or MDI including rosiglitazone (1 μM; Sigma-Aldrich). After 48-hour exposure to the MDI adipogenic differentiation cocktail or MDI plus rosiglitazone, cells were maintained in DMEM with 10% FBS and 1 mg/ml insulin until day 7 for harvest.
Immunofluorescence
Passage one wildtype and immorto APs were plated at 50% confluence on chamber slides (Lab-Tek II: Thermoscientific). Cells were fixed for 15 minutes with 3.7% formaldehyde, permeabilized with 0.25% Triton X in PBS (PBST) and blocked with 1% BSA in PBST for 30 minutes. A monoclonal biotin mouse anti-SV40 (BD Biosciences) antibody diluted 1 in 100 in PBST with 1% BSA was incubated overnight at 4°C. After washing a Rhodamine (TRITC)-Streptavidin secondary antibody (Jackson ImmunoResearch) diluted 1 in 200 was incubated for one hour at room temperature. After washing, the slides were mounted using Vectashield hardset mounting medium with 4',6-diamidino-2-phenylindole (DAPI, VECTOR Laboratories). Cells were imaged on a Zeiss Axioplan 2 fluorescent microscope with Zeiss Axiovision 4.2.
CyQuant cell proliferation
Cells were plated in black walled 96-well plates (approximately 5,000 cells/well) and cultured in DMEM supplemented with 10% FBS for 24 hours. Cell proliferation was analyzed using the CyQUANT kit (Life Technologies), according to the manufacturer's instructions. The plate was incubated for 6 hours, and the fluorescence intensity was measured on a microplate reader at 33°C in the presence of INFγ (Synergy Mx; BioTek) with excitation at 485 nm and emission at 528 nm.
Oil red O staining and extraction
For staining, cells were fixed with 2% formaldehyde and 0.2% glutaraldehyde in PBS for 15 minutes and then rinsed in PBS, water, and 60% isopropanol sequentially. The cells were then stained with oil red O (0.7% in 60% isopropanol; Electron Microscopy Sciences) for 10 minutes and rinsed with 60% isopropanol followed by water. Oil Red O stained cells were directly visualized and imaged using an inverted microscope (Olympus BX61 microscope fitted with Olympus Q Color 5 camera) or EVOS® XL Core Cell Imaging System (Life Technologies).
Quantification of lipid accumulation was determined by Oil Red O extraction by lysis 100% isopropanol with 4% NP40 substitute IGEPAL CA-630, Sigma-Aldrich, MO, USA; I8896; Citation20. Following Oil Red O extraction the lysate was transferred to a 96-well plate and absorbance measured at 500 nm using an Infinite® 200 PRO series microplate reader (Tecan Group, Ltd).
RNA isolation and cDNA synthesis
For gene expression analysis, RNA at pre-confluence, time zero (before differentiation), day 2 and day 7 after adipogenic induction was isolated for passages 0 to 5 from wildtype and immorto cells. Cells were washed with dPBS (Life Technologies) and RNA was isolated using RNeasy Mini kit (Qiagen), according to manufacturers’ instructions. RNA was quantified by a nanodrop spectrophotometer (Thermo Fisher Scientific) and single stranded cDNA was synthesized from total RNA using the High Capacity cDNA reverse Transcription Kit (Applied Biosystems, Life Technologies) according to the manufacturer's instructions.
Quantitative real-time PCR
Quantitative RT-PCR (qPCR) was performed on the cDNA using the Sybr green method of quantification on a Roche Lightcycler 480 using a SYBR FAST quantitative PCR kit (Kapa Biosystems; KK4611) at pre-confluence and day zero (before being induced with the differentiation cocktails). Gene expression was analyzed for each passage number, using the forward and reverse primers indicated for each gene, compared to the cyclophilin house keeping gene (Table S1). cDNA across all treatment and passage groups for the indicated time points were pooled and used to generate a 6-point standard curve that was subsequently used to quantify relative mRNA transcript levels represented by mRNA abundance in arbitrary units (a.u).
Statistical analysis
Immorto and wildtype data was compared using a t-test or 2-way analysis of variance (ANOVA) with Dunnett's multiple comparisons test using GraphPad Prism version 6.00 for Mac, (Prism: GraphPad Software, Inc.). Comparisons are indicated in figure legends. Data are expressed as mean ± SEM, and P < 0.05 was considered as statistically significant.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website
995510_Supplementary_Materials.zip
Download Zip (10.4 KB)Acknowledgments
CC and MSR conceived experiments and analyzed data. CC and MB carried out experiments. All authors were involved in writing the paper and had final approval of the submitted version.
Funding
This work was supported by American Diabetes Association Award 7-12-JF-46, DERC pilot project grant DK045735 and NIDDK grant DK090489 to MSR, and an EMBO long-term fellowship to CC.
References
- Wang QA, Tao C, Gupta RK, Scherer PE. Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nat Med 2013; 19:133844; PMID:23995282; http://dx.doi.org/10.1038/nm.3324
- Cinti S. The adipose organ at a glance. Dis Model Mech 2012; 5:588-94; PMID:22915020; http://dx.doi.org/10.1242/dmm.009662
- Halberg N, Wernstedt-Asterholm I, Scherer PE. The adipocyte as an endocrine cell. Endocrinol Metab Clin North Am 2008; 37:753-68, x-xi; PMID:18775362; http://dx.doi.org/10.1016/j.ecl.2008.07.002
- Cristancho AG, Lazar MA. Forming functional fat: a growing understanding of adipocyte differentiation. Nat Rev Mol Cell Biol 2011; 12:722-34; PMID:21952300; http://dx.doi.org/10.1038/nrm3198
- Church C, Horowitz M, Rodeheffer MS. WAT is a functional adipocyte? Adipocyte 2012; 1:38-45; PMID:23700509; http://dx.doi.org/10.4161/adip.19132
- Poulos SP, Dodson MV, Hausman GJ. Cell line models for differentiation: preadipocytes and adipocytes. Exp Biol Med (Maywood) 2010; 235:118593; PMID:20864461; http://dx.doi.org/10.1258/ebm.2010.010063
- Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol 2006; 7:88596; PMID:17139329; http://dx.doi.org/10.1038/nrm2066
- Berry R, Rodeheffer MS. Characterization of the adipocyte cellular lineage in vivo. Nat Cell Biol 2013; 15:302-8; PMID:23434825; http://dx.doi.org/10.1038/ncb2696
- Rodeheffer MS, Birsoy K, Friedman JM. Identification of white adipocyte progenitor cells in vivo. Cell 2008; 135:2409; PMID:18835024; http://dx.doi.org/10.1016/j.cell.2008.09.036
- Berry R, Jeffery E, Rodeheffer MS. Weighing in on adipocyte precursors. Cell Metab 2014; 19:820; PMID:24239569; http://dx.doi.org/10.1016/j.cmet.2013.10.003
- Lee Y-H, Petkova AP, Mottillo EP, Granneman JG. In vivo identification of bipotential adipocyte progenitors recruited by β3-adrenoceptor activation and high-fat feeding. Cell Metab 2012; 15:48091; PMID:22482730; http://dx.doi.org/10.1016/j.cmet.2012.03.009
- Jha KK, Banga S, Palejwala V, Ozer HL. SV40-Mediated immortalization. Exp Cell Res 1998; 245:1-7; PMID:9828095; http://dx.doi.org/10.1006/excr.1998.4272
- Noble M. Production and growth of conditionally immortal cell lines from the H-2KbtsA58 transgenic mouse. Methods Mol Biol 1999; 97:13958; PMID:10443363
- Obinata M. Conditionally immortalized cell lines with differentiated functions established from temperature-sensitive T-antigen transgenic mice. Genes to Cells 1997; 2(4):235-44. PMID:9224657; http://dx.doi.org/10.1046/j.1365-2443.1997.1160314.x
- Ali SH, DeCaprio JA. Cellular transformation by SV40 large T antigen: interaction with host proteins. Semin Cancer Biol 2001; 11:15-23; PMID:11243895; http://dx.doi.org/10.1006/scbi.2000.0342
- Ahuja D, Sáenz-Robles MT, Pipas JM. SV40 large T antigen targets multiple cellular pathways to elicit cellular transformation. Oncogene 2005; 24:7729-45; PMID:16299533; http://dx.doi.org/10.1038/sj.onc.1209046
- Jat PS, Noble MD, Ataliotis P. Direct derivation of conditionally immortal cell lines from an H-2Kb-tsA58 transgenic mouse. 1991. 88, 5096-100; PMID:1711218
- Schulze DH, Pease LR, Obata Y. Identification of the cloned gene for the murine transplantation antigen H-2Kb by hybridization with synthetic oligonucleotides. Mol Cell Biol 1983; 3(4):750-5; PMID:6855774
- Cherington V, Brown M, Paucha E. Separation of simian virus 40 large-T-antigen-transforming and origin-binding functions from the ability to block differentiation. Mol Cell Biol 1988; 8(3):1380-4; PMID:2835674
- Church CD, Berry R, Rodeheffer MS. Isolation and study of adipocyte precursors. Methods Enzymol 2014; 537:31-46; PMID:24480340; http://dx.doi.org/10.1016/B978-0-12-411619-1.00003-3
- Tang QQ. Mitotic clonal expansion: A synchronous process required for adipogenesis. Proc Natl Acad Sci 2002; 100:44-9; http://dx.doi.org/10.1073/pnas.0137044100
- Jefcoate CR, Wang S, Liu X. Methods that resolve different contributions of clonal expansion to adipogenesis in 3T3-L1 and C3H10T1/2 cells. Methods Mol Biol 2008; 456:17393; PMID:18516561; http://dx.doi.org/10.1007/978-1-59745-245-8_13
- May T, Hauser H, Wirth D. Transcriptional control of SV40 T-antigen expression allows a complete reversion of immortalization. Nucleic Acids Res 2004; 32:552938; PMID:15486202; http://dx.doi.org/10.1093/nar/gkh887
- Boquest AC, Shahdadfar A, Frønsdal K, Sigurjonsson O, Tunheim SH, Collas P, Brinchmann JE. Isolation and transcription profiling of purified uncultured human stromal stem cells: alteration of gene expression after in vitro cell culture. Mol Biol Cell 2005; 16:113141; PMID:15635089; http://dx.doi.org/10.1091/mbc.E04-10-0949
- Shay JW, Wright WE, Werbin H. Defining the molecular mechanisms of human cell immortalization. Biochim Biophys Acta 1991; 1072:1-7; PMID:1850299
- Darimont C. Immortalization of human preadipocytes. Biochimie 2003; 85:1231-3; PMID:14739075; http://dx.doi.org/10.1016/j.biochi.2003.10.015
- Forest C, Czerucka D, Negrel R, Ailhaud G. Establishment of a human cell line after transformation by a plasmid containing the early region of the SV40 genome. Cell Biol Int Rep 1983; 7:73-81; PMID:6299591; http://dx.doi.org/10.1016/0309-1651(83)90106-6
- Counter CM, Hahn WC, Wei W, Caddle SD, Beijersbergen RL, Lansdorp PM, Sedivy JM, Weinberg RA. Dissociation among in vitro telomerase activity, telomere maintenance, and cellular immortalization. Proc Natl Acad Sci USA 1998; 95:14723-8; PMID:9843956; http://dx.doi.org/10.1073/pnas.95.25.14723
- Sedivy JM. Can ends justify the means?: telomeres and the mechanisms of replicative senescence and immortalization in mammalian cells. Proc Natl Acad Sci USA 1998; 95:9078-81; PMID:9689036; http://dx.doi.org/10.1073/pnas.95.16.9078
- Darimont C, Zbinden I, Avanti O. Reconstitution of telomerase activity combined with HPV-E7 expression allow human preadipocytes to preserve their differentiation capacity after immortalization. Cell Death Differ 2003; 10 (9): 1025-31; PMID:12934077; http://dx.doi.org/10.1038/sj.cdd.4401273
- Zebisch K, Voigt V, Wabitsch M, Brandsch M. Protocol for effective differentiation of 3T3-L1 cells to adipocytes. Anal Biochem 2012; 425:8890; PMID:22425542; http://dx.doi.org/10.1016/j.ab.2012.03.005
- Morganstein DL, Christian M, Turner JJO, Parker MG, White R. Conditionally immortalized white preadipocytes: a novel adipocyte model. J Lipid Res 2008; 49:679-85; PMID:18046046; http://dx.doi.org/10.1194/jlr.D700029-JLR200
- Rodriguez A-M, Elabd C, Delteil F, Astier J, Vernochet C, Saint-Marc P, Guesnet J, Guezennec A, Amri E-Z, Dani C, et al. Adipocyte differentiation of multipotent cells established from human adipose tissue. Biochem Biophys Res Commun 2004; 315:25563; PMID:14766202; http://dx.doi.org/10.1016/j.bbrc.2004.01.053
- Mizuno H, Tobita M, Uysal AC. Concise review: Adipose-derived stem cells as a novel tool for future regenerative medicine. Stem Cells 2012; 30:804-10; PMID:22415904; http://dx.doi.org/10.1002/stem.1076
- Harrison MM, Jenkins BV, O'Connor-Giles KM, Wildonger J. A CRISPR view of development. Genes Dev 2014; 28:1859-72; PMID:25184674; http://dx.doi.org/10.1101/gad.248252.114
- Wang H, Yang H, Shivalila CS, Dawlaty MM. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 2013; 153(4):910-8; PMID:23643243; http://dx.doi.org/10.1016/j.cell.2013.04.025
- Ussar S, Lee KY, Dankel SN, Boucher J, Haering MF, Kleinridders A, Thomou T, Xue R, Macotela Y, Cypess AM, Tseng YH, Mellgren G, Kahn CR. ASC-1, PAT2, and P2RX5 are cell surface markers for white, beige, and brown adipocytes. Sci Transl Med 2014; 6:247ra103; PMID:25080478; http://dx.doi.org/10.1126/scitranslmed.3008490
- Noble M, Groves AK, Ataliotis P, Ikram Z, Jat PS. The H-2KbtsA58 transgenic mouse: A new tool for the rapid generation of novel cell lines. Transgenic Res 1995; 4:215-25; PMID:7655511; http://dx.doi.org/10.1007/BF01969114
- PvuII SI. Localization of transgenes and genotyping of H-2kb-tsA58 transgenic mice. Biotechniques 2005; (1):38, 40-42
