ABSTRACT
Lipid droplets are essential for both the storage and retrieval of excess cellular nutrients, and their biology is regulated by a diverse range of cellular proteins, some of which function at the lipid droplet. Numerous studies have characterized lipid droplet proteomes in different organisms and cell types, and RNAi whole genome screening studies have examined the genetic regulation of lipid storage in C. elegans and D. melanogaster. While tumor protein D52 (TPD52) did not emerge from earlier studies as a strong candidate, exogenous expression of human TPD52 in cultured cells resulted in significantly increased numbers of lipid droplets, and oleic acid supplementation increased TPD52 detection at both lipid droplets and the Golgi apparatus. These results suggest that direct testing of proteins that are infrequently but recurrently identified in proteomic and RNAi screening studies may identify novel lipid droplet regulators. While the analysis of these possibly lower-abundance or itinerant lipid droplet proteins may be more technically challenging, such proteins could facilitate a more detailed interrogation of emerging aspects of lipid droplet biology.
The lipid droplet (LD) has progressed from being considered an inert cellular organelle functioning as a storage depot to one that is understood to be highly responsive to the metabolic requirements of the cell.Citation1 Indeed, the long evolutionary history of LDs suggests that they represent a basic prerequisite for organism survival.Citation1 In humans, most excess fatty acids are stored and mobilised from large unilocular LDs within adipocytes, which can be up to 100 μm in diameter.Citation2 Nonetheless, the storage capacity of adipose tissue is frequently exceeded in conditions such as obesity, leading to aberrant lipid accumulation in smaller LDs within skeletal muscle and liver cells.Citation2 As dysfunctional lipid storage plays a key role in conditions as diverse as diabetes, inflammation, obesity and cancer, adipocytes and LDs are being increasingly investigated as drug targets.Citation3
The LD is unique among cellular organelles in that it is bound by a phospholipid monolayer.Citation1 This amphipathic phospholipid coating serves to emulsify the hydrophobic core of triglycerides and cholesterol esters within an aqueous cytosol.Citation4 Many proteins have been reported to be associated with LDs,Citation5,6 where the amphipathic nature of the membrane (hydrophilic facing the cytosol, hydrophobic at the core) is predicted to represent an important determinant of how proteins associate with the LD.Citation4 Some LD proteins may be regular residents, while others may drop in and out, and also function within other cell compartments.Citation1,5 While it is perhaps tempting to conclude that the major regulatory proteins have been identified, Kamili et al. recently reported a new protein regulating LD biology.Citation7 This protein is tumor protein D52 (TPD52), a protein first discovered through its overexpression in cancer some 20 years ago.Citation8
Historical background
Since its identification, the functions of tumor protein D52 (TPD52) have been largely a mystery. TPD52 represents one of 4 paralogues in mammals (, ).Citation9 The family includes 3 genes (TPD52, TPD52L1, and TPD52L2) with broad but incompletely overlapping tissue expression, and one gene (TPD52L3) whose expression is largely restricted to the testis ().Citation9 The functions of TPD52-like proteins have been difficult to predict, as their sequences have been poorly predictive of function. TPD52 overexpression in different cancer types has been broadly reported, as have regulatory functions within signaling pathways.Citation9,10 However, these types of results can be demonstrated for many cellular proteins, so apart from the analysis of TPD52 functions in secretory processes,Citation11,12 progress in understanding TPD52′s molecular functions has been slow.Citation10 A lack of structural information has also made it difficult to predict how TPD52 contributes to macromolecular complexes, and what additional functions it might have.
Figure 1. Schematic diagram showing sequence features of the indicated human TPD52-like proteins. Protein names are shown at left and Uniprot identifiers and amino acid lengths of each isoform are shown at right. Blue bars represent coiled-coil motifs, yellow bars represent PEST domains, whereas purple and green bars represent alternatively-spliced domains. The green bar includes a 14-3-3 binding motif, whereas the purple bar represents an alternatively-spliced region termed insert 2 (Ins 2), which is present in some TPD52L2 isoforms, but not other TPD52-like proteins.Citation10
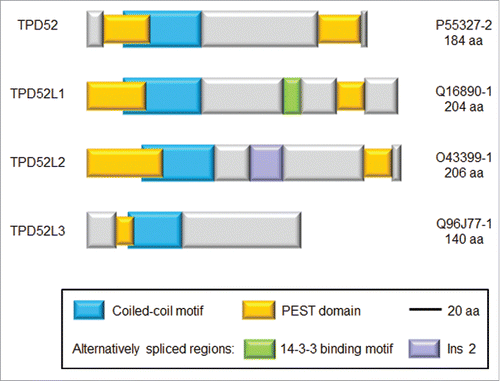
Table 1. Summary of TPD52-like protein functions as relevant to lipid droplets.
Early evidence
While Kamili et al.Citation7 were the first to demonstrate a LD function for TPD52, these results were not entirely unanticipated. However, individual studies linking TPD52 and cellular lipid storage had not drawn previous attention within the literature. The mouse Tpd52 ortholog was reported to be upregulated 5-fold in obese versus lean mouse adipose tissue,Citation13 and knock-down of the ancestral C. elegans TPD52 ortholog F13E6.1 was subsequently associated with reduced lipid storage.Citation14 However, this latter result was not supported by several whole genome RNAi screening studies in D. melanogaster,Citation15-17 although a proteomics study reported a moderate association between the ancestral D. melanogaster TPD52 ortholog CG5174 and LDs.Citation18 In both these C. elegans and D. melanogaster studies,Citation14,18 TPD52 orthologues were identified as anonymous proteins, and the nomenclature used suggested no link to mammalian TPD52 proteins (). Perhaps the closest “near miss” was the identification of TPD52 as a perilipin-1 (PLIN1) binding partner through yeast 2-hybrid screening.Citation19 PLIN1 is a member of the perilipin (PLIN) protein family (previously referred to as the PAT protein family), which are well characterized LD-associated proteins.Citation1 The same yeast 2-hybrid screening that identified TPD52 also identified CGI58,Citation19 a major regulator of PLIN1 function.Citation20 However, when GFP-tagged TPD52 was expressed in 3T3-L1 cells, GFP-TPD52 showed a perinuclear sub-cellular localization, contrasting with the obvious LD co-localization of GFP-tagged CGI58.Citation19 The authors may therefore have considered TPD52 as a screening false-positive, and there was no further published follow up of TPD52 as a PLIN1 partner.
Our group reconsidered these and other results following the publication of Kourtidis et al. in 2010, which described the reliance of ERBB2-positive breast cancer cell lines upon chromosome 17-encoded regulators of lipid metabolism.Citation21 At the time, we were analyzing the effects of TPD52 knock-down in breast cell lines.Citation22 Like Kourtidis et al.,Citation21 we noted that TPD52 knock-down decreased the survival of ERBB2-positive breast cancer cell lines,Citation22 which were characterized by higher levels of LD staining.Citation21 Breast and prostate cancers have been shown to accumulate cytoplasmic LDs,Citation23-25 and ERBB2 expression in breast cancer has been associated with a transcriptional program upregulating fatty acid synthase.Citation26 TPD52 also has a long history of being identified and studied as an androgen-induced gene/ protein,Citation27,28 and androgen treatment of prostate cancer cells increases cytoplasmic LDsCitation25 and regulates key anabolic enzymes.Citation29 Careful searches of the literature identified previous links between TPD52 and lipid storageCitation13,14,18,19,30,31 which were bolstered by numerous proteomic studies reporting the TPD52 paralogue TPD52L2 as a LD-associated protein ().Citation32-37 Armed with these collective results, we examined whether TPD52 expression led to increased cellular lipid storage, and whether TPD52 represented be a bona fide partner of PLIN proteins.Citation7
New findings
The approaches taken by Kamili et al.Citation7 were straightforward. Existing cell line models were examined to determine whether TPD52 expression status altered LD numbers, and this was the case in the 2 series of cell lines available (mouse Balb/c 3T3 cells and human MDA-MB-231 cells).Citation7 However, 3T3 cells expressing a related TPD52 protein (TPD52-like 1, or TPD52L1) () showed LD numbers similar to those in vector controls and parental cells.Citation7 Correspondingly, to our best knowledge, TPD52L1 has not been identified within any reported LD proteome (). We then studied TPD52-expressing 3T3 cell lines in more detail, and found that these contained more LDs following oleic acid supplementation than did control cells.Citation7 Two of 3 TPD52-expressing 3T3 cell lines also showed significantly increased lipogenesis from both de novo synthesis and exogenous uptake.Citation7 TPD52 partially co-localized with Golgi markers and Plin2/Adrp-coated LDs, with co-localization becoming more pronounced following oleic acid supplementation.Citation7 Interestingly TPD52 showed detectable interactions with both PLIN2/ADRP and PLIN3/TIP47 in the yeast 2-hybrid system (),Citation7 supporting the earlier identification of TPD52 as a PLIN1 partner.Citation19 However, as TPD52L1 did not detectably interact with PLIN proteins (), isoform-specific interactions between TPD52 and PLIN proteins may therefore underpin a function for TPD52 in promoting intracellular lipid storage.Citation7
How does TPD52 associate with lipid droplets?
While direct associations with PLINs may mediate TPD52 localization at LDs,Citation7 such associations do not exclude TPD52 also directly associating with the LD. Amphipathic helices may underpin protein associations with LDs and other organelles,Citation4,38 and although TPD52-like proteins include predicted amphipathic helices, their significance is unknown. Other hydrophobic LD targeting motifs identified in lipid-droplet associated proteinsCitation39,40 are not predicted in TPD52-like proteins. As TPD52L1 expression in 3T3 cells did not increase LD numbers and TPD52L1 did not detectably bind either PLIN2/ADRP or PLIN3/TIP47 in yeast 2-hybrid analyses,Citation7 mapping molecular differences between TPD52 and TPD52L1 may explain why TPD52 (and possibly TPD52L2) associates with LDs. Both TPD52 and TPD52L2 were identified as cholesterol-interacting proteins in HeLa cells, with both selectively binding a trans-sterol probe vs. a non-steroidal neutral lipid probe.Citation41 Furthermore, both TPD52 and TPD52L2 have been reported to bind the phosphoinositide PI(3,4,5)P3,Citation42 raising the possibility that a subset of TPD52-like proteins may bind lipid species directly.
Recent studies of protein binding to LDs have suggested a protein crowding model, where space is created as LDs expand, and proteins are crowded off as LDs shrink.Citation43 If TPD52 binds LDs directly or indirectly, increased detection of TPD52 at the LD surface under conditions where droplets are expanding would be in accordance with a crowding model ().Citation43 TPD52 binding to PLIN proteins would also be expected to result in increased TPD52 detection at the LD under conditions requiring lipid storage. However, the fact that TPD52 has been only infrequently identified as a component of the LD proteomeCitation44 agrees with only a small proportion of TPD52 being detected at LDs using fractionation studies, in marked contrast to Plin2/Adrp.Citation7 This disparity between the relative enrichment of Plin2/Adrp and TPD52 at LDsCitation7 argues against PLIN binding being the major determinant of TPD52 localization at the LD.
Figure 2. Possible mechanisms by which TPD52 might promote lipid storage in LDs, considering the predicted low abundance of TPD52 at the LD. TPD52 (shown in red) may traffic positive lipid storage regulators (shown in green) to the LD (1), and then may transiently bind the LD (2), prior to TPD52 release (3). TPD52 may (also) be recruited to bind and thereby remove negative storage regulators (shown in blue) from the LD surface (4). When bound to the LD, either in association with a positive storage regulator (5) or directly (6), TPD52 may also reduce the binding of negative storage regulators (7) by increasing local protein crowding.
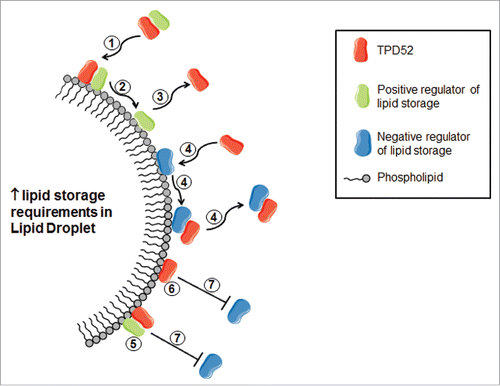
TPD52 functions at the lipid droplet
The actual function of TPD52 at the LD is also a matter of conjecture. Detection of TPD52 at LDs significantly increased in TPD52-expressing 3T3 cells treated with oleic acid,Citation7 which could reflect active TPD52 recruitment in response to increased storage requirements (), but could also more passively reflect increased LD numbers. The relatively small amount of TPD52 detected within the LD fraction of oleic acid-treated cellsCitation7 suggests a transient association, consistent with TPD52 trafficking other proteins to/ from the LD surface (). TPD52 could also compete within other LD proteins for binding, and thereby regulate the local abundance of other positive or negative regulators of lipid storage ().
In addition to the widely recognized differences in LD functions in different tissues, there are also multiple LD sub-types.Citation1,45 For example, Wifling et al. identified LD populations that either remained static or expanded in response to oleic acid supplementation.Citation45 Furthermore, the expanding LD sub-population was characterized by glycerol-3-phosphate acyltransferase-4 co-localization, a rate limiting enzyme in triglyceride synthesis.Citation45 The ability to synthesize triglyceride directly at the LD surface may allow for rapid storage, thereby avoiding lipotoxicity.Citation45 Infrequent detection of TPD52 within the LD proteomeCitation44 may also reflect that TPD52 associates with a subset of LDs within the cell, although a detailed analysis of TPD52 detection at LDs according to both size and responses to lipid loading will be required to determine whether this is true. As TPD52 expression in 3T3 cells was also associated with increased lipogenesis,Citation7 TPD52 could directly facilitate triglyceride synthesis, either by trafficking and/or tethering components of the synthetic machinery to the LD surface, and/or by regulating the expression or activity of specific enzymes.
TPD52 was also prominently detected at the Golgi in TPD52-expressing 3T3 cells, and TPD52 localization with Gm130 and Arl1 detection significantly increased following oleic acid treatment.Citation7 Whereas the formation of LDs within the endoplasmic reticulum has been the subject of intense research and discussion,Citation4 the role of the Golgi in LD biology has been studied to a lesser extent. However, a number of Golgi-associated proteins have been shown through RNAi screening approaches to regulate LD biology,Citation15,16 and other LD regulators can be detected at both the Golgi and LDs.Citation46,47 Further studies will be required to determine whether TPD52 functions at the Golgi are related to its LD functions.
Lipid droplets and cancer
Finally, Kamili et al.Citation7 also discussed the possible relevance of TPD52′s function to cancer. A number of cancer-related phenotypes have been ascribed to TPD52, including increased cell proliferation and invasive capacity and reduced apoptosis and DNA repair capacity.Citation10 There is therefore no shortage of explanations for why cancer cells might benefit from TPD52 overexpression, but these explanations are relatively generic and would not be expected to favor one cancer cell type over others. Where knock-down studies have been conducted in different breast cell lines, reduced TPD52 expression commonly reduced the survival of ERBB2-positive breast cancer cell lines,Citation21,22 suggesting a particular function in this cell type. ERBB2-positive breast cancer cell lines are characterized by increased LDs,Citation21 and ERBB2-positive breast cancers more frequently express proteins associated with lipogenesis or lipid storage.Citation48 Increased TPD52 expression could therefore favor the survival of cell lines with increased lipogenic activity by contributing to the increased lipid storage capacity that these cells require.Citation7 Again, further research will be necessary to determine whether there is an association between TPD52 expression and abundant LDs in cancer cell lines and the consequences of TPD52 knock-down in cancer cell lines with scarce versus abundant LDs.
Concluding remarks
Proteomic and RNAi screening approaches have collectively identified many LD-associated and -regulatory proteins.Citation5,6,14-18,32-27,43 Although TPD52 did not emerge as a strong candidate from these collective studies, the direct expression of TPD52 in cultured cells resulted in significantly increased LD numbers.Citation7 This finding suggests that there may be other important regulators of LDs identified through RNAi screening and proteomics approaches that are yet to be prioritised for further study. As demonstrated by Kamili et al.,Citation7 the direct testing of proteins that are infrequently but nonetheless recurrently identified by screening studies may identify bona fide LD regulators. While the analysis of possibly lower-abundance or itinerant LD proteins is likely to be more technically challenging, such proteins may provide a means to explore functional differences between LD subtypes and to subsequently exploit such differences for therapy. Understanding how proteins such as TPD52 “drop in” on the LD may therefore provide unexpected insights into the functions of other LD-associated proteins and how many LD regulators exert their functions from a distance within the cell.Citation6
Abbreviations
| LD | = | lipid droplet |
| PLIN | = | perilipin |
| TPD52 | = | tumor protein D52 |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- Beller M, Thiel K, Thul PJ, Jäckle H. Lipid droplets: a dynamic organelle moves into focus. FEBS Lett 2010; 584:2176-82; PMID:20303960; http://dx.doi.org/10.1016/j.febslet.2010.03.022
- Krahmer N, Farese RV Jr, Walther TC. Balancing the fat: lipid droplets and human disease. EMBO Mol Med 2013; 5:905-15; PMID:23740690; http://dx.doi.org/10.1002/emmm.201100671
- Nawrocki AR, Scherer PE. Keynote review: the adipocyte as a drug discovery target. Drug Discov Today 2005; 10:1219-30; PMID:16213414
- Thiam AR, Farese RV Jr, Walther TC. The biophysics and cell biology of lipid droplets. Nat Rev Mol Cell Biol 2013; 14:775-86; PMID:24220094; http://dx.doi.org/10.1038/nrm3699.
- Yang L, Ding Y, Chen Y, Zhang S, Huo C, Wang Y, Yu J, Zhang P, Na H, Zhang H, et al. The proteomics of lipid droplets: structure, dynamics, and functions of the organelle conserved from bacteria to humans. J Lipid Res 2012; 53:1245-53; PMID:22534641; http://dx.doi.org/10.1194/jlr.R024117
- Krahmer N, Hilger M, Kory N, Wilfling F, Stoehr G, Mann M, Farese RV Jr, Walther TC. Protein correlation profiles identify lipid droplet proteins with high confidence. Mol Cell Proteomics 2013; 12:1115-26; PMID:23319140; http://dx.doi.org/10.1074/mcp.M112.020230
- Kamili A, Roslan N, Frost S, Cantrill LC, Wang D, Della-Franca A, Bright RK, Groblewski GE, Straub BK, Hoy AJ, et al. TPD52 expression increases neutral lipid storage within cultured cells. J Cell Sci 2015; 128:3223-38; PMID:26183179; http://dx.doi.org/10.1242/jcs.167692
- Byrne JA, Tomasetto C, Garnier JM, Rouyer N, Mattei MG, Bellocq JP, Rio MC, Basset P. A screening method to identify genes commonly overexpressed in carcinomas and the identification of a novel complementary DNA sequence. Cancer Res 1995; 55:2896-903; PMID:7796418
- Boutros R, Fanayan S, Shehata M, Byrne JA. The tumor protein D52 family: many pieces, many puzzles. Biochem Biophys Res Commun 2004; 325:1115-21; PMID:15555543
- Byrne JA, Frost S, Chen Y, Bright RK. Tumor protein D52 (TPD52) and cancer-oncogene understudy or understudied oncogene? Tumour Biol 2014; 35:7369-82; PMID:24798974; http://dx.doi.org/10.1007/s13277-014-2006-x
- Chew CS, Chen X, Zhang H, Berg EA, Zhang H. Calcium/calmodulin-dependent phosphorylation of tumor protein D52 on serine residue 136 may be mediated by CAMK2delta6. Am J Physiol Gastrointest Liver Physiol 2008; 295:G1159-72; PMID:18832449; http://dx.doi.org/10.1152/ajpgi.90345.2008
- Messenger SW, Thomas DD, Falkowski MA, Byrne JA, Gorelick FS, Groblewski GE. Tumor protein D52 controls trafficking of an apical endolysosomal secretory pathway in pancreatic acinar cells. Am J Physiol Gastrointest Liver Physiol 2013; 305:G439-52; PMID:23868405; http://dx.doi.org/10.1152/ajpgi.00143.2013
- Nadler ST, Stoehr JP, Schueler KL, Tanimoto G, Yandell BS, Attie AD. The expression of adipogenic genes is decreased in obesity and diabetes mellitus. Proc Natl Acad Sci USA 2000; 97:11371-6; PMID:11027337
- Ashrafi K, Chang FY, Watts JL, Fraser AG, Kamath RS, Ahringer J, Ruvkun, G. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 2003; 421:268-72; PMID:12529643
- Guo Y, Walther TC, Rao M, Stuurman N, Goshima G, Terayama K, Wong JS, Vale RD, Walter P, Farese RV. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature 2008; 453:657-61; PMID:18408709; http://dx.doi.org/10.1038/nature06928
- Beller M, Sztalryd C, Southall N, Bell M, Jäckle H, Auld DS, Oliver B. COPI complex is a regulator of lipid homeostasis. PLoS Biol 2008; 6:e292; PMID:19067489; http://dx.doi.org/10.1371/journal.pbio.0060292
- Pospisilik JA, Schramek D, Schnidar H, Cronin SJ, Nehme NT, Zhang X, Knauf C, Cani PD, Aumayr K, Todoric J, et al. Drosophila genome-wide obesity screen reveals hedgehog as a determinant of brown versus white adipose cell fate. Cell 2010; 140:148-60; PMID:20074523; http://dx.doi.org/10.1016/j.cell.2009.12.027
- Cermelli S, Guo Y, Gross SP, Welte MA. The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr Biol 2006; 16:1783-95; PMID:16979555
- Yamaguchi T, Omatsu N, Omukae A, Osumi T. Analysis of interaction partners for perilipin and ADRP on lipid droplets. Mol Cell Biochem 2006; 284:167-73; PMID:16532261
- Yamaguchi T, Omatsu N, Matsushita S, Osumi T. CGI-58 interacts with perilipin and is localized to lipid droplets. Possible involvement of CGI-58 mislocalization in Chanarin-Dorfman syndrome. J Biol Chem 2004; 279:30490-7; PMID:15136565
- Kourtidis A, Jain R, Carkner RD, Eifert C, Brosnan MJ, Conklin DS. An RNA interference screen identifies metabolic regulators NR1D1 and PBP as novel survival factors for breast cancer cells with the ERBB2 signature. Cancer Res 2010; 70:1783-92; PMID:20160030; http://dx.doi.org/10.1158/0008-5472.CAN-09-1550
- Roslan N, Bieche I, Bright RK, Lidereau R, Chen Y, Byrne JA. TPD52 represents a survival factor in ERBB2-amplified breast cancer cells. Mol Carcinog 2014; 53:807-19; PMID:23661506; http://dx.doi.org/10.1002/mc.22038
- Fisher ER, Gregorio R, Kim WS, Redmond C. Lipid in invasive cancer of the breast. Am J Clin Pathol 1977; 68:558-61; PMID:920651
- Bässler R, Eckardt C. Histopathology and frequency of invasive breast cancer with lipid synthesis. In: Zander J, Baltzer J, editors. Early breast cancer: histopathology, diagnosis and treatment. Berlin: Springer-Verlag; 2005. p. 39-47
- Swinnen JV, Verhoeven G. Androgens and the control of lipid metabolism in human prostate cancer cells. J Steroid Biochem Mol Biol 1998; 65:191-8; PMID:9699873
- Kumar-Sinha C, Ignatoski KW, Lippman ME, Ethier SP, Chinnaiyan AM. Transcriptome analysis of HER2 reveals a molecular connection to fatty acid synthesis. Cancer Res 2003; 63:132-9, PMID:12517789
- Wang R, Xu J, Saramäki O, Visakorpi T, Sutherland WM, Zhou J, Sen B, Lim SD, Mabjeesh N, Amin M, et al. PrLZ, a novel prostate-specific and androgen-responsive gene of the TPD52 family, amplified in chromosome 8q21.1 and overexpressed in human prostate cancer. Cancer Res 2004; 64:1589-94; PMID:14996714
- Rubin MA, Varambally S, Beroukhim R, Tomlins SA, Rhodes DR, Paris PL, Hofer MD, Storz-Schweizer M, Kuefer R, Fletcher JA, et al. Overexpression, amplification, and androgen regulation of TPD52 in prostate cancer. Cancer Res 2004; 64:3814-22; PMID:15172988
- Massie CE, Lynch A, Ramos-Montoya A, Boren J, Stark R, Fazli L, Warren A, Scott H, Madhu B, Sharma N, et al. The androgen receptor fuels prostate cancer by regulating central metabolism and biosynthesis. EMBO J 2011; 30:2719-33; PMID:21602788; http://dx.doi.org/10.1038/emboj.2011.158
- Clement K, Viguerie N, Poitou C, Carette C, Pelloux V, Curat CA, Sicard A, Rome S, Benis A, Zucker JD, et al. Weight loss regulates inflammation-related genes in white adipose tissue of obese subjects. FASEB J 2004; 18:1657-69; PMID:15522911
- Keller MP, Choi Y, Wang P, Davis DB, Rabaglia ME, Oler AT, Stapleton DS, Argmann C, Schueler KL, Edwards S et al. A gene expression network model of type 2 diabetes links cell cycle regulation in islets with diabetes susceptibility. Genome Res 2008; 18:706-16; PMID:18347327; http://dx.doi.org/10.1101/gr.074914.107
- Brasaemle DL, Dolios G, Shapiro L, Wang R. Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3-L1 adipocytes. J Biol Chem 2004; 279:46835-42; PMID:15337753
- Umlauf E, Csaszar E, Moertelmaier M, Schuetz GJ, Parton RG, Prohaska R. Association of stomatin with lipid bodies. J Biol Chem 2004; 279:23699-709; PMID:15024010
- Sato S, Fukasawa M, Yamakawa Y, Natsume T, Suzuki T, Shoji I, Aizaki H, Miyamura T, Nishijima M. Proteomic profiling of lipid droplet proteins in hepatoma cell lines expressing hepatitis C virus core protein. J Biochem 2006; 139: 921-30; PMID:16751600
- Cho SY, Shin ES, Park PJ, Shin DW, Chang HK, Kim D, Lee HH, Lee JH, Kim SH, Song MJ, et al. Identification of mouse Prp19p as a lipid droplet-associated protein and its possible involvement in the biogenesis of lipid droplets. J Biol Chem 2007; 282:2456-65; PMID:17118936
- Wan HC, Melo RC, Jin Z, Dvorak AM, Weller PF. Roles and origins of leukocyte lipid bodies: proteomic and ultrastructural studies. FASEB J 2007; 21:167-78; PMID:17135363
- Hodges BD, Wu CC. Proteomic insights into an expanded cellular role for cytoplasmic lipid droplets. J Lipid Res 2010; 51:262-73; PMID:19965608; http://dx.doi.org/10.1194/jlr.R003582
- Drin G, Antonny B. Amphipathic helices and membrane curvature. FEBS Lett 2010; 584:1840-7; PMID:19837069; http://dx.doi.org/10.1016/j.febslet.2009.10.022
- Zehmer JK, Bartz R, Liu P, Anderson RG. Identification of a novel N-terminal hydrophobic sequence that targets proteins to lipid droplets. J Cell Sci 2008; 121:1852-60; PMID:18477614; http://dx.doi.org/10.1242/jcs.012013
- Ingelmo-Torres M, González-Moreno E, Kassan A, Hanzal-Bayer M, Tebar F, Herms A, Grewal T, Hancock JF, Enrich C, Bosch M, et al. Hydrophobic and basic domains target proteins to lipid droplets. Traffic 2009; 10:1785-801; PMID:19874557; http://dx.doi.org/10.1111/j.1600-0854.2009.00994.x
- Hulce JJ, Cognetta AB, Niphakis MJ, Tully SE, Cravatt BF. Proteome-wide mapping of cholesterol-interacting proteins in mammalian cells. Nat Methods 2013; 10:259-64; PMID:23396283; http://dx.doi.org/10.1038/nmeth.2368
- Catimel B, Yin MX, Schieber C, Condron M, Patsiouras H, Catimel J, Robinson DE, Wong LS, Nice EC, Holmes AB, et al. PI(3,4,5)P3 interactome. J Proteome Res 2009; 8:3712-26; PMID:19463016; http://dx.doi.org/10.1021/pr900320a
- Kory N, Thiam AR, Farese RV Jr, Walther TC. Protein crowding is a determinant of lipid droplet protein composition. Dev Cell 2015; 34:351-63; PMID:26212136; http://dx.doi.org/10.1016/j.devcel.2015.06.007
- Saka HA, Thompson JW, Chen YS, Dubois LG, Haas JT, Moseley A, Valdivia RH. Chlamydia trachomatis infection leads to defined alterations to the lipid droplet proteome in epithelial cells. PLoS One 2015; 10:e0124630; PMID:25909443;http://dx.doi.org/10.1371/journal.pone.0124630
- Wilfling F, Wang H, Haas JT, Krahmer N, Gould TJ, Uchida A, Cheng JX, Graham M, Christiano R, Fröhlich F, et al. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev Cell 2013; 24: 384-99; PMID:23415954; http://dx.doi.org/10.1016/j.devcel.2013.01.013.
- Bouvet S, Golinelli-Cohen MP, Contremoulins V, Jackson CL. Targeting of the Arf-GEF GBF1 to lipid droplets and Golgi membranes. J Cell Sci 2013; 126:4794-805; PMID:23943872; http://dx.doi.org/10.1242/jcs.134254
- Gannon J, Fernandez-Rodriguez J, Alamri H, Feng SB, Kalantari F, Negi S, Wong AH, Mazur A, Asp L, Fazel A, et al. ARFGAP1 is dynamically associated with lipid droplets in hepatocytes. PLoS One 2014; 9:e111309; PMID:25397679; http://dx.doi.org/10.1371/journal.pone.0111309
- Kim S, Lee Y, Koo JS. Differential expression of lipid metabolism-related proteins in different breast cancer subtypes. PLoS One 2015; 10:e0119473; PMID:25751270; http://dx.doi.org/10.1371/journal.pone.0119473
