ABSTRACT
The present study aimed to identify genes exhibiting concomitant obesity-dependent changes in DNA methylation and gene expression in adipose tissues in the mouse using diet-induced obese (DIO) C57BL/6J and genetically obese ob/ob mice as models. Mature adipocytes were isolated from epididymal and inguinal adipose tissues of ob/ob and DIO C57BL/6J mice. DNA methylation was analyzed by MeDIP-sequencing and gene expression by microarray analysis. The majority of differentially methylated regions (DMRs) were hypomethylated in obese mice. Global methylation of long interspersed elements indicated that hypomethylation did not reflect methyl donor deficiency. In both DIO and ob/ob mice, we observed more obesity-associated methylation changes in epididymal than in inguinal adipocytes. Assignment of DMRs to promoter, exon, intron and intergenic regions demonstrated that DIO-induced changes in DNA methylation in C57BL/6J mice occurred primarily in exons, whereas inguinal adipocytes of ob/ob mice exhibited a higher enrichment of DMRs in promoter regions than in other regions of the genome, suggesting an influence of leptin on DNA methylation in inguinal adipocytes. We observed altered methylation and expression of 9 genes in epididymal adipocytes, including the known obesity-associated genes, Ehd2 and Kctd15, and a novel candidate gene, Irf8, possibly involved in immune type 1/type2 balance. The use of 2 obesity models enabled us to dissociate changes associated with high fat feeding from those associated with obesity per se. This information will be of value in future studies on the mechanisms governing the development of obesity and changes in adipocyte function associated with obesity.
Introduction
Nutrient excess as well as deficit during fetal development may predispose to obesity and diabetes in humans and animals, and mouse studies have shown that this effect can be transmitted to subsequent generations through epigenetic changes such as histone modifications and DNA methylation in the germ line.Citation1,2 Obesity and accompanying insulin resistance may also affect DNA methylation in adipose tissue leading to phenotypic changes in the adipocytes.Citation3,4
DNA methylation is implicated in the regulation of metabolism, and differential methylation has been identified in the promoter of several genes associated with obesity development and adipocyte function. Thus, dexamethasone treatment of mice leads to decreased DNA methylation at the Cebpa promoter inducing a shift in the preference of bone marrow stromal cells to favor adipocyte over osteoblast development.Citation5 Moreover, methylation of the cAMP response elements in the Ucp1 promoter has been suggested to repress expression of Ucp1 in white adipocytes.Citation6 Intake of high fat diet has been shown to increase methylation of the leptin promoter in retroperitoneal adipocytes in rats, and this was associated with lower circulating leptin levels.Citation7 A recent study also demonstrated that long-term high fat feeding induced hypermethylation of both the Leptin and the Pparg promoter in gonadal, but not subcutaneous adipose tissueCitation8 Finally, it has been shown that the adiponectin promotor is hypermethylated in adipocytes from diet-induced obese (DIO) C57BL/6J mice and db/db mice. This was associated with decreased insulin sensitivity, which was relieved by treatment with the DNMT inhibitor RG108.Citation9
The majority of studies investigating DNA methylation in mouse models of obesity has focused on specific genes.Citation6,7,9 Only few studies have performed genome-wide DNA methylation analysis in adipose tissue or isolated mature adipocytes from mice.Citation10,11
In this study, we compared obesity-associated changes in DNA methylation of mature adipocytes from DIO C57BL/6J and genetically obese (ob/ob) mice, representing 2 widely used models for diet- and genetically induced obesity, respectively, to identify obesity associated changes. The aim was to identify genes exhibiting concomitant obesity-dependent changes in DNA methylation and gene expression in adipose tissue.
Results
The distribution of differentially methylated regions (DMRs) differs between diet-induced obese C57BL/6J and ob/ob mice
To identify changes in methylation patterns associated with genetically determined obesity and diet-induced obesity we isolated mature adipocytes from 2 obesity models. Ob/ob mice on a chow diet were chosen as a model for genetically induced obesity, whereas diet-induced obesity was induced by high fat feeding of C57BL/6J mice for 15 weeks. Age-matched chow fed C57BL/6J mice were used as lean controls in both experiments. Both DIO mice and ob/ob mice were significantly heavier than their corresponding controls, and importantly, the 2 models had similar body masses at sacrifice (Fig. S1).
DNA methylation in mature adipocytes from inguinal and epididymal fat pads was assessed using MeDIP-seq. On average, 170 million paired-end reads per sample were obtained from sequencing. After filtering PCR duplicates, mapped reads without proper pairing, and poor quality alignments (Mapping quality <30), approx. 35% of reads from each sample were used for further analyses (Fig. S2). Assignment of DMRs to promoter, exon, intron and intergenic regions demonstrated that changes in DNA methylation associated with DIO in C57BL/6J mice were primarily enriched in exons (Fig. S3). This tendency was less pronounced in ob/ob mice. Compared to epididymal adipocytes in ob/ob and both epididymal and inguinal adipocytes in DIO mice, inguinal adipocytes of ob/ob mice exhibited a higher enrichment of DMRs in promoter regions than in other regions of the genome, suggesting a specific influence of leptin on DNA methylation in the more plastic inguinal adipocytes.
Obesity is associated with hypomethylation
Differential methylation as represented in volcano plots () revealed that hypomethylation was dominating in epididymal and inguinal adipose tissue from both obesity models (details are summarized in , and DMR region details for individual comparisons are listed in Supplementary Tables ST1a-d, available at http://www.cbs.dtu.dk/suppl/Sonne2016/). Nutritional factors influencing the supply of methyl donors are known to affect DNA methylation.Citation12 Therefore, amino acid and homocysteine levels were measured in plasma samples from the DIO mice. Amino acid profiling showed that the plasma levels of 3 amino acids, 1-methyl-L-histidine, L-methionine and arginine were significantly lower (p-value < 0.01) in obese than in lean mice (Table ST2). By contrast, we detected no significant difference in the level of total homocysteine in the plasma samples from obese and lean mice. To further investigate whether the observed obesity related hypomethylation reflected a general decrease in methylation, global DNA methylation was assessed based on methylation of LINE repeats. We observed no changes in global methylation in obese mice indicating that the observed hypomethylation was not a result of methyl donor insufficiency (Fig. S4).
Figure 1. DNA methylation in diet-induced obese and genetically obese ob/ob mice. Volcano plots showing mean methylation differences between mature adipocytes from inguinal and epididymal adipose tissue from obese vs lean mice. Ob/ob (n = 4) or DIO (n = 3). Significantly different methylation sites are shown as blue spots, non-significant are shown as red spots (adjusted p-value <0.001). DIO: Diet-induced obese.
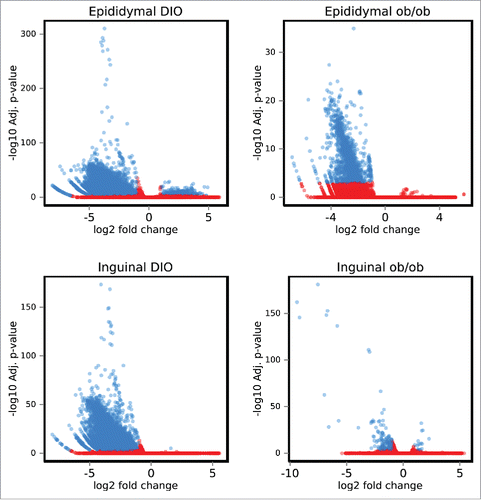
Table 1. Differentially methylated regions in epididymal and inguinal adipocytes from diet-induced and ob/ob mice. Proportion of hyper- and hypo-methylated regions and number of genes mapped within the +/−10kb region of the DMRs. DMRs: Differentially methylated regions. DIO: Diet-induced obese.
DMRs are predominantly shared between inguinal and epididymal tissues in the DIO mice
The mechanism by which DNA methylation modulates transcription is not completely understood, but it may affect distant regions as well as regions proximal to the promoter.Citation13 DMRs were assigned to genes using a distance-based strategy (+/− 10kb of gene boundaries) as described in the methods. Comparison of DMR-associated genes revealed that the highest number of shared genes (2004) were found between inguinal and epididymal adipocytes in the DIO mice, whereas only a few of the identified DMR-associated genes were shared between adipocytes from the 2 models ().
Figure 2. Differentially methylated regions in diet-induced obese and genetically obese ob/ob mice. [A] Genes harboring differentially methylated regions in adipocytes in inguinal and epididymal adipose tissue of diet-induced obese C57BL/6J mice (n = 3) and genetically obese ob/ob mice (n = 4). The highest overlap of 2004 genes was found between inguinal and epididymal adipose tissue from DIO mice (adjusted p-value <0.001). DIO: Diet-induced obese. [B] Venn diagram showing the number of overlapping genes harboring differentially methylated regions in diet-induced obese male C57BL/6J mice (n = 3) and genetically obese male ob/ob mice (n = 4). [A] epididymal adipose tissue and [B] inguinal adipose tissue. Of the 55 overlapping genes with obesity-associated methylation sites as well as altered expression in epididymal adipocytes (adjusted p-value <0.1), 9 genes with abs. log2FC in gene expression in epididymal tissue >1 are enlisted in the figure. DIO: Diet-induced obese. [C] Top 3 motifs predicted in the DMRs based on 16 hypomethylated genes shared by inguinal adipocytes in genetically obese male ob/ob mice (n = 3) and diet-induced obese male C57BL/6J mice (n = 3). [D] Three of the top 7 motifs predicted in the DMRs (see Table S4 for a full list), based on 302 hypomethylated genes shared by epididymal adipocytes in genetically obese male ob/ob mice (n = 3) and diet-induced obese male C57BL/6J mice (n = 3). DIO: Diet-induced obese. Letter size reflects the prevalence of the nucleotide at the position in the motif.
![Figure 2. Differentially methylated regions in diet-induced obese and genetically obese ob/ob mice. [A] Genes harboring differentially methylated regions in adipocytes in inguinal and epididymal adipose tissue of diet-induced obese C57BL/6J mice (n = 3) and genetically obese ob/ob mice (n = 4). The highest overlap of 2004 genes was found between inguinal and epididymal adipose tissue from DIO mice (adjusted p-value <0.001). DIO: Diet-induced obese. [B] Venn diagram showing the number of overlapping genes harboring differentially methylated regions in diet-induced obese male C57BL/6J mice (n = 3) and genetically obese male ob/ob mice (n = 4). [A] epididymal adipose tissue and [B] inguinal adipose tissue. Of the 55 overlapping genes with obesity-associated methylation sites as well as altered expression in epididymal adipocytes (adjusted p-value <0.1), 9 genes with abs. log2FC in gene expression in epididymal tissue >1 are enlisted in the figure. DIO: Diet-induced obese. [C] Top 3 motifs predicted in the DMRs based on 16 hypomethylated genes shared by inguinal adipocytes in genetically obese male ob/ob mice (n = 3) and diet-induced obese male C57BL/6J mice (n = 3). [D] Three of the top 7 motifs predicted in the DMRs (see Table S4 for a full list), based on 302 hypomethylated genes shared by epididymal adipocytes in genetically obese male ob/ob mice (n = 3) and diet-induced obese male C57BL/6J mice (n = 3). DIO: Diet-induced obese. Letter size reflects the prevalence of the nucleotide at the position in the motif.](/cms/asset/95c8ef6f-7dcf-4de2-ab51-c5b72f9f6e06/kadi_a_1320002_f0002_oc.gif)
Among the DMRs found in DIO and ob/ob mice, 306 genes in epididymal adipocytes and 16 genes in inguinal adipocytes contained DMRs in both models. We term such sites obesity associated (, Table ST3a and ST3b, available at http://www.cbs.dtu.dk/suppl/Sonne2016/). Interestingly, a subset of these revealed opposite methylation patterns, underlining the difference between the 2 obesity models. Of the 306 identified DMRs in epididymal adipocytes, 302 were hypomethylated. A motif search of the 16 and 302 hypomethylated regions identified 3 () and 7 significant motifs, respectively (Table ST4). Of the 7 significant motifs in the epididymal adipocytes motif 3, 4 and 6 harbor binding sites for transcription factors Zic1, a reported marker for classical brown adipocytes,Citation14 Myf6, a myocyte marker,Citation15 and the cell cycle regulators E2f2/E2f3,Citation16 respectively ().
DNA methylation changes in epididymal fat from both models are associated with changes in gene expression of known obesity-related genes
To examine if the obesity-associated changes in DNA methylation were accompanied by changes in gene expression, a second experiment was performed to isolate RNA from mature adipocytes from inguinal and epididymal fat pads from DIO mice and corresponding lean controls. As in the first experiments, DIO mice gained significantly more weight than their RD littermates and exhibited significantly higher masses of inguinal and epididymal white, as well as interscapular brown adipose tissue, and liver (Fig. S5). In this experiment, we found 1135 genes differentially expressed in mature adipocytes from epididymal tissue of DIO vs control mice, with a fold change >2 and adjusted p-value <0.1. In line with the observed hypomethylation patterns, expression of the majority, 757 genes, was upregulated while expression of 378 genes was downregulated in DIO mice (Figure S6a, Table ST5, available at http://www.cbs.dtu.dk/suppl/Sonne2016/). Comparing obese mice with the lean control mice, 411 genes were differentially expressed in mature adipocytes from inguinal adipose tissue (Figure S6b, Table ST6, available at http://www.cbs.dtu.dk/suppl/Sonne2016/). Of these, expression of 304 genes was upregulated and expression of 107 genes downregulated in the DIO mice.
Of the obesity-associated methylation sites shared between genetically obese and DIO mice, 55 genes in epididymal adipocytes and one gene in inguinal adipocytes also showed altered expression (adjusted p-value < 0.1). Of the 55 genes in epididymal adipocytes, 9 showed a fold change of more than 2. These were all hypomethylated and expression of 5 genes (Arhgap6, Irf8, Prcp, Setd6, Shank3) was upregulated whereas expression of 4 genes (Ehd2, Kctd15, Pde1a, Reep6) was downregulated (Table ST7, available at http://www.cbs.dtu.dk/suppl/Sonne2016/).
Among the genes downregulated in epididymal adipocytes, a few have earlier been described in the context of obesity. The Ehd2 gene is involved in trafficking of SLC2A4 to the plasma membrane, a process that is impaired in hypertrophic adipocytes independent of inflammation.Citation17 In the Kctd15 risk gene, SNPs were found to be associated with BMI (P = 2.6E–07, β = 0.06)Citation18 and with obesity and obesity-related traits (P < 0.005, odds ratio = 1.54).Citation19 The upregulated genes include Prcp, previously associated with obesity. PRCP-deficient mice have decreased fat mass and improved glucose tolerance and insulin sensitivity.Citation20 In line with this, elevated levels of serum PRCP have been described in obese subjects.Citation21
DNA methylation changes in diet-induced obesity associate with expression of genes involved in adipogenesis
As the overlap in obesity-associated DMRs between epididymal and inguinal adipocytes was higher in DIO mice than in ob/ob mice (), results obtained from DIO mice were used for further analyses integrating DNA methylation and gene expression data. The analyses identified subsets of genes exhibiting the canonical negative correlation between methylation and gene expression, where hypomethylation of gene was associated with increased expression (, green box). On the other hand, a subset of genes exhibited unexpected correlations between methylation and gene expression, where hypomethylation in various gene regions was associated with decreased expression (, red box), a phenomenon described previously in transformed cells.Citation22
Figure 3. Correlation between DNA methylation and gene expression changes. [A] Box plot showing the median methylation fold changes in the 3 classes of gene expression, upregulated (log2(FC)>1, green), downregulated (log2(FC)<−1, red) and genes with no significant difference in gene expression (gray). The methylation levels on the y-axis were quantified as hypermethylated in diet-induced obesity if methylation level was greater than zero where as hypomethylated if methylation level was less than zero. The box represents the distribution of methylation states in each gene expression class, where the horizontal line denotes median methylation status of the group and the box represents the first and third quantiles. The dots represent above third quantile and below first quantile. [B] Number of overlapping genes exhibiting differences in gene expression (n = 5) and DNA methylation (n = 3) in epididymal and inguinal adipocytes from diet-induced obese C57BL/6J mice. Venn diagram represents the overlap between genes with differential gene expression along with differential methylation between inguinal and epididymal adipocytes from DIO C57BL/6J mice. DIO: Diet-induced obese.
![Figure 3. Correlation between DNA methylation and gene expression changes. [A] Box plot showing the median methylation fold changes in the 3 classes of gene expression, upregulated (log2(FC)>1, green), downregulated (log2(FC)<−1, red) and genes with no significant difference in gene expression (gray). The methylation levels on the y-axis were quantified as hypermethylated in diet-induced obesity if methylation level was greater than zero where as hypomethylated if methylation level was less than zero. The box represents the distribution of methylation states in each gene expression class, where the horizontal line denotes median methylation status of the group and the box represents the first and third quantiles. The dots represent above third quantile and below first quantile. [B] Number of overlapping genes exhibiting differences in gene expression (n = 5) and DNA methylation (n = 3) in epididymal and inguinal adipocytes from diet-induced obese C57BL/6J mice. Venn diagram represents the overlap between genes with differential gene expression along with differential methylation between inguinal and epididymal adipocytes from DIO C57BL/6J mice. DIO: Diet-induced obese.](/cms/asset/9a422020-08d7-4c71-8e50-e7dd3e28881e/kadi_a_1320002_f0003_oc.gif)
In total, we identified 103 genes in epididymal adipocytes and 39 genes in inguinal adipocytes with concomitant differential methylation (adjusted p-value < 0.001) and expression (adjusted p-value < 0.1) (). Of these, 24 genes were shared between the 2 adipose tissues (Table ST8, available at http://www.cbs.dtu.dk/suppl/Sonne2016/). Among the shared downregulated genes was Lipe, encoding one of the major enzymes involved in lipolysis in adipocytes. Further, expression of Aacs, encoding a ketone-using enzyme, providing acetyl-CoA used for synthesis of fatty acids and cholesterol, was downregulated, and knockout of Aacs in mice leads to suppressed expression of adipocyte markers like Pparg and Cebpa.Citation23 Furthermore, Nrp2, a lymphatic vessel developmental gene was hypomethylated and demonstrated increased gene expression in both inguinal and epididymal adipocytes possibly reflecting neovascularization associated with adipose tissue growth.Citation24
Genes exhibiting tissue-specific regulation included Zinc-finger nuclear protein (Zfp521), which acts as a repressor of adipogenesis.Citation25 Surprisingly, we found Zfp521 expression upregulated in epididymal adipose tissue in obese mice. In the inguinal tissue, we observed upregulation of Pkig expression, possibly favoring adipocyte differentiation over osteogenesisCitation26 and increased expression of Cpt1a, reported to promote adipocyte differentiation by preserving insulin sensitivity.Citation27
Discussion
MeDIP sequencing of DNA from mature adipocytes demonstrated profound changes in patterns and levels of DNA methylation associated with both diet-induced and genetically determined obesity. Interestingly, DMRs were predominantly hypomethylated in the obese state. Global hypomethylation has in previous studies been associated with methyl donor insufficiency due to lack of essential nutrients such as folate, vitamin B6 and B12, riboflavin or choline in the diet.Citation28 In the current study, lower serum levels of 1-methyl-L-histidine, L-methionine and arginine were observed in DIO mice compared with lean controls, but homocysteine levels were unchanged. Furthermore, no differences in methylation of LINE repeat regions indicated that the observed hypomethylation was not global, and probably not a result of methyl donor insufficiency. Hence, it is likely that obesity leads to specific changes in DNA methylation, and the majority of differentially methylated regions are demethylated leading to increased DNA accessibility. This is in line with a previous study reporting global hypomethylation in adipocytes of post-obese vs never-obese women.Citation29
Comparison of the 2 obesity models identified a subset of 306/16 genes that were differentially methylated in mature adipocytes from epididymal/inguinal adipose tissue, irrespective of whether the excess fat mass was due to intake of a high fat diet (diet-induced obesity) or chow overeating (ob/ob). Interestingly, we found more differentially methylated genes in the diet-induced obesity model than the ob/ob model. This seemed inversely correlated with tissue expansion, as a higher proportion of the weight gain in ob/ob mice is due to increased fat mass.Citation30 However, it has been shown that adipose tissues from DIO mice are more prone to inflammation than ob/ob mice,Citation31 and inflammation is more pronounced in visceral compared with subcutaneous adipose tissue.Citation31 Since we observed more changes in epididymal than inguinal adipose tissue, we suggest that the differential methylation may reflect the inflammatory state of the tissue. In line with this, a large proportion of the DMRs is associated with pro-inflammatory genes expressed in the mature adipocytes themselves, including Ccl2 which is expressed in mature adipocytes, or residual immune cells remaining in the fraction containing the mature adipocytes (Table ST9).
Previous studies of obesity-related methylation changes in mice are not directly comparable. Fan et al. used whole epididymal fat padsCitation10 while Multhaup et al. did not specify the adipose tissue for collection of mature adipocytes.Citation11 Similar to our findings, Fan et al. found more hypo- than hyper-methylated regions in epididymal tissue after high fat feeding. The diet-associated genes identified by Fan et al.,Citation10 Kcnh2, Mboat7, Fzr1, Kctd5, Trpm4 and Ssr2 were also hypomethylated in mature adipocytes from both epididymal and inguinal adipose tissue in our study and Hmg20a was hypermethylated in epididymal adipose tissue in both studies.
Multhaup et al. used a more complicated cross-species setup, comparing DNA methylation changes in mature adipocytes from DIO vs lean mice to subcutaneous adipocytes from obese and lean humans.Citation11 In line with the observed hypermethylation of Tmcc3 in epididymal adipose tissue of DIO mice in our study, Multhaup et al. observed hypermethylation of Tmcc3 in high fat fed mice and obese humans. However, this was not accompanied by significant changes in gene expression. Comparing the findings by Multhaup et al. to our results, 16 DMRs displayed the same direction of methylation changes in epididymal tissue in DIO.Citation11
Comparison of the regions that were differentially methylated in epididymal and inguinal adipose depots in the 2 mouse models of obesity allowed us to dissociate changes in methylation patterns associated with HF feeding from those associated with obesity. In epididymal adipose tissue, we identified a subset of 9 genes, which were differentially methylated and associated with changes in gene expression in both obesity models. Of these Ehd2, Kctd15 and Prcp have previously been associated with obesity.Citation17,18,21 The gene encoding the transcription factor, IRF8, which is necessary for production of IgG2a and mounting a type 1 immune responseCitation32 was hypomethylated and exhibited upregulated expression in obesity. This is in line with the finding that visceral adipose tissue inflammation and insulin resistance is dominated by a type 1 response.Citation33 Interestingly, a type 1 rather than a type 2 response dominates in C57BL/6J mice,Citation34 which may explain why C57BL/6J mice are more prone to inflammation, obesity and insulin resistance. Furthermore, IRFs are suggested to play a role in adipocyte differentiation and Irf8 has been shown to have a higher expression in mature adipocytes as compared with the stromal-vascular fraction and F4/80+ macrophages.Citation35
One problem associated with the present study and previous studies relates to the presence of cells from the stromal vascular fraction, including immune cells, in the isolated mature adipocytes. While elaborate FACS-based protocols minimizing the presence of cells from the stromal vascular fraction in the fraction representing the mature adipocytes have been developed,Citation36 they still do not guarantee the complete absence of cells from the stromal vascular fraction. To completely rule out such contamination, analyses based on single isolated adipocytes would seem to be required. We used available published data and BioGPSCitation37 (http://biogps.org) to examine to what extent the genes exhibiting concurrent changes in DNA methylation and expression represented genes expressed in mature adipocyte. This survey showed that all these genes with the exception of Arhgap6 are reported to be expressed in mature adipocytes, but also showed that Irf8, Prcp, Setd6, Shank3, Nrp2 Zfp521, Pkig, and Cpt1a are expressed in cell of the stromal vascular fraction that may contaminate the mature adipocytes. However, the use of 2 distinct models of obesity in this study would point to the mature adipocytes as a prominent contributor to the observed changes in DNA methylation and gene expression reported in the present study.
Thus, in summary we provide information on the correlation between changes in DNA methylation patterns and gene expression in a preparation of mature adipocytes from epididymal and inguinal adipose tissues in relation to obesity. By comparing DIO and genetically obese mice, we were able to dissociate changes associated with high fat feeding from those associated with obesity per se. This information will be of value in future studies on the mechanisms governing the development of obesity and changes in adipocyte function associated with obesity. In general, obesity was associated with pronounced global DNA hypomethylation, with epididymal fat exhibiting more obesity-associated DMRs than inguinal fat. Among genes exhibiting concurrent changes in methylation and gene expression we identified several known obesity-associated genes, and in addition, our analysis pointed to the transcription factor Irf8 as a new candidate gene possibly involved in the function of adipose tissue in obese mice through hypomethylation driven expression changes.
Methods
Experimental design (animal model and DNA isolation)
Diet-induced obesity model: Four-week old male C57BL/6J (wt) mice obtained from the Jackson Laboratory were fed a regular diet (RD: ∼20 E% fat Mouse diet 20, 5058, Picolab) or a high fat diet (HFD: 60 E% fat, D12492, Research Diets Inc.) for 15 weeks (n = 3). Genetically obese model: Nine-week old male ob/ob and wild type (wt) C57BL/6J mice (n = 4) were obtained from the Jackson Laboratory and fed a chow diet corresponding to the regular diet in the diet-induced obesity model.
Mature adipocytes were isolated from epididymal and inguinal adipose tissues from lean and obese mice by digesting with collagenase D (1.5U/mL) and dispase II (2.4U/mL) in 4% BSA in PBS supplemented with 10mM CaCl2 at 37°C with constant agitation for 15 or 25 minutes for eWAT and iWAT, respectively, the completely digested sample was washed twice with 4% BSA in PBS and the floating layer of mature adipocytes collected. DNA was isolated using the DNeasy kit (Qiagen) and used for MeDIP sequencing.
A separate experiment was performed to investigate global gene expression differences in mature adipocytes corresponding to methylation changes. Twenty 4-week old male C57BL/6J mice were obtained from the Jackson Laboratory and fed either RD (n = 10) or a HFD (n = 10) as described above. After 15 weeks of feeding, plasma was collected for quantification of amino acids and metabolites. Mature adipocytes were collected from epididymal and inguinal adipose tissue as described above. Total RNA was extracted using Trizol LS (Invitrogen), DNAse treated (Qiagen) and LiCl precipitated.
MeDIP-seq library preparation
A library was prepared from 10 μg of DNA as described previously.Citation38 Briefly, DNA was fragmented. End repair, <A> base addition and adaptor ligation were performed using Illumina's Paired-End DNA Sample Prep kit following the manufacturer's instructions. Adaptor-ligated DNA was immunoprecipitated by anti-5mC, and MeDIP products were validated by qPCR using SYBR green mastermix (Applied Biosystems) and primers for positive and negative control regions supplied in the MeDIP kit (Diagenode). MeDIP DNA was purified with ZYMO DNA Clean & Concentrator-5 column following the manufacturer's instructions and amplified by adaptor-mediated PCR in a final reaction volume of 50 μL. Amplification quality and quantity were evaluated using Agilent 2100 bioanalyzer and DNA 1000 chips (Agilent Technology). Paired-end libraries were constructed and subjected to Illumina HiSeq2000 sequencing.
MeDIP-Seq data analysis
The paired-end reads from the MeDIP sequencing were checked for quality using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The quality checked reads were mapped to the mouse reference genome build mm9 using Bowtie2Citation39 for each sample independently. Mapped reads were filtered for mapping quality 30 and sorted using Picard (http://picard.sourceforge.net) and SAMtools,Citation40 whereas duplicates were removed using Picard MarkDuplicates (http://picard.sourceforge.net). Aligned reads were filtered for missing mates in the alignments. Using the mapped reads, the correlation between replicates was checked using Spearman correlation coefficient. Mapped data in BAM format were further analyzed to find differentially methylated regions (DMRs) between the ob/ob vs wt and HFD vs RD in epididymal and inguinal adipose tissue. In the MEDIPSCitation41 package of R, reads mapped to the genome were extended to 300 nucleotides to account for all CpGs in the region. The genome was divided into non-overlapping bins of 250 nucleotides during this analysis and reads mapped per region were counted. Relative methylation scores (rms) were calculated for each bin of the genome by counting the number of mapped reads. Normalization was applied on this count data to convert it to reads per million. MEDIPS internally uses the edgeR package to identify differentially methylated regions between the samples. EdgeR uses a negative binomial distribution (especially useful for discrete count data) to identify the DMRs between the 2 states under comparison, and thus, calculates mean methylation values (rpm, rms, ams), log fold changes, variances and p-values by comparing 2 sample sets. The DMRs were mapped and assigned to a gene if they were placed within 10kB +/− of the gene boundary.
Microarray analysis
The quality and quantity of RNA were determined using a Bioanalyzer nano kit (Agilent Technologies) and Qubit RNA BR Assay (Life Technologies), respectively. RNA from the 5 mice with the best RNA quality (RIN > 6.7) was chosen for subsequent analysis. Gene expression profiles were determined using the Mouse Agilent 4 × 44 v2 gene expression arrays (Agilent Technologies).
The single color microarray data was analyzed using the limma package.Citation42 Background correction and normalization of the data was done based on the negative controls. After normalization, we used the Bayesian method in the limma package to identify genes that exhibited differential expression between RD and HFD mice. Gene expression levels were determined as the median expression value of the probes corresponding to a given gene. Differential expression was filtered based on an FDR corrected p-value of less than 0.1 (corresponding to an FDR of 10%).
Integrative methods
The regions exhibiting differential methylation (DMRs) between RD and HFD were mapped to genes with differential gene expression (DGE). If the DMR fell within the gene boundary or +/− 10 KB of the gene boundary, the DMR was considered likely to affect gene expression. The genes with DMRs and DGE were only considered for further analysis if the fold change in expression between the 2 conditions was greater than 2.
Functional analysis
Using the MEME suite,Citation43 we performed de novo motif analysis of genomic regions, which were shared between DIO and ob/ob mice in epididymal and inguinal adipose tissues.
Determination of free amino acids and homocysteine in plasma
Plasma samples were collected from RD and HFD fed mice at termination and profiled for levels of amino acids. Plasma samples were first deproteinized by adding 10% sulfosalicylic acid (1:1 v/v) and centrifuged. The supernatants were filtered and the concentration of free amino acids were determined by using a Biochrom 30+ Amino Acid Analyzer as described previously.Citation44 For the analysis of total homocysteine, DL-dithiothreitol (DTT) was added to a final concentration of 1.2%. Samples were deproteinized and further analyzed as described for free amino acids.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Author contributions
SK, KK, JW, RG and LM designed and supervised the study. SK, SBS, MDD and LSM performed the experiments, RY and YG did the data analyses and SBS, RY, SK, LM and KK wrote the manuscript.
KADI_A_1320002_Supplemental.zip
Download Zip (2.4 MB)Funding
This work was supported by the Benzon foundation (SBS), the Danish Council for Strategic Research (KK), the Danish and Swedish childhood cancer foundations (RY and RG) and NIH grant DK097441 to SK.
References
- Dunn GA, Bale TL. Maternal high-fat diet effects on third-generation female body size via the paternal lineage. Endocrinology 2011; 152(6):2228-36; PMID:21447631; https://doi.org/10.1210/en.2010-1461
- Jimenez-Chillaron JC, Isganaitis E, Charalambous M, Gesta S, Pentinat-Pelegrin T, Faucette RR, Otis JP, Chow A, Diaz R, Ferguson-Smith A, et al. Intergenerational transmission of glucose intolerance and obesity by in utero undernutrition in mice. Diabetes 2009; 58(2):460-8; PMID:19017762; https://doi.org/10.2337/db08-0490
- Benton MC, Johnstone A, Eccles D, Harmon B, Hayes MT, Lea RA, Griffiths L, Hoffman EP, Stubbs RS, Macartney-Coxson D. An analysis of DNA methylation in human adipose tissue reveals differential modification of obesity genes before and after gastric bypass and weight loss. Genome Biol 2015; 16:8; PMID:25651499; https://doi.org/10.1186/s13059-014-0569-x
- Arner P, Sahlqvist AS, Sinha I, Xu H, Yao X, Waterworth D, Rajpal D, Loomis AK, Freudenberg JM, Johnson T, et al. The epigenetic signature of systemic insulin resistance in obese women. Diabetologia 2016; 59(11):2393-405; PMID:27535281; https://doi.org/10.1007/s00125-016-4074-5
- Li J, Zhang N, Huang X, Xu J, Fernandes JC, Dai K, Zhang X. Dexamethasone shifts bone marrow stromal cells from osteoblasts to adipocytes by c/ebpalpha promoter methylation. Cell Death Dis 2013; 4:e832; https://doi.org/10.1038/cddis.2013.348
- Kiskinis E, Hallberg M, Christian M, Olofsson M, Dilworth SM, White R, Parker MG. Rip140 directs histone and DNA methylation to silence ucp1 expression in white adipocytes. EMBO J 2007; 26(23):4831-40; PMID:17972916; https://doi.org/10.1038/sj.emboj.7601908
- Milagro FI, Campion J, Garcia-Diaz DF, Goyenechea E, Paternain L, Martinez JA. High fat diet-induced obesity modifies the methylation pattern of leptin promoter in rats. J Physiol Biochem 2009; 65(1):1-9; PMID:19588726; https://doi.org/10.1007/BF03165964
- Zwamborn RA, Slieker RC, Mulder PC, Zoetemelk I, Verschuren L, Suchiman HE, Toet KH, Droog S, Slagboom PE, Kooistra T, et al. Prolonged high-fat diet induces gradual and fat depot-specific DNA methylation changes in adult mice. Sci Rep 2017; 7:43261; PMID:28256596; https://doi.org/10.1038/srep43261
- Kim AY, Park YJ, Pan X, Shin KC, Kwak SH, Bassas AF, Sallam RM, Park KS, Alfadda AA, Xu A, et al. Obesity-induced DNA hypermethylation of the adiponectin gene mediates insulin resistance. Nat Commun 2015; 6:7585; PMID:26139044; https://doi.org/10.1038/ncomms8585
- Fan C, Dong H, Yan K, Shen W, Wang C, Xia L, Zhan D, Qi K. Genome-wide screen of promoter methylation identifies novel markers in diet-induced obese mice. Nutr Hosp 2014; 30(1):42-52; PMID:25137261
- Multhaup ML, Seldin MM, Jaffe AE, Lei X, Kirchner H, Mondal P, Li Y, Rodriguez V, Drong A, Hussain M. Mouse-human experimental epigenetic analysis unmasks dietary targets and genetic liability for diabetic phenotypes. Cell Metab 2015; 21(1):138-49; PMID:25565211; https://doi.org/10.1016/j.cmet.2014.12.014
- Adaikalakoteswari A, Finer S, Voyias PD, McCarthy CM, Vatish M, Moore J, Smart-Halajko M, Bawazeer N, Al-Daghri NM, McTernan PG, et al. Vitamin b12 insufficiency induces cholesterol biosynthesis by limiting s-adenosylmethionine and modulating the methylation of srebf1 and ldlr genes. Clin Epigenetics 2015; 7(1):14; PMID:25763114; https://doi.org/10.1186/s13148-015-0046-8
- Slieker RC, Bos SD, Goeman JJ, Bovée J, Talens RP, van der Breggen R, Suchiman H, Lameijer EW, Putter H, van den Akker EB. Identification and systematic annotation of tissue-specific differentially methylated regions using the illumina 450k array. Epigenetics Chromatin 2013; 6(1):26; PMID:23919675; https://doi.org/10.1186/1756-8935-6-26
- Walden TB, Hansen IR, Timmons JA, Cannon B, Nedergaard J. Recruited vs. Nonrecruited molecular signatures of brown, “brite,” and white adipose tissues. Am J Physiol Endocrinol Metab 2012; 302(1):E19-31; PMID:21828341; https://doi.org/10.1152/ajpendo.00249.2011
- Perry R, Rudnick MA. Molecular mechanisms regulating myogenic determination and differentiation. Front Biosci 2000; 5(D750–767):148
- Fajas L, Landsberg RL, Huss-Garcia Y, Sardet C, Lees JA, Auwerx J. E2fs regulate adipocyte differentiation. Dev Cell 2002; 3(1):39-49; PMID:12110166; https://doi.org/10.1016/S1534-5807(02)00190-9
- Kim JI, Huh JY, Sohn JH, Choe SS, Lee YS, Lim CY, Jo A, Park SB, Han W, Kim JB. Lipid-overloaded enlarged adipocytes provoke insulin resistance independent of inflammation. Mol Cell Biol 2015; 35(10):1686-99; PMID:25733684; https://doi.org/10.1128/MCB.01321-14
- Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, Heid IM, Berndt SI, Elliott AL, Jackson AU, Lamina C. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet 2009; 41(1):25-34; PMID:19079261; https://doi.org/10.1038/ng.287
- Lv D, Zhang DD, Wang H, Zhang Y, Liang L, Fu JF, Xiong F, Liu GL, Gong CX, Luo FH. Genetic variations in sec 16b, mc4r, map2k5 and kctd15 were associated with childhood obesity and interacted with dietary behaviors in chinese school-age population. Gene 2015; 560(2):149-55; PMID:25637721; https://doi.org/10.1016/j.gene.2015.01.054
- Jeong JK, Szabo G, Raso GM, Meli R, Diano S. Deletion of prolyl carboxypeptidase attenuates the metabolic effects of diet-induced obesity. Am J Physiol Endocrinol Metab 2012; 302(12):E1502-10; PMID:22454290; https://doi.org/10.1152/ajpendo.00544.2011
- Xu S, Lind L, Zhao L, Lindahl B, Venge P. Plasma prolylcarboxypeptidase (angiotensinase c) is increased in obesity and diabetes mellitus and related to cardiovascular dysfunction. Clin Chem 2012; 58(7):1110-5; PMID:22539806; https://doi.org/10.1373/clinchem.2011.179291
- Wagner JR, Busche S, Ge B, Kwan T, Pastinen T, Blanchette M. The relationship between DNA methylation, genetic and expression inter-individual variation in untransformed human fibroblasts. Genome Biol 2014; 15(2):R37; PMID:24555846; https://doi.org/10.1186/gb-2014-15-2-r37
- Hasegawa S, Ikeda Y, Yamasaki M, Fukui T. The role of acetoacetyl-coa synthetase, a ketone body-utilizing enzyme, in 3t3-l1 adipocyte differentiation. Biol Pharm Bull 2012; 35(11):1980-5; https://doi.org/10.1248/bpb.b12-00435
- Rupnick MA, Panigrahy D, Zhang CY, Dallabrida SM, Lowell BB, Langer R, Folkman MJ. Adipose tissue mass can be regulated through the vasculature. Proc Natl Acad Sci U S A 2002; 99(16):10730-5; PMID:12149466; https://doi.org/10.1073/pnas.162349799
- Kang S, Akerblad P, Kiviranta R, Gupta RK, Kajimura S, Griffin MJ, Min J, Baron R, Rosen ED. Regulation of early adipose commitment by zfp521. PLoS Biol 2012; 10(11):e1001433; PMID:23209378; https://doi.org/10.1371/journal.pbio.1001433
- Chen X, Hausman BS, Luo G, Zhou G, Murakami S, Rubin J, Greenfield EM. Protein kinase inhibitor gamma reciprocally regulates osteoblast and adipocyte differentiation by downregulating leukemia inhibitory factor. Stem Cells 2013; 31(12):2789-99; PMID:23963683; https://doi.org/10.1002/stem.1524
- Gao X, Li K, Hui X, Kong X, Sweeney G, Wang Y, Xu A, Teng M, Liu P, Wu D. Carnitine palmitoyltransferase 1a prevents fatty acid-induced adipocyte dysfunction through suppression of c-jun n-terminal kinase. Biochem J 2011; 435(3):723-32; PMID:21348853; https://doi.org/10.1042/BJ20101680
- Crider KS, Yang TP, Berry RJ, Bailey LB. Folate and DNA methylation: A review of molecular mechanisms and the evidence for folate's role. Adv Nutr 2012; 3(1):21-38; PMID:22332098; https://doi.org/10.3945/an.111.000992
- Dahlman I, Sinha I, Gao H, Brodin D, Thorell A, Rydén M, Andersson D, Henriksson J, Perfilyev A, Ling C. The fat cell epigenetic signature in post-obese women is characterized by global hypomethylation and differential DNA methylation of adipogenesis genes. Int J Obes 2015; 39(6):910-9
- Breslow MJ, Min-Lee K, Brown DR, Chacko VP, Palmer D, Berkowitz DE. Effect of leptin deficiency on metabolic rate in ob/ob mice. Am J Physiol 1999; 276(3 Pt 1):E443-9; PMID:10070008
- Kim SJ, Choi Y, Choi YH, Park T. Obesity activates toll-like receptor-mediated proinflammatory signaling cascades in the adipose tissue of mice. J Nutr Biochem 2012; 23(2):113-22; PMID:21414767; https://doi.org/10.1016/j.jnutbio.2010.10.012
- Luda KM, Joeris T, Persson EK, Rivollier A, Demiri M, Sitnik KM, Pool L, Holm JB, Melo-Gonzalez F, Richter L, et al. Irf8 transcription-factor-dependent classical dendritic cells are essential for intestinal t cell homeostasis. Immunity 2016; 44(4):860-74; PMID:27067057; https://doi.org/10.1016/j.immuni.2016.02.008
- McLaughlin T, Liu LF, Lamendola C, Shen L, Morton J, Rivas H, Winer D, Tolentino L, Choi O, Zhang H. T-cell profile in adipose tissue is associated with insulin resistance and systemic inflammation in humans. Arterioscler Thromb Vasc Biol 2014; 34(12):2637-43; PMID:25341798; https://doi.org/10.1161/ATVBAHA.114.304636
- Mills CD, Kincaid K, Alt JM, Heilman MJ, Hill AM. M-1/m-2 macrophages and the th1/th2 paradigm. J Immunol 2000; 164(12):6166-73; https://doi.org/10.4049/jimmunol.164.12.6166
- Eguchi J, Yan QW, Schones DE, Kamal M, Hsu CH, Zhang MQ, Crawford GE, Rosen ED. Interferon regulatory factors are transcriptional regulators of adipogenesis. Cell Metab 2008; 7(1):86-94; PMID:18177728; https://doi.org/10.1016/j.cmet.2007.11.002
- Majka SM, Miller HL, Helm KM, Acosta AS, Childs CR, Kong R, Klemm DJ. Analysis and isolation of adipocytes by flow cytometry. Methods Enzymol 2014; 537:281-96; PMID:24480352
- Wu C, Jin X, Tsueng G, Afrasiabi C, Su AI. Biogps: Building your own mash-up of gene annotations and expression profiles. Nucleic Acids Res 2016; 44(D1):D313-16; PMID:26578587; https://doi.org/10.1093/nar/gkv1104
- Li N, Ye M, Li Y, Yan Z, Butcher LM, Sun J, Han X, Chen Q, Zhang X, Wang J. Whole genome DNA methylation analysis based on high throughput sequencing technology. Methods 2010; 52(3):203-12; PMID:20430099; https://doi.org/10.1016/j.ymeth.2010.04.009
- Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Methods 2012; 9(4):357-9; PMID:22388286; https://doi.org/10.1038/nmeth.1923
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The sequence alignment/map format and samtools. Bioinformatics 2009; 25(16):2078-9; PMID:19505943; https://doi.org/10.1093/bioinformatics/btp352
- Lienhard M, Grimm C, Morkel M, Herwig R, Chavez L. Medips: Genome-wide differential coverage analysis of sequencing data derived from DNA enrichment experiments. Bioinformatics 2014; 30(2):284-6; PMID:24227674; https://doi.org/10.1093/bioinformatics/btt650
- Smyth GK. Limma: Linear models for microarray data. Bioinformatics and computational biology solutions using r and bioconductor. Springer 2005. p. 397-420
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. Meme suite: Tools for motif discovery and searching. Nucleic Acids Res 2009; 37:W202-8. gkp335
- Liaset B, Julshamn K, Espe M. Chemical composition and theoretical nutritional evaluation of the produced fractions from enzymic hydrolysis of salmon frames with protamex™. Process Biochem 2003; 38(12):1747-59; https://doi.org/10.1016/S0032-9592(02)00251-0
