ABSTRACT
Obesity is a chronic metabolic disorder characterized by the accumulation of excess fat in the body. Preventing and controlling obesity by inhibiting the adipogenic differentiation of mesenchymal stem cells (MSCs) and thereby avoiding the increase of white adipose tissue is safe and effective. Recent studies have demonstrated that Sprouty proteins (SPRYs) are involved in cell differentiation and related diseases. However, the role and mechanism of SPRY4 in MSC adipogenic differentiation remain to be explored. Here, we found that SPRY4 positively correlates with the adipogenic differentiation of human adipose-derived MSCs (hAMSCs). Via gain- and loss-of-function experiments, we demonstrated that SPRY4 promotes hAMSC adipogenesis both in vitro and in vivo. Mechanistically, SPRY4 functioned by activating the MEK–ERK1/2 pathway. Our findings provide new insights into a critical role for SPRY4 as a regulator of adipogenic differentiation, which may illuminate the underlying mechanisms of obesity and suggest the potential of SPRY4 as a novel treatment option.
Introduction
Obesity has become a major public health challenge, and the number of obese people has increased dramatically over the past few decades [Citation1–3]. In essence, obesity is caused by an increase in the mass of white adipose tissue (WAT), including an abnormal increase in the number and volume of adipocytes [Citation4]. Adipogenesis is a process in which preadipocytes differentiate into mature adipocytes, leading to WAT expansion [Citation5]. Therefore, adipogenesis can be used as a handle to control WAT accumulation, which offers an effective strategy for treating and preventing obesity [Citation6]. MSCs are multipotent cells that can differentiate into chondroblasts, osteoblasts and adipocytes [Citation7]. As they are a major source of adipocytes, understanding the specific mechanism of MSC adipogenesis is of great importance. Multiple signalling pathways, in concert with various transcription factors, regulate MSC adipogenic differentiation precisely [Citation8–10]. However, the important functional regulators involved in MSC adipogenesis remains to be well defined.
SPRYs are highly conserved protein families and were originally identified as novel FGFR signalling antagonists during tracheal development in Drosophila. They were later proven to be inhibitors of receptor tyrosine kinase (RTK) signalling [Citation11]. SPRYs have varied biological significance, including the regulation of cell proliferation, differentiation, cytoplasmic calcium concentration and cell migration [Citation11,Citation12]. Recently, it was reported that SPRYs play a critical role in adipogenesis. SPRY1 was upregulated after weight loss interventions, and SPRY1 depletion in human adipogenic stromal cells by RNA interference reduced the expression of the early adipogenic transcription factor CCAAT/enhancer binding protein-β (C/EBPβ), with the abrogation of adipogenesis [Citation13]. CRISPR/Cas9-mediated SPRY1 knockout in human adipogenic stromal cells also leads to impaired adipogenesis [Citation14]. However, conflicting results have been observed. Fat-specific deletion of SPRY1 in mouse model induced total body fat accumulation, while SPRY1 gain of function led to reduced total body fat and hypertrophy of abdominal fat cells [Citation15]. In vitro studies have shown SPRY1 suppresses adipocyte differentiation in the bone marrow MSCs [Citation15]. Gonadal WAT expanded in SPRY1 knockout mice, and loss of SPRY1 promotes adipocyte differentiation [Citation16]. Downregulaion of SPRY4 had different effects on MSC fate determination. SPRY4 suppression enhanced MSC osteogenic differentiation and bone formation, while inhibited chondrogenic differentiation and had no effect on the adipogenic differentiation of MSC in vitro [Citation17]. However, a recent study found that SPRY4 inhibits osteogenic differentiation and induced adipogenic differentiation of mouse marrow stromal progenitor cells (including marrow-derived MSCs) in vitro and in vivo. Mechanistic studies suggested SPRY4 plays a key role in stem cell lineage commitments through inactivating ERK1/2-Wnt/β-catenin regulatory loop [Citation18].
Previously, we found that SPRY4 regulates the osteogenic differentiation of bone marrow-derived MSCs and that it was implicated in the pathogenesis of adolescent idiopathic scoliosis [Citation19]. The present study was conducted to explore whether SPRY4 affects the adipogenic differentiation of hAMSCs.
Material and methods
Antibodies and key reagents
Antibodies against Peroxisome proliferator-activated receptor gamma (PPARγ), CCAAT/enhancer binding protein-α (C/EBPα), fatty acid binding protein 4 (FABP4), and lipoprotein lipase (LPL) were from Abcam (Cambridge, MA, USA). Antibodies against SPRY4 and GAPDH were from Proteintech (Wuhan, China). Antibodies against phosphorylated (p)-ERK1/2, ERK1/2 and Perilipin 1 were from Cell Signalling Technology (Danvers, MA, USA). U0126 was from Selleck Chemicals (Houston, TX, USA).
Isolation, culture and adipogenic differentiation of hAMSCs
hAMSCs were isolated and expanded as previously described [Citation20]. Briefly, human adipose tissues were collected from donors undergoing liposuction surgery. The cells were isolated, and then cultured in a humidified incubator at 37°C with 5% CO2. Passage 3 hAMSCs were used in the following experiments. All experiments followed the procedures approved by the Ethics Committee of the Chinese Academy of Medical Sciences and Peking Union Medical College (NO.2019002) .
For adipogenic differentiation, hAMSCs were seeded at a density of 3 × 105 cells in 6-well plates with basic growth medium. After the cells had reached 90% confluence, the culture medium was replaced with adipogenic medium composed of DMEM(Gibco) supplemented with 10% foetal bovine serum (Gibco), 1 μM dexamethasone (Sigma-Aldrich), 0.5 mM isobutylmethylxanthine (Sigma-Aldrich), and 1 mM L-ascorbic acid (Sigma-Aldrich). The medium was changed every other day during adipogenic differentiation.
Oil red O staining and quantification
As previously described [Citation21], oil red O staining was performed after the 10-day adipogenic differentiation. Briefly, cells were washed twice with phosphate-buffered saline (PBS) and fixed in 4% paraformaldehyde (500 μL/well) for 10 min. Then, the cells were stained with filtered oil red O solution (stock solution: 1 mg/mL in isopropanol; working solution: 60% oil red O stock solution mixed with 40% double-distilled water) for 30 min at 37°C. After staining, the cells were washed with double-distilled water to remove the unbound dye and were subsequently imaged. For quantification of oil red O staining, 100–200 μL isopropanol was added to each well, and the OD value was measured at a wavelength of 510 nm with an ELISA microplate reader.
Small interfering RNA (siRNA) transfection and virus infection
For transient knockdown of SPRY4, hAMSCs were transfected with siRNAs via Lipofectamine 2000 (Life Technology) according to the manufacturer’s recommendations. The SPRY4 siRNAs and negative control (NC) synthesized by GenePharma (Shanghai, China) are listed in Supplementary Table S1. For stable knockdown in the animal experiments in vivo, the same siRNA sequences were inserted into LV3-pGLV-H1-GFP+Puro lentiviral vector, then lentivirus short hairpin RNAs (shRNAs) expression vector was constructed and packaged. For overexpression, the coding sequence of SPRY4 was cloned into a lentivirus vector (pEZ-Lv225), which was packaged by GeneCopoeia (Guangzhou, China). To obtain stable cell lines, hAMSCs were infected with lentivirus (multiplicity of infection [MOI] = 10) for 24 h, followed by treatment with 1 μg/mL puromycin (Sigma-Aldrich).
Heterotopic lipid formation in vivo
hAMSCs with SPRY4 overexpression, SPRY4 knockdown, or the corresponding controls (generated by lentivirus infection) were incubated in adipogenic medium for 4 days. Then, 2 × 106 cells were mixed with 150 µL Matrigel (BD Biosciences), and then injected subcutaneously into two symmetrical sites on the upper dorsal surface of 6-week-old BALB/c nu/nu female mice (n = 5 per group). After 8 weeks, the cells/Matrigel implants were obtained and fixed in 4% paraformaldehyde. All animal experiments were performed with the approval of the Ethics Committee of the Chinese Academy of Medical Sciences and Peking Union Medical College.
HE staining and Immunofluorescence
Heterotopic lipid formation was observed using haematoxylin eosin (HE) staining, and immunofluorescence for Perilipin 1 was used to observe the efficiency of hAMSC adipogenic differentiation in vivo. Briefly, paraffin sections with a thickness of 3 μm were prepared according to standard protocols, and then dewaxed in xylene, rehydrated through decreasing concentrations of ethanol, and washed in PBS. For HE staining, the sections were stained with haematoxylin and eosin. After staining, sections were dehydrated through increasing concentrations of ethanol and xylene and mounted. For immunofluorescence, tissue antigens were retrieved by citric acid buffer (PH6.0) microwave antigen retrieval (Servicebio, Wuhan, China) at first. Then, the sections were blocked for 30 min at room temperature in 10% BSA (Servicebio), perilipin 1 was detected by incubation with anti- perilipin 1 antibody (Cell Signalling Technology) at 4°C overnight. Sections were rinsed three times in PBS and incubated with Goat anti-Rabbit fluorescent secondary antibody (Servicebio) for 1 h at room temperature the next day, and then cell nucleus were stained with DAPI (Servicebio) for 5 mins at room temperature. After rinsing three times in PBS, expression of perilipin 1 protein was observed under fluorescence microscope.
RNA extraction and qRT-PCR
Total RNA was extracted from hAMSCs cultured in vitro using TRIzol (Invitrogen, Waltham, MA, USA) and then treated with DNase I (Ambion, Austin, TX, USA) at 37°C for 30 min. cDNA was synthesized using a reverse transcription kit (Takara, Otsu, Japan) according to the manufacturer’s instructions. qRT-PCR was conducted with Hieff™ qPCR SYBR® Green Master Mix (Yeasen, Shanghai, China) using QuantStudio 3 (Applied Biosystems, Foster City, CA, USA). The relative expression of each gene was evaluated using the comparative threshold cycle (2−ΔΔCt) method and normalized to the mRNA level of GAPDH. Supplementary Table S1 shows the primers used in this study.
Western blotting
The cells were washed twice with cold PBS and then lysed with RIPA lysis buffer containing 1 mM protease inhibitor cocktail (Beyotime, Beijing, China). Total protein was collected and quantified with a BCA protein assay kit (Beyotime). Western blotting was performed as previously described [Citation19]. Briefly, protein fractions (15–20 μg) were separated by SDS-PAGE gels and then transferred to 0.45 µm PVDF membranes (Millipore, Billerica, MA, USA). After blocking with 5% BSA for 1 h, the membranes were incubated with specific primary antibodies at 4°C overnight. Then, the membranes were incubated with HRP-conjugated secondary antibodies (Neobioscience, Shenzhen, China) for 1 h at room temperature after washing with TBST. The target protein bands were visualized by a chemiluminescence ECL reagent (Millipore).
Statistical analysis
The experiments were repeated more than three times; all data were analysed by GraphPad Prism 7 (GraphPad Software Inc., La Jolla, CA, USA) and expressed as the mean ± standard deviation (SD). Statistical significance between two groups or among multiple groups was analysed using Student’s t-tests (two-sided) and one-way analysis of variance (ANOVA), respectively. Differences were considered statistically significant at * p < 0.05, ** p < 0.01 and *** p < 0.001.
Results
SPRY4 positively correlated with hAMSC adipogenic differentiation capacity
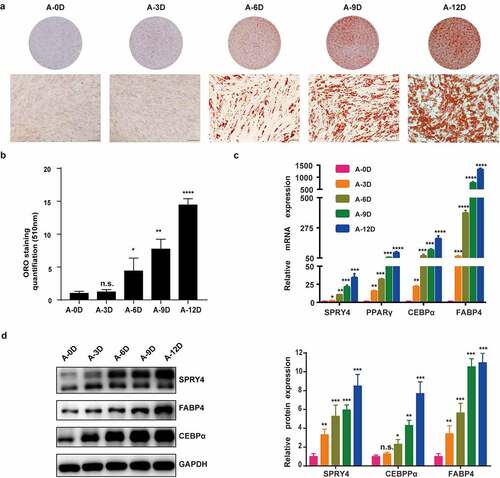
We determined that all hAMSCs used in this study met the minimum standard for MSCs by the International Society for Cellular Therapy (Fig. S1). When passage 3 hAMSCs were 80–90% confluent, the culture medium was replaced by adipogenic medium for up to 12 days. The hAMSC adipogenic differentiation capacity was detected with oil red O staining, qRT-PCR and western blotting. The oil red O staining-positive rate increased gradually from 0 to 12 days (), and quantitative analysis confirmed this result (). qRT-PCR and western blotting also showed that the adipogenic transcription factors and marker genes PPARγ, C/EBPα and FABP4 were upregulated during adipogenic differentiation within 12 days (). Consistently, SPRY4 mRNA and protein levels were increased during adipogenic differentiation from day 0 to day 12 (). These data indicate that SPRY4 is positively associated with hAMSC adipogenic differentiation.
Figure 1. SPRY4 positively correlated with hAMSC adipogenic differentiation capacity.
Downregulation of SPRY4 impaired hAMSC adipogenic differentiation in vitro
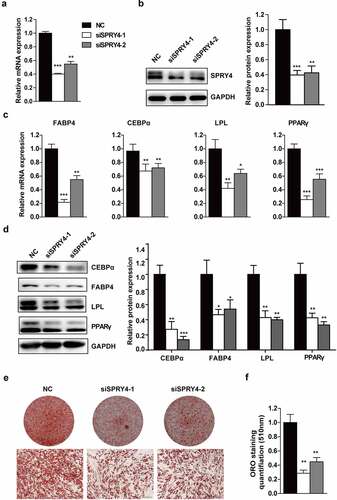
To investigate the role of SPRY4 in hAMSC adipogenic differentiation, we silenced SPRY4 using two independent siRNAs. Compared to the NC, SPRY4 mRNA and protein levels were greatly downregulated in the knockdown groups (). Subsequently, the transfected hAMSCs were induced to differentiate into the adipogenic lineage. The mRNA levels of PPARG, CEBPA, FABP4 and LPL were significantly downregulated with SPRY4 knockdown (). Meanwhile, expression of the adipogenic markers at protein level was attenuated (). Consistently, oil red O staining-positive cells and quantification () further indicated that SPRY4 knockdown resulted in adipogenesis delay. These results prove that SPRY4 knockdown impairs hAMSC adipogenic differentiation in vitro.
Figure 2. Downregulation of SPRY4 impaired hAMSC adipogenic differentiation in vitro.
Ectopic expression of SPRY4 promoted hAMSC adipogenic differentiation in vitro
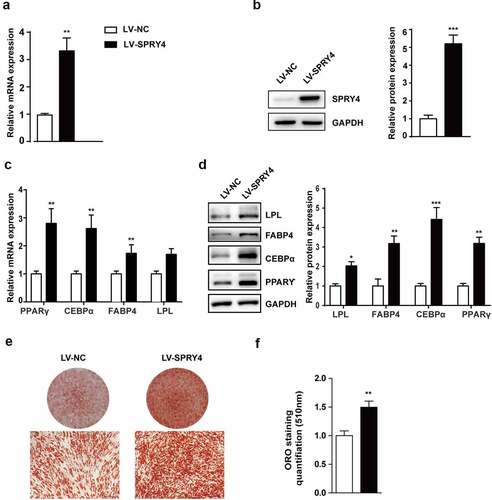
To elucidate the effect of SPRY4 on adipogenic differentiation, we used lentivirus to overexpress SPRY4. qRT-PCR and western blotting showed that SPRY4 expression was upregulated effectively after the lenti-SPRY4 virus infection (). Next, the lentivirus-infected hAMSCs were induced towards the adipogenic lineage. Ectopic expression of SPRY4 significantly increased PPARγ, C/EBPα, FABP4 and LPL expression at both mRNA and protein level (). Furthermore, oil red O staining and quantification showed that more lipid droplets formed after lenti-SPRY4 infection, suggesting that there were more adipocytes and that hAMSC adipogenesis had been promoted (). Altogether, these data demonstrate that SPRY4 is a positive regulator of hAMSC adipogenic differentiation in vitro.
Figure 3. Ectopic expression of SPRY4 promoted hAMSC adipogenic differentiation in vitro.
SPRY4 accelerated hAMSC heterotopic lipid formation in vivo
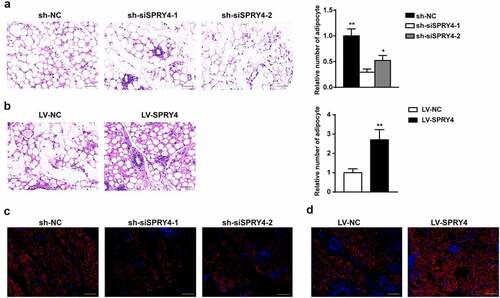
To assess the potential effect of SPRY4 in vivo, we used a heterotopic lipid formation model in nude mice. hAMSCs were incubated in adipogenic medium for 4 days after treatment with sh-NC, sh-siSPRY4-1, sh-siSPRY4-2, LV-NC, or LV-SPRY4 lentivirus. After pre-induction, the same number of hAMSCs were mixed with Matrigel and then injected subcutaneously into the upper dorsal surface of the mice. Eight weeks later, the preadipocytes/Matrigel implants were harvested for HE staining and immunofluorescence for perilipin 1. Perilipin 1, a major lipid droplet-binding protein, is a specific marker of adipogenic differentiation due to its high expression in adipocytes [Citation22]. HE staining revealed that SPRY4 depletion (sh-siSPRY4-1 and sh-siSPRY4-2 groups) clearly reduced the number of adipocytes as compared to the sh-NC group, while more adipocytes were observed in the LV-SPRY4 group than in the LV-NC group (). Immunofluorescence demonstrated that the SPRY4-silenced group had fewer Perilipin 1+ adipocytes than the control group, whereas more were detected in the SPRY4-overexpressing group (). Collectively, SPRY4 overexpression in the hAMSCs accelerated heterotopic lipid formation in vivo, while SPRY4 knockdown had the opposite effect.
Figure 4. SPRY4 accelerated hAMSC heterotopic lipid formation in vivo.
MEK–ERK1/2 pathway inhibition suppressed hAMSC adipogenic differentiation
SPRY4 belongs to the SPRY family, which can regulate the MEK–ERK1/2 pathway [Citation23]. To clarify the molecular mechanism by which SPRY4 regulates adipogenic differentiation, we detected ERK1/2 activation with western blotting. Fig. S2 shows that the knockdown of SPRY4 dramatically decreased the phosphorylation level of ERK1/2. Conversely, SPRY4 overexpression increased them.
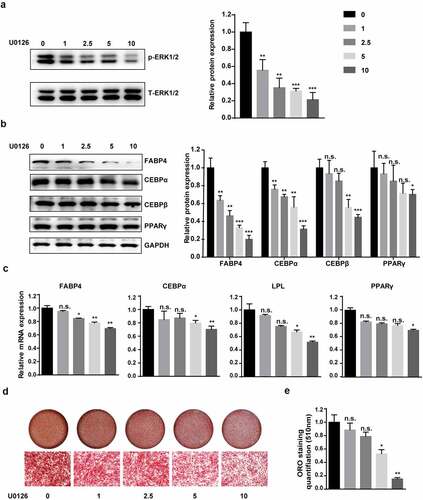
However, the role of the MEK–ERK1/2 pathway in adipogenic differentiation and obesity has been the subject of contradictory reports [Citation24–27]. To explore the correlation between the MEK–ERK1/2 signalling pathway and hAMSC adipogenic differentiation, we blocked the pathway using U0126, an ERK signalling pathway-specific inhibitor. We found that U0126 could decrease the phosphorylation level of ERK 1/2 in a concentration dependent manner (). During adipogenic differentiation, qRT-PCR and western blotting showed that U0126 notably downregulated C/EBPα, FABP4 and LPL expression in a concentration-dependent manner (). Consistent with these results, the gradually decreasing oil red O staining-positive cells and quantification confirmed that U0126 treatment resulted in impaired adipogenesis (). These results prove that inhibiting the MEK–ERK1/2 pathway suppresses hAMSC adipogenic differentiation in a concentration-dependent manner.
Figure 5. MEK–ERK1/2 pathway inhibition suppressed adipogenic differentiation.
SPRY4 regulated hAMSC adipogenic differentiation via the MEK–ERK1/2 pathway
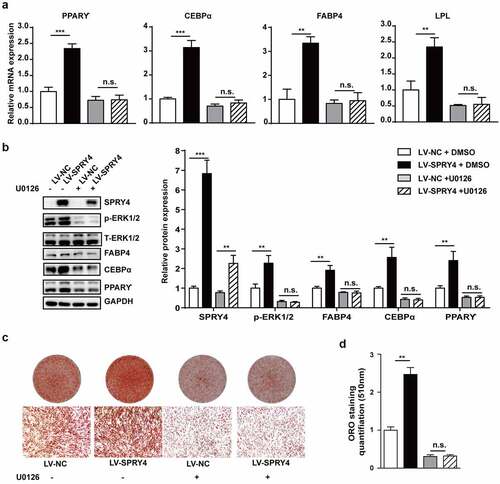
Next, we attempted to determine whether the MEK–ERK1/2 pathway is responsible for the adipogenic-promoting effects of SPRY4. hAMSCs were infected with lentivirus to overexpress SPRY4, and then induced towards adipogenic lineage treating with 10 μM U0126. shows that U0126 almost blocked SPRY4 enhancement of the phosphorylation level of ERK1/2. Ectopic expression of SPRY4 increased PPARγ, C/EBPα, FABP4 and LPL expression, while U0126 abolished these results (). Oil red O staining and quantification also proved that blocking the MEK–ERK1/2 pathway abrogated the adipogenic-promoting abilities of SPRY4 (). Together, these data demonstrate that SPRY4 regulates hAMSC adipogenic differentiation via the MEK–ERK1/2 signalling pathway.
Figure 6. SPRY4 functioned by activating the MEK–ERK1/2 pathway.
Discussion
Obesity is a major risk factor for a variety of diseases. The main feature of obesity is the massive increase in white adipose tissue, while the growth of adipose tissue involves an increase in adipocyte size and the formation of new adipocytes [Citation28]. Therefore, inhibiting the fat-forming process is the fundamental approach for treating and preventing obesity. MSCs are original progenitor cells with multilineage differentiation potential and reproductive activity. MSCs can not only differentiate into osteoblasts, chondrocytes and adipocytes of the mesoderm, but can also differentiate across the germ layer under proper induction conditions [Citation10]. As the common progenitor cells of adipocytes, MSCs are considered the promising target for obesity therapy. Many studies have examined the potential applications of MSCs for treating obesity, and some results are encouraging [Citation29–31], which suggests that obesity can be avoided by controlling MSC adipogenic differentiation. However, MSC adipogenic differentiation is a highly complex process, and the specific molecular mechanisms remain ill-defined.
SPRYs were found in genetic screening for trachea [Citation32] and eye [Citation33] developmental regulators in Drosophila. In mammalians, four different Spry members (Spry1–4) have been identified. Spry1, Spry2 and Spry4 can be detected in various tissues and organs, whereas Spry3 is only expressed in mouse testis and adult brain [Citation34]. SPRYs have recently been implicated in MSC differentiation [Citation15,Citation18,Citation35], making them promising therapeutic targets in related diseases. Herein, we show that SPRY4 positively correlates with the adipogenic differentiation capacity of hAMSCs. Further study revealed that knockdown of SPRY4 significantly inhibited the expression of the adipogenic transcription factors PPARγ and C/EBPα, strongly impairing the hAMSC adipogenesis process in vitro. Besides, we show that the ectopic expression of SPRY4 had the opposite effect. Consistently, a recent study has shown that SPRY4 expression was induced in bone marrow stromal cells (BMSCs) during adipogenic differentiation [Citation18]. SPRY4 promoted BMSC adipogenic differentiation by inactivating ERK–Wnt–β-catenin signalling [Citation18]. Mandl et al. found that SPRY1 is essential for the initiation of adipogenesis. Knockdown of SPRY1 by shRNAs inhibited adipogenesis by augmenting ERK signalling and preventing C/EBPβ expression [Citation13]. In addition, they later reported that adipogenesis was abrogated in human adipose stem/progenitor cells with SPRY1 knockout by CRISPR/Cas9-mediated genome editing [Citation14]. Therefore, we and others show that downregulating SPRYs prevents adipogenesis.
SPRYs are mainly considered a major class of negative feedback loop modulators of RTK-induced ERK1/2 activation [Citation12,Citation36]. However, we reveal an active effect of SPRY4 on the MEK–ERK1/2 pathway during hAMSC adipogenic differentiation. Knockdown of SPRY4 dramatically decreased ERK1/2 phosphorylation, while SPRY4 overexpression increased ERK1/2 phosphorylation. In fact, SPRYs do not always inhibit ERK1/2 activation [Citation37–39]. SPRY1 and SPRY2 increased EGF-mediated ERK activation in endothelial HEK293, CHO and HeLa cells by binding c-Cbl protein and thereby weakening EGF receptor ubiquitination and degradation [Citation40–43]. Therefore, the role of SPRYs in the RTK-mediated MAPK–ERK signalling pathway seems to be cell-specific and context-dependent. In addition, SPRYs have many binding partners, including different effectors of the MAPK signalling pathway. The intersection points where SPRYs interfere with the MAPK pathway and their interplay with other interacting molecules may partly explain the dual but opposite effects on MAPK activation [Citation37,Citation44].
Although the effect of the MEK–ERK1/2 signalling pathway on adipogenic differentiation has been addressed, the results obtained so far appear contradictory [Citation24,Citation25,Citation45]. On the one hand, changes in molecular expression in the same cell type have different effects on the ERK signalling pathway and subsequently on adipogenic differentiation. Downregulating CTRP6 suppressed the differentiation of 3T3-L1 preadipocytes by inhibiting the ERK1/2 signalling pathway [Citation46], while inhibition of the MEK–ERK1/2 signalling pathway by PD98059 had no inhibitory effect on 3T3-L1 preadipocyte adipogenesis [Citation47]. Moreover, adipogenic differentiation was impaired in 3T3-L1 preadipocytes containing hyperactivated MEK1 or constitutively expressing active ERK1/2 [Citation47]. Consequently, the contradiction may be that ERK1/2 is necessary for initiating the differentiation process of preadipocytes, and afterwards, this signal pathway needs to be shut off for adipocyte maturation to continue [Citation25]. Based on this line of thought, interventions in the ERK1/2 signalling pathway at different stages of the differentiation process have different effects on adipogenic differentiation even within the same cell type. On the other hand, various stimuli inversely regulate ERK1/2 phosphorylation in different cell types, which leads to contradictory effects on adipogenic differentiation. Phloretin promoted adipogenesis in mouse marrow stromal ST2 cells by inhibiting ERK1/2 [Citation48]. Erythropoietin inhibited the adipogenic commitment of human BMSCs (hBMSCs) in vitro by increasing ERK1/2 phosphorylation level [Citation49]. hAMSC adipogenic differentiation was inhibited by maintaining continuous ERK1/2 phosphorylation with FGF2 at a concentration of >10 ng/mL [Citation50]. These results suggest that the role of the MEK–ERK1/2 signalling pathway on adipocyte differentiation depends on the intervention period, type of stimulation and cell type involved. Our results show that blocking ERK1/2 phosphorylation from the initial stage of induction suppresses hAMSC adipogenic differentiation in a concentration-dependent manner. Then, we verified whether SPRY4 regulates hAMSC adipogenic differentiation through the MEK–ERK1/2 signalling pathway. As shown in , the enhancement of adipogenesis by SPRY4 was almost abolished by U0126, thereby confirming our hypothesis. Certainly, in order to find more precise targets for obesity treatment, further studies are required to elucidate the specific mechanism by which SPRY4 activates the ERK1/2 signalling pathway. Moreover, researches showed that white mature adipocytes are plastic cells able to reversibly transdifferentiate towards fibroblast-like cells maintaining stem cell gene signatures under appropriate paracrine or endocrine stimuli; this could be important to curb adipogenesis and the number of adipocytes [Citation51]. Thus, further studies are needed to find out whether SPRY4 is involved in adipocytes transdifferentiate towards stem cells.
Conclusion
We have elucidated the role of the SPRY4–ERK1/2 interaction in hAMSC adipogenic differentiation. We demonstrated that SPRY4 is positively associated with hAMSC adipogenic differentiation. Functional analyses revealed that of SPRY4 promotes hAMSC adipogenic differentiation in vitro and enhances ectopic fat formation in vivo effectively. Importantly, the molecular mechanism studies show that inhibiting the MEK–ERK1/2 pathway inhibits hAMSC adipogenic differentiation. SPRY4 increased ERK1/2 phosphorylation and activated the MEK–ERK1/2 pathway, thereby regulating the expression of genes involved in hAMSC adipogenesis. Our findings reveal the contribution of SPRY4 to hAMSC adipogenesis, and further identification of the cell type-specific signalling pathways in hAMSC adipogenesis, will help to expand the treatment options for combating obesity and the related health complications.
Author contributions
NL designed, performed most of the experiments and wrote the paper; YC, HW and XY performed some experiments and analyzed the data; JL organized, designed the paper, and RC initiated the study and analyzed the data.
Supplemental Material
Download Zip (2.6 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/21623945.2022.2123097
Additional information
Funding
References
- van Dijk Sj, Molloy PL, Varinli H, et al. Epigenetics and human obesity [J]. Int J Obes (Lond). 2015;39(1):85–97.
- Ng M, Fleming T, Robinson M, et al. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980-2013: a systematic analysis for the global burden of disease study 2013 [J]. Lancet. 2014;384(9945):766–781.
- Malik VS, Willett WC, Hu FB. Global obesity: trends, risk factors and policy implications [J]. Nat Rev Endocrinol. 2013;9(1):13–27.
- Zhang Y, Zheng Y, Fu Y, et al. Identification of biomarkers, pathways and potential therapeutic agents for white adipocyte insulin resistance using bioinformatics analysis [J]. Adipocyte. 2019;8(1):318–329.
- Vishvanath L, Gupta RK. Contribution of adipogenesis to healthy adipose tissue expansion in obesity [J]. J Clin Invest. 2019;129(10):4022–4031.
- Sim MO, Lee HJ, Jeong DE, et al. 6’-O-acetyl mangiferin from Iris rossii Baker inhibits lipid accumulation partly via AMPK activation in adipogenesis [J]. Chem Biol Interact. 2019;311:108755.
- Wei Z, Chen N, Guo H, et al. Bone marrow mesenchymal stem cells from leukemia patients inhibit growth and apoptosis in serum-deprived K562 cells [J]. J Exp Clin Cancer Res. 2009;28(1):141.
- Almalki SG, Agrawal DK. Key transcription factors in the differentiation of mesenchymal stem cells [J]. Differentiation. 2016;92(1–2):41–51.
- Tang QQ, Lane MD. Adipogenesis: from stem cell to adipocyte [J]. Annu Rev Biochem. 2012;81(1):715–736.
- Golpanian S, Wolf A, Hatzistergos KE, et al. Rebuilding the damaged heart: mesenchymal stem cells, cell-based therapy, and engineered heart tissue [J]. Physiol Rev. 2016;96(3):1127–1168.
- Kim HJ, Bar-Sagi D. Modulation of signalling by Sprouty: a developing story [J]. Nat Rev Mol Cell Biol. 2004;5(6):441–450.
- Mason JM, Morrison DJ, Basson MA, et al. Sprouty proteins: multifaceted negative-feedback regulators of receptor tyrosine kinase signaling [J]. Trends Cell Biol. 2006;16(1):45–54.
- Mandl M, Wagner SA, Hatzmann FM, et al. Sprouty1 is a weight-loss target gene in human adipose stem/progenitor cells that is mandatory for the initiation of adipogenesis [J]. Cell Death Dis. 2019;10(6):411.
- Mandl M, Wagner SA, Hatzmann FM, et al. Sprouty1 prevents cellular senescence maintaining proliferation and differentiation capacity of human adipose stem/progenitor cells [J]. J Gerontol A Biol Sci Med Sci. 2020;75(12):2308–2319.
- Urs S, Venkatesh D, Tang Y, et al. Sprouty1 is a critical regulatory switch of mesenchymal stem cell lineage allocation [J]. Faseb J. 2010;24(9):3264–3273.
- Yang X, Pande S, Koza RA, et al. Sprouty1 regulates gonadal white adipose tissue growth through a PDGFRα/β-Akt pathway [J]. Adipocyte. 2021;10(1):574–586.
- Park S, Arai Y, Kim BJ, et al. Suppression of SPRY4 promotes osteogenic differentiation and bone formation of mesenchymal stem cell [J]. Tissue Eng Part A. 2019;25(23–24):1646–1657.
- Tian L, Xiao H, Li M, et al. A novel Sprouty4-ERK1/2-Wnt/β-catenin regulatory loop in marrow stromal progenitor cells controls osteogenic and adipogenic differentiation [J]. Metabolism. 2020;105:154189.
- Li J, Li N, Chen Y, et al. SPRY4 is responsible for pathogenesis of adolescent idiopathic scoliosis by contributing to osteogenic differentiation and melatonin response of bone marrow-derived mesenchymal stem cells [J]. Cell Death Dis. 2019;10(11):805.
- Huang S, Wang S, Bian C, et al. Upregulation of miR-22 promotes osteogenic differentiation and inhibits adipogenic differentiation of human adipose tissue-derived mesenchymal stem cells by repressing HDAC6 protein expression [J]. Stem Cells Dev. 2012;21(13):2531–2540.
- Li H, Fan J, Fan L, et al. MiRNA-10b reciprocally stimulates osteogenesis and inhibits adipogenesis partly through the TGF-β/SMAD2 signaling pathway [J]. Aging Dis. 2018;9(6):1058–1073.
- Sztalryd C, Brasaemle DL. The perilipin family of lipid droplet proteins: gatekeepers of intracellular lipolysis [J]. Biochim Biophys Acta Mol Cell Biol Lipids. 2017;1862(10 Pt B):1221–1232.
- Christofori G. Split personalities: the agonistic antagonist Sprouty [J]. Nat Cell Biol. 2003;5(5):377–379.
- Aubert J, Belmonte N, Dani C. Role of pathways for signal transducers and activators of transcription, and mitogen-activated protein kinase in adipocyte differentiation [J]. Cell Mol Life Sci. 1999;56(5–6):538–542.
- Bost F, Aouadi M, Caron L, et al. The role of MAPKs in adipocyte differentiation and obesity [J]. Biochimie. 2005;87(1):51–56.
- Lee M, Sorn SR, Lee Y, et al. Salt induces adipogenesis/lipogenesis and inflammatory adipocytokines secretion in adipocytes [J]. Int J Mol Sci. 2019;20(1):160.
- Ning X, He J, Shi X, et al. Regulation of adipogenesis by quinine through the ERK/S6 pathway [J]. Int J Mol Sci. 2016;17(4):504.
- Gregoire FM, Smas CM, Sul HS. Understanding adipocyte differentiation [J]. Physiol Rev. 1998;78(3):783–809.
- Zhu XY, Ma S, Eirin A, et al. Functional plasticity of adipose-derived stromal cells during development of obesity [J]. Stem Cells Transl Med. 2016;5(7):893–900.
- Louwen F, Ritter A, Kreis NN, et al. Insight into the development of obesity: functional alterations of adipose-derived mesenchymal stem cells [J]. Obes Rev. 2018;19(7):888–904.
- Matsushita K, Dzau VJ. Mesenchymal stem cells in obesity: insights for translational applications [J]. Lab Invest. 2017;97(10):1158–1166.
- Hacohen N, Kramer S, Sutherland D, et al. sprouty encodes a novel antagonist of FGF signaling that patterns apical branching of the Drosophila airways [J]. Cell. 1998;92(2):253–263.
- Casci T, Vinós J, Freeman M. Sprouty, an intracellular inhibitor of Ras signaling [J]. Cell. 1999;96(5):655–665.
- Doriguzzi A, Haigl B, Gsur A, et al. The increased Sprouty4 expression in response to serum is transcriptionally controlled by Specific protein 1 [J]. Int J Biochem Cell Biol. 2015;64:220–228.
- Joo A, Long R, Cheng Z, et al. Sprouty2 regulates endochondral bone formation by modulation of RTK and BMP signaling [J]. Bone. 2016;88:170–179.
- Cabrita MA, Christofori G. Sprouty proteins, masterminds of receptor tyrosine kinase signaling [J]. Angiogenesis. 2008;11(1):53–62.
- Masoumi-Moghaddam S, Amini A, Morris DL. The developing story of Sprouty and cancer [J]. Cancer Metastasis Rev. 2014;33(2–3):695–720.
- Guy GR, Wong ES, Yusoff P, et al. Sprouty: how does the branch manager work? [J]. J Cell Sci. 2003;116(Pt 15):3061–3068.
- Li X, Wheldon L, Heath JK. Sprouty: a controversial role in receptor tyrosine kinase signalling pathways [J]. Biochem Soc Trans. 2003;31(Pt 6):1445–1446.
- Impagnatiello MA, Weitzer S, Gannon G, et al. Mammalian sprouty-1 and −2 are membrane-anchored phosphoprotein inhibitors of growth factor signaling in endothelial cells [J]. J Cell Biol. 2001;152(5):1087–1098.
- Egan JE, Hall AB, Yatsula BA, et al. The bimodal regulation of epidermal growth factor signaling by human Sprouty proteins [J]. Proc Natl Acad Sci U S A. 2002;99(9):6041–6046.
- Hall AB, Jura N, DaSilva J, et al. hSpry2 is targeted to the ubiquitin-dependent proteasome pathway by c-Cbl [J]. Curr Biol. 2003;13(4):308–314.
- Rubin C, Litvak V, Medvedovsky H, et al. Sprouty fine-tunes EGF signaling through interlinked positive and negative feedback loops [J]. Curr Biol. 2003;13(4):297–307.
- Cabrita MA, Christofori G. Sprouty proteins: antagonists of endothelial cell signaling and more [J]. Thromb Haemost. 2003;90(4):586–590.
- Moseti D, Regassa A, Kim WK. Molecular regulation of adipogenesis and potential anti-adipogenic bioactive molecules [J]. Int J Mol Sci. 2016;17(1):124.
- Wu WJ, Mo DL, Zhao CZ, et al. Knockdown of CTRP6 inhibits adipogenesis via lipogenic marker genes and Erk1/2 signalling pathway [J]. Cell Biol Int. 2015;39(5):554–562.
- de Mora J F, Porras A, Ahn N, et al. Mitogen-activated protein kinase activation is not necessary for, but antagonizes, 3T3-L1 adipocytic differentiation [J]. Mol Cell Biol. 1997;17(10):6068–6075.
- Takeno A, Kanazawa I, Notsu M, et al. phloretin promotes adipogenesis via mitogen-activated protein kinase pathways in mouse marrow stromal ST2 cells [J]. Int J Mol Sci. 2018;19(6):1772.
- Liu GX, Zhu JC, Chen XY, et al. Inhibition of adipogenic differentiation of bone marrow mesenchymal stem cells by erythropoietin via activating ERK and P38 MAPK [J]. Genet Mol Res. 2015;14(2):6968–6977.
- Kim S, Ahn C, Bong N, et al. Biphasic effects of FGF2 on adipogenesis [J]. PLoS One. 2015;10(3):e0120073.
- Maurizi G, Petäistö T, Maurizi A, et al. Key-genes regulating the liposecretion process of mature adipocytes [J]. J Cell Physiol. 2018;233(5):3784–3793.
