ABSTRACT
It is well established that the anticancer immune response determines the success of anthracycline-based adjuvant chemotherapy of breast cancer. This effect is in part due to the capacity of anthracyclines to induce immunogenic cell death (ICD), a cell death modality that is preceded by autophagy and followed by HMGB1 release. Recent data on 1,798 mammary carcinoma specimens indicate that patients harboring neoplastic cells that lack immunohistochemical signs of autophagy or that have lost HMGB1 expression have indeed a poor prognosis.
Over the last decade it has become increasingly clear that chemotherapy and radiotherapy of mammary both act as a sort of cryptic immunotherapies, meaning that they exert their beneficial effects by reinstating anticancer immunosurveillance. Indeed, the density and composition of the immune infiltrate at diagnosis constitutes an independent prognostic feature that predicts the outcome of neoadjuvant chemotherapy. Thus, a high-density of tumor infiltrating CD8+ cytotoxic T lymphocytes as well as a low-density of CD68+ macrophages constitute positive prognostic features.Citation1,2 This notion has recently been confirmed in a meta-analysis of the transcriptomes from 1,045 breast cancers showing that a CXCL13-centered, highly reproducible metagene signature reflecting the intratumoral presence of interferonγ-producing T cells predicted the pathological complete response to neadjuvant chemotherapy.Citation3 In addition, chemotherapy induces dynamic changes in the composition of the immune infiltrate that are already detectable after the first treatment cycle. An improvement of the ratio between CD8+ cytotoxic T lymphocytes and FOXP3+ regulatory T cells also predicts favorable treatment outcome,Citation4 supporting the notion that chemotherapy must induce changes that favor the anticancer immune response to be efficient.
What are then the mechanisms through which anthracyclines, which are widely used for breast cancer chemotherapy, can stimulate an anticancer immune response? One plausible scenario consists in the induction of ICD in a fraction of cancer cells. This would then convert the tumor into a sort of vaccine that elicits a potent cellular immune response against residual tumor cells. ICD is only induced by some specific anticancer agents including anthracyclines that have the property to stimulate a cascade events that render cancer cells immunogenic.Citation5-7 These immunogenic alterations include the induction of premortem autophagy, which is required for tumor cells to release ATP as a chemotactic agent acting on purinergic receptors expressed on myeloid cells including immature dendritic cells. Indeed, cancers that have manipulated to lack the expression of essential autophagy-related genes (and that hence are unable to mount an autophagic response to anthracycline-based chemotherapy) escape from immunosurveillanceCitation8 and are totally chemoresistant.Citation9 Another immunogenic alteration that must be induced by chemotherapy is the release of high molecular group B1 (HMGB1), an abundant chromatin binding protein that exits dead cells to interact with toll-like receptor -4 (TLR4) on dendritic cells. Ligation of TLR4 then stimulates dendritic cells to present tumor-associated antigens. Breast cancer patients who bear TLR4 loss-of-function alleles exhibit poor prognosis after adjuvant chemotherapy, supporting the clinical importance of the HMGB1/TLR4 interaction.Citation1
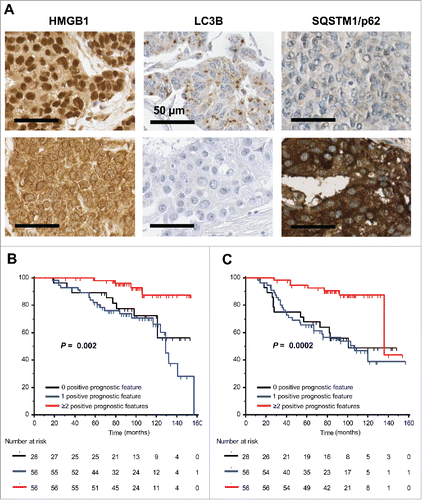
Based on these premises, we investigated autophagy and HMGB1 expression on a large series of tumor specimens from breast cancer patients that were treated with anthracycline-based adjuvant chemotherapy.Citation10 The staining with HMGB1 revealed a large heterogeneity between samples from intense nuclear staining of the totality of carcinoma cells to a total absence of nuclear staining (). Similarly, there was a major heterogeneity in the intensity and the subcellular distribution of LC3B, a protein that is known to be diffusely distributed in the cell when autophagy is inactive, but relocates to cytoplasmic puncta when autophagy is active (). The absence and presence of multiple LC3B+ autophagosomes correlates with high and low expression, respectively, of the autophagic substrate STQSM1/p62 (). Subsequent statistical analyses revealed that the absence of LC3B puncta in >90% of the cancer cells (meaning the inactivation of the autophagic process), as well as the absence of nuclear HMGB1 staining in >50% of the tumor cells (implying the impossibility of releasing HMGB1 in response to chemotherapy) had a significant negative impact on overall and progression-free patient survival. Especially those patients whose tumors lack both autophagy and HMGB1 expression had a poor prognosis.Citation10 Moreover, the combined analysis of all positive prognostic features (nuclear HMGB1 expression in >50% of the carcinoma cells, LC3B punta in >10% of the neoplastic cells, STQSM1 staining below the median value) yielded an excellent stratification of the training cohort (140 patients), both with respect to overall survival () and with respect to metastasis-free survival ().
Figure 1. Impact of the HMGB1, LC3B and STQSM1/p62 staining patterns on overall and progression-free survival. (A) Representative immunohistological staining patterns of HMGB1, LC3B and STQSM1/p62 in breast cancer patients. For each marker two different patients are shown. Nuclear HMGB1 staining in >50% of the cancer cells, the presence of LC3B puncta in >10% of the neoplastic cells, and a low STQSM1 staining (<median value) were considered as positive prognostic features. (B, C) Impact of prognostic features determined as in A on overall survival (B) or metastasis-free survival (C) in a cohort of 140 breast cancer patients that were stratified into three categories bearing 0, 1 or ≥2 positive prognostic features.
Altogether, these observations constitute the first evidence that parameters linked to ICD including autophagy and HMGB1 expression do influence the fate of breast cancer patients.Citation10 Previous attempts to study ICD-related gene expression profiles failed,Citation3 most likely because ICD heavily relies on post-transcriptional processes. Hence, it is essential to study ICD by suitable immunohistochemical methods (). It will be important to correlate these histo-morphological parameters in subsequent studies with the density, location and composition of the immune infiltrate. Moreover, it will be interesting to learn whether other parameters linked to ICD (such as endoplasmic reticulum stress and expression of type-1 interferon-regulated genes) may allow for a further refinement of risk stratification. Altogether, this strategy should yield optimal biomarkers for personalizing chemotherapeutic regimens by combining them with suitable immunotherapeutic measures.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- Zitvogel L, Galluzzi L, Smyth MJ, Kroemer G. Mechanism of action of conventional and targeted anticancer therapies: reinstating immunosurveillance. Immunity 2013; 39:74-88; PMID:23890065; http://dx.doi.org/10.1016/j.immuni.2013.06.014
- Kroemer G, Galluzzi L, Zitvogel L. Immunological effects of chemotherapy in spontaneous breast cancers. Oncoimmunology 2013; 2:e27158; PMID:24498568; http://dx.doi.org/10.4161/onci.27158
- Stoll G, Enot D, Mlecnik B, Galon J, Zitvogel L, Kroemer G. Immune-related gene signatures predict the outcome of neoadjuvant chemotherapy. Oncoimmunology 2014; 3:e27884; PMID:24790795; http://dx.doi.org/10.4161/onci.27884
- Senovilla L, Vitale I, Martins I, Tailler M, Pailleret C, Michaud M, Galluzzi L, Adjemian S, Kepp O, Niso-Santano M et al. An immunosurveillance mechanism controls cancer cell ploidy. Science 2012; 337:1678-84; PMID:23019653; http://dx.doi.org/10.1126/science.1224922
- Vacchelli E, Senovilla L, Eggermont A, Fridman WH, Galon J, Zitvogel L, Kroemer G, Galluzzi L. Trial watch: Chemotherapy with immunogenic cell death inducers. Oncoimmunology 2013; 2:e23510; PMID:23687621; http://dx.doi.org/10.4161/onci.23510
- Sukkurwala AQ, Adjemian S, Senovilla L, Michaud M, Spaggiari S, Vacchelli E, Baracco EE, Galluzzi L, Zitvogel L, Kepp O, Kroemer G. Screening of novel immunogenic cell death inducers within the NCI Mechanistic Diversity Set. Oncoimmunology 2014; 3:e28473; PMID:25050214; http://dx.doi.org/10.4161/onci.28473
- Kepp O, Senovilla L, Vitale I, Vacchelli E, Adjemian S, Agostinis P, Apetoh L, Aranda F, Barnaba V, Bloy N et al. Consensus guidelines for the detection of immunogenic cell death. Oncoimmunology 2014; 3:e955691; PMID:25941621; http://dx.doi.org/10.4161/21624011.2014.955691
- Rao S, Tortola L, Perlot T, Wirnsberger G, Novatchkova M, Nitsch R, Sykacek P, Frank L, Schramek D, Komnenovic V et al. A dual role for autophagy in a murine model of lung cancer. Nat Commun 2014; 5:3056; PMID:24445999; http://dx.doi.org/10.1038/ncomms4056
- Michaud M, Xie X, Bravo-San Pedro JM, Zitvogel L, White E, Kroemer G. An autophagy-dependent anticancer immune response determines the efficacy of melanoma chemotherapy. Oncoimmunology 2014; 3:e944047; PMID:25610726; http://dx.doi.org/10.4161/21624011.2014.944047
- Ladoire S, Penault-Llorca F, Senovilla L, Dalban C, Enot D, Locher C, Prada N, Poirier-Colame V, Chaba K, Arnould L et al. Combined evaluation of LC3B puncta and HMGB1 expression predicts residual risk of relapse after adjuvant chemotherapy in breast cancer. Autophagy 2015 Oct 3; 11(10):1878-90; PMID:26506894; http://dx.doi.org/10.1080/15548627.2015.1082022
