ABSTRACT
Scavenger receptor class B type I (SR-B1) binds pathogen-associated molecular patterns participating in the regulation of the inflammatory reaction but there is no information regarding potential interactions between SR-B1 and the interferon system. Herein, we report that SR-B1 ligands strongly regulate the transcriptional response to interferon α (IFNα) and enhance its antiviral and antitumor activity. This effect was mediated by the activation of TLR2 and TLR4 as it was annulled by the addition of anti-TLR2 or anti-TLR4 blocking antibodies. In vivo, we maximized the antitumor activity of IFNα co-expressing in the liver a SR-B1 ligand and IFNα by adeno-associated viruses. This gene therapy strategy eradicated liver metastases from colon cancer with reduced toxicity. On the other hand, genetic and pharmacological inhibition of SR-B1 blocks the clathrin-dependent interferon receptor recycling pathway with a concomitant reduction in IFNα signaling and bioactivity. This effect can be applied to enhance cancer immunotherapy with oncolytic viruses. Indeed, SR-B1 antagonists facilitate replication of oncolytic viruses amplifying their tumoricidal potential. In conclusion, SR-B1 agonists behave as IFNα enhancers while SR-B1 inhibitors dampen IFNα activity. These results demonstrate that SR-B1 is a suitable pharmacology target to enhance cancer immunotherapy based on IFNα and oncolytic viruses.
Abbreviations
| AAV-L37 | = | adeno-associated vector encoding L37pA peptide |
| AAV-Luc | = | adeno-associated vector encoding luciferase |
| AAV-IFN | = | adeno-associated vector encoding mouse interferon alpha |
| AAV | = | adeno-associated virus |
| apoA-I | = | apolipoprotein A-I |
| BSA | = | bovine serum albumin |
| CPZ | = | chlorpromazine |
| EMCV | = | encephalomyocarditis virus |
| ERK | = | extracellular signal-regulated kinase |
| HDLs | = | high density lipoproteins |
| IFNα | = | interferon alpha |
| LPS | = | lipopolysaccharide |
| MEFs | = | mouse embryonic fibroblasts |
| NDV-GFP | = | Newcastle disease virus encoding green fluorescent protein |
| NDV-IL2 | = | Newcastle disease virus encoding interleukin 2 |
| SAA | = | serum amyloid A |
| SR-B1 | = | scavenger receptor class B type I |
Introduction
Scavenger receptor class B type I (SR-B1) is a transmembrane glycoprotein known to bind and internalize a broad variety of ligands.Citation1,2 Most of the SR-B1 ligands form micellar particles containing either negatively charged phospholipids or anionic class A amphipathic α-helixes.Citation3 Among the best characterized SR-B1 ligands are lipoproteins, particularly high density lipoproteins (HDLs), and therefore the major protein component of HDL: apolipoprotein A-1 (Apo A-1). SR-B1 is involved in cholesterol homeostasis both at cellular and systemic levels via the selective uptake of cholesteryl esters and other lipids from HDLs and by promoting bidirectional movements of unesterified cholesterol.Citation4 More recently, further studies have highlighted that SR-B1 also plays a role in innate immunity by binding pathogen-associated molecular patterns or damage-associated molecular patterns and internalizing pathogens.Citation5 SR-B1 has been shown to modulate the macrophage response to lipopolysaccharide (LPS) response, to mediate LPS clearance by hepatocytes, to attenuate neutrophil activation and to modulate stress-induced glucocorticoid production by adrenal glands.Citation6-8 SR-B1 deficiency in mice is associated with enhanced production of proinflammatory cytokines and autoimmunity.Citation9 Apo A-I mimetic peptides such as L37pA attenuate inflammation in models of cardiac ischemia/reperfusion injury and sepsis.Citation10,11 However, some SR-B1 ligands such as serum amyloid A (SAA), glycated or oxidated Apo A-I or dysfunctional HDLs have been shown to promote inflammation.Citation12-14 Thus, SR-B1 plays a dual role in inflammation and may represent a novel target to design new immunotherapies against cancer.
Interferon α (IFNα) is an essential player in innate immunityCitation15 and links innate immune responses by acting as a signal-3 cytokine in the immunological synapse.Citation16 It is an essential cytokine in anti-viral and antitumor immunity but deregulation of its production may promote the development of autoimmune disorders such as lupus, thyroiditis, diabetes mellitus type I, autoimmune dermatitis, psoriasis and Sjogren's syndrome.Citation17 The potent immunostimulatory, antiviral and antineoplastic activities of pharmacological doses of the recombinant IFNα led to the clinical use of this cytokine for the treatment of viral infections and several type of cancers such as hairy cell leukemia, malignant melanoma, AIDS-related Kaposi's sarcoma and follicular non-Hodgkin's lymphoma.Citation18. In spite of a large clinical experience, systemic side effects associated to the use of high pharmacological doses have limited the clinical utility of IFNα and is being replaced by novel targeted small molecules that are better tolerated.Citation19,20 Drugs that potentiate IFNα activity, but limit its toxic effects could take advantage of the huge antiviral and antitumor properties of IFNα. IFNα also plays an important role on the antitumor efficacy of gene therapy-based oncolytic viruses. On one hand, IFNα is induced upon oncolytic virus injection, limiting the replication of the vector, but on the other hand this cytokine is essential for inducing an effector antitumor immune response. Thus, transient blockade of this pathway may enhance viral replication within the tumor without affecting the induction of the antitumor immune responses.Citation21,22 Newcastle disease virus (NDV) is a promising oncolytic vector that can be engineered to over-express pro-inflammatory cytokines. These armed NDVs are able to achieve eradication of tumors in several preclinical models.Citation23 However, NDV are potent inductors of IFNα and strategies to block IFNα can benefit NDV-based antitumor therapies.
In this study, we found that SR-B1 is a critical modulator of IFNα bioactivity. SR-B1 agonists or antagonists may be used as immunomodulators with different therapeutic applications.
Results
IFNα activity is modulated by L37pA: In vitro studies
It has been reported that the ApoA-I mimetic peptide L37pA binds SR-B1 and mediates anti-inflammatory effects.Citation24 We investigated whether this peptide influenced IFNα bioactivity in a mouse L929 fibroblast cell line. We found that in the presence of L37pA significantly lower doses of IFNα were required to protect fibroblasts from a lethal challenge with encephalomyocarditis virus (EMCV) (). Similarly, the cytotoxic effect on mouse fibroblasts induced by higher IFNα doses was greatly amplified by L37pA (). In isolation, this peptide lacked any antiviral or cytotoxic activity (data not shown). The observed synergy of the combination IFNα plus L37pA was abrogated by mutating the peptide sequence to disrupt the amphipathic α-helix structure ().
Figure 1. ApoAI mimetic peptide L37pA potentiates the antiviral and cytotoxic activity of IFNα. (A) Cytopathic effect reduction assay in L929 cells pretreated overnight with 2-fold serial dilutions of IFNα starting from 125 U/mL with L37pA (200 µg/mL), L37mut (200 µg/mL) or with no peptide. Cells viability was quantified 24 h after EMCV infection. (Extra sum-of-squares F test ***p < 0.001). (B) Cytotoxicity assay in L929 cells incubated for 3 d with IFNα (1500 U/mL) and L37pA (200 µg/mL), L37mut or with no peptide (one way ANOVA followed by Dunnett's multiple comparison test. ***p < 0.001). (C) Quantitative real time PCR analysis of interferon-stimulated genes after 3 h of stimulation of L929 cells with IFNα (200 U/mL), L37pA (200 µg/mL) or the combination. Data are expressed as mean + SEM (one way ANOVA, followed by Dunnett's multiple comparison test. **p < 0.01, ***p < 0.001).
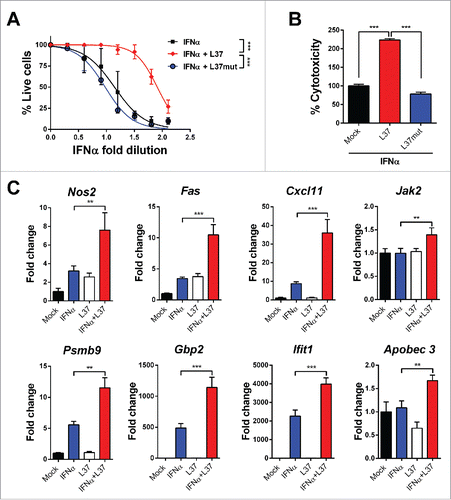
Microarray analysis of L929 cells treated with IFNα, with or without L37pA showed upregulation (fold change > 0.69) of 196 transcripts related to inflammation and IFNα response (data not shown). Among those with the highest fold change, eight were validated by qPCR. These included Nos2 (inducible nitric oxide synthase), Fas (a death receptor molecule mediating pro-apoptotic effects), Cxcl11 (also called interferon-gamma-inducible protein 9 involved in the attraction of activated T cells), Jak2 (a non-receptor tyrosine kinase involved in signaling of type II cytokine receptors including interferon receptors), Gbp2 (Interferon-induced guanylate-binding protein 2), Ifit1 (interferon-induced antiviral RNA-binding protein which inhibits the expression of viral mRNA) and Apobec3 (a cytidine deaminase with important functions in innate antiviral immunity). Fas and Cxcl11 showed the highest upregulation upon combined treatment and were selected as a gene signature of the IFNα/L37pA synergy in subsequent experiments ().
The synergy between IFNα and L37pA is not exclusive to L929, as Cxcl11 and Fas were also induced by the combined treatment in other mouse cell lines, such as 3T3 fibroblasts and CT-26 murine colon cancer (Fig. S1A). More interestingly, the synergy was also observed in human cell lines such as human monocytes, hepatic HepaRG and fibroblast BJ cells (Fig. S1B).
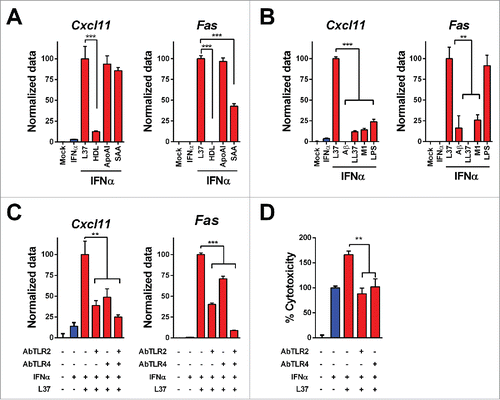
To address whether the lipidated or delipidated status of endogenous SR-B1 ligands might determine their ability to enhance IFNα response, we analyzed the effect of HDLs, delipidated ApoA-I and SAA in L929 cells. HDLs combined with IFNα failed to upregulate Cxcl11 and Fas transcripts but delipidated ApoA-I and SAA synergized with IFNα in the induction of both transcripts () indicating that the lipid composition of SR-B1 ligands is critical for IFNα potentiation. Finally, we evaluated the activity of Toll-like receptor (TLR) ligands in this experimental setting. Of note, TLR ligands such as Alzheimer amyloid β peptide (TLR2 and TLR4); LL37 (TLR9); phenol-soluble modulin 1 (TLR2) and LPS (TLR4 ligand) failed to upregulate Cxcl11 in combination with IFNα () although the latter was able to enhance Fas expression ().
Figure 2. Mechanisms of IFNα and L37pA synergy. We determined the expression of Cxcl11 and Fas as readout of the effect of IFNα plus L37pA using quantitative real time RT-PCR in L929 cells treated as follows: (A) Cells were stimulated with IFNα (200 U/mL) for 3 h alone or in combination with L37pA (200 µg/mL), high density lipoprotein (HDL) (5 µg/mL), apolipoprotein A-I (ApoA-I) (30 µg/mL) or serum amyloid A (SAA) (30 µg/mL). (B) Cells were stimulated with IFNα (200 U/mL) for 3 h in combination with L37PA (200 µg/mL), the Alzheimer amyloid β peptide (Aβ) (200 µg/mL), cathelicidin (LL37) (200 µg/mL), phenol-soluble modulin 1 (M1) (200 µg/mL) or LPS (160 µg/mL). (C) Cells were pretreated with neutralizing antibodies against TLR2 (5 µg/mL), TLR4 (40 µg/mL) or with the combination for 1 h. Then, cells were treated with IFNα (200 U/mL) alone or plus L37pA (200 µg/mL) for 3 h. (D) Cytotoxicity assay in L929 cells incubated for 3 d with IFNα (1500 U/mL) and L37pA (200 µg/mL), and pretreated with neutralizing antibodies against TLR2 (5 µg/mL), TLR4 (5 µg/mL) for 1 h. Data are expressed as mean + SEM (one way ANOVA, followed by Dunnett's multiple comparison test. **p < 0.01, ***p < 0.001).
TLR2 and TLR4 mediate the enhancement of IFNα bioactivity induced by SR-B1 agonists
As members of the scavenger receptor class B family, such as CD36, have been shown to form complexes with other transmembrane proteins including TLR, we studied the role of the latter molecules in the amplification of IFNα response when cells were co-stimulated with this cytokine plus L37pA. We found that blocking antibodies to TLR-2 or TLR-4 inhibited the effect of IFNα/L37pA on their target genes (). Moreover, these blocking antibodies also abrogated the activity of L37pA on IFNα-induced cytotoxicity (). Both TLR can interact forming heterodimersCitation25,26 and indeed we detected these complexes in L929 cells used in this study (Fig. S2A). The mitogen-activated protein kinase pathway is a common signaling pathway activated by all TLRs.Citation27 L37pA was able to induce the phosphorylation of extracellular signal-regulated kinase (ERK) and p38 (Fig. S2B). Our data implicate TLRs as mediators of the enhancing effects of L37pA on IFNα.
Activity of the combination of IFNα and L37pA: In vivo studies
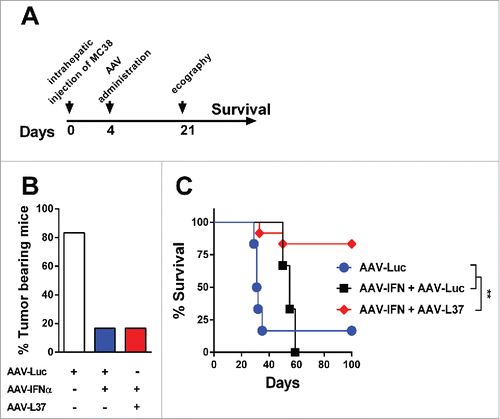
We then assessed whether SR-B1 ligation may affect IFNα in vivo. To this end, we transduced the liver of C57BL/6 mice with an adeno-associated viral vector (AAV) encoding IFNα (AAV-IFN) plus AAV expressing luciferase (AAV-Luc) or the peptide L37pA (AAV-L37) or only with AAV-Luc (at a dose equal to the combination therapy). At week 2, after vector administration IFNα serum levels were similar in mice given AAV-IFN/AAV-Luc and in those treated with AAV-IFN/AAV-L37 (data not shown). At week 4, however, while in the former group serum IFNα continued to rise until it reached toxic levels causing lethal pancytopenia (), in mice that received AAV-IFN/AAV-L37 serum IFNα dropped markedly as result of the almost complete disappearance of viral genomes from the liver (). Consistently, at this time point no hematological toxicity was observed in the last group of mice which showed 100% long-term survival in sharp contrast with the 100% mortality by week 8 in all mice given AAV-IFN/AAV-Luc (). Despite the elimination of AAV genomes from the liver in the AAV-IFN/AAV-L37 group, the amount of interferon-sensitive genes (ISG) transcripts were higher than both AAV-IFN/AAV-Luc and AAV-Luc groups (). AAV clearance occurred in the presence of normal serum transaminases (Fig. S3) and in animals lacking T cells, B cells or NK cells () supporting a non-cytopathic cell autonomous mechanism for viral elimination. The high dose of AAVs used transduces more than 90% hepatocytes and a cytopathic elimination of the infected hepatocytes would have been reflected in elevated serum levels of the transaminases. In order to test whether the self-limited system formed by the combination of AAV-IFN and AAV-L37 could be used to treat liver metastasis, we inoculated MC38 colon cancer cells in the liver of mice. Four days later, the AAVs were administered and 21 d after tumor inoculation, the presence of hepatic tumors was assessed by echography (). The AAV-IFN in combination with AAV-Luc or AAV-L37 displayed a potent antitumor effect eradicating tumors in more than 80% of mice (). However, in the case of AAV-IFN treatment, the high levels of IFNα expressed to achieve such a potent antitumor effect were toxic and led to the death of all mice. Thus, the combined treatment was the only option that successfully eradicated tumors ().
Figure 3. Co-expression of L37pA and IFNα eliminates AAV genomes in the liver. C57BL/6 mice were treated with 1 × 1012 vg AAV-Luc (n = 6), 5 × 1011 AAV-Luc and 5 × 1011 AAV-IFN (n = 6) or 5 × 1011 AAV-L37 and 5 × 1011 AAV-IFN (n = 6). Four weeks after virus administration, we determined the following parameters: (A) IFNα serum levels were measured by ELISA. (B) White blood cells (WBC), platelets (PLT) and red blood cells (RBC). (C) Viral genomes in the liver were determined by quantitative real time PCR. (D) ISGs expression in the liver. Data are expressed as mean + SEM (one way ANOVA, followed by Bonferroni's multiple comparison test. **p < 0.01, ***p < 0.001). (E) Kaplan–Meier plot representing the survival of C57BL/6 mice treated with 5 × 1011 AAV-Luc and 5 × 1011 AAV-IFN (n = 6) and the C57BL/6 mice (n = 6), Rag1 knockout mice (n = 5), Rag2γc knockout mice (n = 7), SR-B1 knockout mice (n = 3) treated with 5 × 1011 AAV-L37 and 5 × 1011 AAV-IFN. (Log rank test. ***p < 0.001). (F) Viral genomes in the liver were determined by quantitative real time PCR in mice that survived. Data is normalized to the viral genomes of wild type mice treated with 5 × 1011 AAV-Luc and 5 × 1011 AAV-L37 (one way ANOVA, followed by Dunnett's multiple comparison test. **p < 0.01). Results were confirmed with at least two independent experiments.
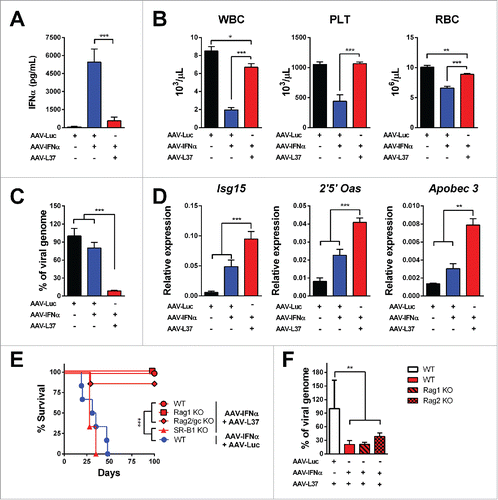
Figure 4. Antitumor effect in mice treated with AAV-IFN and AAV-L37. C57BL/6 mice were treated with 1 × 1012 vg AAV-Luc (n = 6), 5 × 1011 AAV-Luc and 5 × 1011 AAV-IFN (n = 6) or 5 × 1011 AAV-L37 and 5 × 1011 AAV-IFN (n = 12). (A) Experimental protocol. (B) 21 d after tumor inoculation, presence of tumors in the liver was assessed by echography and the percentage of tumor bearing animals is represented. (C) Survival is represented with a Kaplan–Meier plot (Log rank test. **p < 0.01). Results were confirmed with at least two independent experiments.
IFNα activity is abolished by disrupting SR-B1
To determine whether IFNα bioactivity was dependent on functional SR-B1, we knocked-down this molecule using specific siRNA (). We found that IFNα-mediated upregulation of the ISGs Isg15, Usp18 and 2′5′OAS was significantly impaired in SR-B1 deficient cells (). Further supporting the role of SR-B1 on IFNα bioactivity, we found that mouse embryonic fibroblasts (MEFs) from SR-B1 knockout mice required higher doses of IFNα to achieve the same protection against EMCV than wild type cells to a lethal challenge with this virus (Fig. S4A) and that the induction of ISGs such as Pbms8, Tap2, Irf1 and Isg15 was dampened in SR-B1 deficient cells (Fig. S4B).We then tested if known SR-B1 antagonists also affected cell response to IFNα. We used three different SR-B1 chemical inhibitors: block lipid transport 1 (BLT-1)Citation28, ITX-5061 (ITX)Citation29 and glyburideCitation30 and we found that upregulation of Isg15, Usp18 and 2′5′OAS by IFNα was blunted in the cells treated with all these SR-B1 antagonists (). A dose-response curve with BTL-1 showed that concentrations in the order of 1 µM were able to dampen IFNα activity (Fig. S5A). The impact of SR-B1 inhibitors on IFNα response was similar in L929 mouse fibroblasts and in other murine cell lines such as MC38 colon cancer cells and RAW macrophages (Fig. S5B). Moreover, BLT-1 inhibited ISGs induction by IFNα in HepaRG human cells and human monocytes (Fig. S5C).
Figure 5. Inhibition of SR-B1 impairs IFNα function. L929 cells were transfected with siRNA targeting SR-B1 (siSR-B1) or control scrambled siRNA (siCTL) for 48 h. (A) SR-B1 mRNA was determined by quantitative real time PCR. Data are expressed as mean + SEM. (t-test. ***p < 0.001). Total SR-B1 protein was detected by immunoblot and β-actin is shown as loading control. (B) The silenced cells were stimulated with IFNα (200 U/mL) for 2 h. Then, ISGs expression was determined using quantitative real time PCR. mRNA levels were normalized to Rplpo. (C) Quantitative real time PCR analysis of ISGs expression after 3 h of stimulation with IFNα (200 U/mL) in L929 cells pretreated for 1 h with BLT1 (15 µM), ITX-5061 (30 µM) and Glyburide (500 µM). (D) Flow cytometry analysis of the IFNα receptor 1 (IFNAR1) in L929 cells treated with BLT-1 (15 µM) for 3 h. Normalized geometric mean of three samples (left panel) and representative histogram (right panel). Data are expressed as mean + SEM (E) Phosphorylated STAT1 and total STAT1 protein determined by immunoblotting in L929 cells stimulated with IFNα (1500 U/mL) for 30 min after incubation of BLT-1 (15 µM) or DMSO for 1 h. (F) Cytopathic effect reduction assay in mouse L929 cells pretreated overnight with 2-fold serial dilutions of IFNα starting from 125 U/mL in the presence or absence of BLT-1 (15 µM). Cells viability was quantified 24 h after EMCV infection (Extra sum-of-squares F test ***p < 0.001). (G) Cytotoxicity assay in mouse L929 incubated for 3 d with IFNα (1500 U/mL) in the presence or absence of BLT-1 (15 µM). (t-test or one way ANOVA, followed by the Dunnett's Multiple comparison test. *p < 0.05, **p < 0.01, ***p < 0.001).
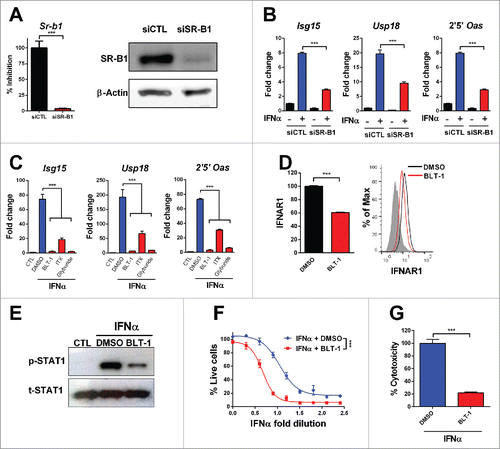
Notably, we observed that the impaired response to IFNα in BLT-1 treated cells was associated with a decreased display of IFNAR1 on the cell membrane () and with reduced activation of STAT-1 upon cell stimulation with IFNα (). Accordingly, BLT-1-treated cells required higher amounts of IFNα to survive a lethal inoculum of EMCV and were less sensitive to the cytotoxic effects of high doses of IFNα (). Finally, we analyzed whether other interferons were blocked by BLT-1. The activity of other type I IFN such as IFNβ was affected by BLT-1 but this compound did not modify the activity of IFNγ, a type II IFN (Fig. S5D and E).
SR-B1 inhibitors block clathrin-dependent endocytosis
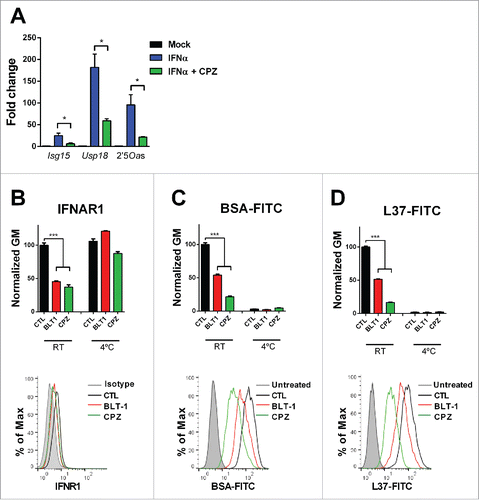
It has been shown that that IFNα signaling requires clathrin-dependent IFNAR endocytosis and recycling.Citation31-33 We confirmed that chlorpromazine (CPZ), a clathrin inhibitor, similarly to BTL-1, reduced ISGs upregulation in IFNα-treated L929 cells () and decreased the expression of IFNAR1 on the cell membrane at 37°C. At 4°C, neither CPZ nor BLT-1 affected the IFNAR1 levels on the cell membrane (). To investigate whether BLT-1 could block clathrin-endocytosis, we analyzed its effect on the internalization of fluorescent-labeled bovine serum albumin (BSA). We found that both BLT-1 and CPZ reduced the internalization of this molecule by clathrin at 37°C (). Moreover, both compounds attenuated the internalization of the SR-B1 ligand L37pA at 37°C (). At 4°C, neither BSA nor L37pA were internalized. In addition, downregulation of SR-B1 by siRNA also decreased internalization of L37pA (Fig. S6A). Taken together, these data indicate that SR-B1 is essential for IFNAR internalization and recycling.
Figure 6. SR-B1 is necessary for the clathrin-mediated endocytosis. (A) Quantitative real time PCR analysis of ISGs expression 3 h after stimulation with IFNα (200 U/mL) in L929 cell pretreated for 1 h with chlorpromazine (CPZ) (10 µM) or BLT1 (15 µM). Data are expressed as mean + SEM (t-test. *p < 0.05). (B) Flow cytometry analysis of the IFNα receptor 1 (IFNAR1) prior treatment with BLT-1 (15 µM) or CPZ (10 µM) for 3 h and flow cytometry analysis in L929 cells treated with BSA-FITC (C) or L37pA-FITC (D) for 1 h prior treatment with CPZ (10 µM) or BLT-1 (15 µM) for 30 min at 37°C or 4°C. The normalized geometric mean (GM) fluorescence (number of replicates = 3) and a representing image at 37°C is shown. Data are expressed as mean + SEM (one way ANOVA, followed by Dunnett's multiple comparison test. ***p < 0.001).
Blockade of SR-B1 function promotes Newcastle disease virus infection
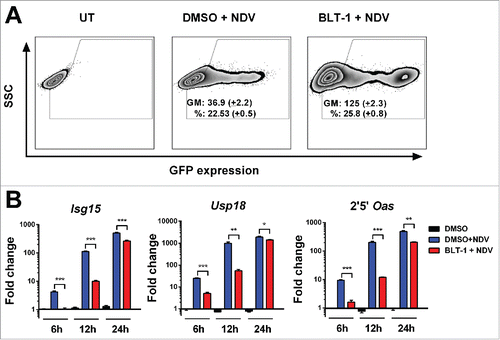
Next, we analyzed whether SR-B1 function was required for endogenous IFNα activity. To this end, we infected L929 mouse fibroblasts with NDV encoding green fluorescent protein (NDV-GFP) and transgene expression was evaluated by flow cytometry. We found that BLT-1 increased the percentage of GFP-expressing cells and the intensity of fluorescence in the infected cells (). In line with these results, NDV infection induced the transcription of several ISGs but blockade of SR-B1 by BLT-1 profoundly altered this process. With this treatment, 6 h after viral infection, we detected reduced transcription of ISGs. 24 h after NDV infection, cells treated or not with BLT-1 expressed high levels of the ISGs (). Thus, the effect of BLT-1 on IFNα signaling is transient. To confirm that SR-B1 was involved in this process, we infected murine L929 fibroblasts with NDV-GFP after silencing SR-B1 with specific siRNA. We noted increased fluorescence intensity and an elevated proportion of GFP-positive cells (Fig. S6B) in association with blunted ISGs expression in SR-B1 deficient cells (Fig. S6C).
Figure 7. BLT-1 increases NDV infection. (A) Representative flow cytometry dot plot 24 h after infection with an MOI of 1 of NDV expressing GFP in L929 cells pretreated 1 h with BLT-1 (15 µM), vehicle (DMSO) or left untreated (UT). The geometric mean (GM) of GFP expression and the percentage of infection (%) are expressed as mean + SEM (number of replicates = 3). (B) Quantitative real time PCR analysis of ISGs mRNA at 6 h, 12 h and 24 h postinfection at an MOI of 1 after treatment with BLT-1 (15 µM) for 1 h. Data are expressed as mean + SEM (number of replicates = 3). (t-test. *p < 0.05, **p < 0.01, ***p < 0.001).
We further studied whether SR-B1 inhibitors might foster intratumoral replication of oncolytic viruses. To this purpose, we inoculated CT-26 or MC-38 colon cancer cells subcutaneously and when mean tumor diameter was superior to 5 mm, we performed intratumoral injection of NDV encoding GFP (NDV-GFP) to CT-26 tumor-bearing mice or NDV encoding interleukin 2 (NDV-IL2) to MC-38 tumor-bearing mice with or without BLT-1. These treatments were repeated 72 h later. We found that BLT-1+NDV-GFP or NDV-IL2 achieved a stronger antitumor effect than the oncolytic virus alone (Fig. S7).
Discussion
In this study, we describe a novel role of SR-B1 agonist and antagonist modulating IFNα biology. Tissue fibroblasts and stromal cells are important sentinels to initiate tissue-adapted immune responses. Upon direct cell damage or recognition of pathogen-associated molecular patterns or damage-associated molecular patterns, these tissue-resident cells release antimicrobial peptides, chemokines or cytokines that modulate the adaptive immune response initiated by tissue-resident dendritic cells.Citation34 Thus, the identification of the mechanism by non-immune cells is of great interest to understand the activation of the immune system and the mechanisms that trigger immune-mediated disorders.
SR-B1 is a promiscuous receptor that binds and internalizes amphipathic molecules. TLR4 and TLR9 ligands are able to bind to SR-B1. In these cases, SR-B1 acts as a scavenger reducing the bioavailability of the TLR ligands and thus limiting the immune response.Citation35,36 Here, we show that SR-B1 can present certain ligands to TLR2 and TLR4. Heterodimers of these molecules have been reported to be activated by ethanolCitation25 and hemoglobin.Citation26 In our study, we used the Apo A-1 mimetic peptide L37pA as a model of the SR-B1 ligand. This peptide is a tandem repetition of a model amphipatic α-helix of 18 aminoacids linked by a proline.Citation37 SR-B1 is able to bind and internalize L37pACitation24 and we confirmed the role of SR-B1 in L37pA internalization using siRNA in L929. It was likely that endogenous ligands were able to trigger the inflammatory response initiated by SR-B1. Indeed, the two most important endogenous ligands of SR-B1, SAA and ApoA-I, were able to trigger this effect. Of note, only delipidated ApoA-I was active indicating an inhibitory effect of the lipid bound to the SR-B1 ligands.
Moreover, the SR-B1—mediated activity was sensitive to the activity of chloroquine (data not shown), a molecule that can inhibit TLR2 activities that require internalization of the TLR.Citation38,39 Finally, L37pA induces the phosphorylation of ERK. ERK is implicated in the signaling of all TLRs and many cytokines including interferon α.Citation27,40 Indeed, in the L929 fibroblasts, IFNα alone phosphorylated this protein and although the combination did not increase the phosphorylation, it is likely that the modification of the kinetics may exert an impact on cytokine-induced transcriptional activity.
The potent effect against AAV exerted when combined with IFNα indicated the great potential of these ligands. AAV infection generates stable episomal viral genomes that persist over time, and this stabilization of the AAV genome is not abrogated even upon treatment with potent immunostimulatory cytokines such as IFNα and interleukin 12. Here, we show that the AAV genome can be eliminated when IFNα is combined with L37pA. Interestingly, the AAV genome elimination was not dependent on T, B, NK cells and was probably due to a phenomenon of intrinsic immunity.Citation41
In addition to the effect of SR-B1 agonist on IFNα biology, we also show the effect of SR-B1 inhibitors on IFNα signaling. Selective uptake of lipids from HDLs by SR-B1 has been shown to be independent of endocytosisCitation42 and only SR-B1 is implicated as this transport can be reproduced in artificial membranes containing only purified SR-B1.Citation43 Moreover, it has been shown that BLT-1 does not affect clathrin-mediated endocytosis analyzing transferrin and epidermal growth factor uptake in Hela cellsCitation28 or transferrin in ldlA[mSR-BI], a Chinese hamster ovary (CHO) cell line that overexpresses SR-B1.Citation44 The interference of BLT-1 with caveolin-mediated endocytosis was evaluated analyzing the entry of cholera toxin into BSC-1 cells and no effect was observed.Citation28 However, at least three isoforms of SR-B1 are produced by alternatively splicing of the mRNA. The isoform SR-B1.2 mediates a clathrin-mediated endocytosis of ligands that accumulate within the transferrin-positive endosomal recycling compartment.Citation45,46 This isoform only differs from SR-B1.1 in the carboxyl-terminal cytoplasmatic tail. SR-B1.1 represents 85–90% of SR-B1 protein present in the liver but in other tissues, SR-B1.2 is the dominant isoform.Citation47,48 In our mouse cell lines, we were able to detect the mRNA of both isoforms (data not shown) and both isoforms were inhibited by the siRNA and by the pharmacologic inhibitors. Our experiments show that SR-B1 not only mediated the internalization of an ApoA-1 mimetic ligand, such as L37pA, but also that interference with SR-B1 activity by genetic or pharmacological targeting reduced clathrin-dependent endocytosis of albumin. This finding may have broad implications. Among these, we focused our research on the effect on interferon α signaling. The type I interferon receptor is composed of IFNAR1 and IFNAR2 subunits.Citation49 Upon ligand binding, the ternary complex is internalized through a clathrin-dependent endocytosis. The endocytosis is required for the activation of STAT1 and for the biological responses induced by IFNα.Citation33 We corroborated this model using CPZ as a clathrin inhibitor. In fact, the activity of this inhibitor was similar to BLT-1 on SR-B1-dependent internalization, clathrin-dependent internalization, abrogation of ISGs induction and decrease of IFNAR1 levels on the cell membrane. Thus, it is likely that SR-B1 activity is required to maintain clathrin-enriched microdomains and drugs that block SR-B1 can affect cell processes mediated by clathrin. In addition, IFNβ activity was inhibited by BLT-1 but it does not affect IFNγ activity. These results are in line with the clathrin-dependent type I IFN activity and clathrin-independent IFNγ activity.Citation33
Interferon α is an important cytokine due to its antiviral, antiproliferative and immunomodulatory activity. However, unbalanced production or unbalanced response may produce deleterious effects leading to autoimmune responses such systemic lupus erythematous and Sjogren's syndrome.Citation17 Moreover, chronic interferon induction may be a strategy used by a diverse range of pathogens, including some viruses to increase host susceptibility.Citation50 Thus, blockade of IFNα signaling is an appealing therapeutic target and we present an alternative strategy to the conventional targets present in the IFN signaling pathway.
In this study, we have focused on other applications of IFNα blockade: the enhancement of the efficacy of oncolytic viruses. IFNα plays a dual role in the biology of oncolytic viruses. While the induction of IFNα may be essential for triggering an antitumor immune response,Citation51,52 the antiviral activity of the released IFNα may limit the spreading of the oncolytic virus.Citation21,22 Thus, a transient inhibition during the initial phase of virus infection may allow optimal virus replication and induction of a strong immune response. That is what we achieved using BLT-1. This SR-B1 inhibitor allowed NDV to infect more cells and express a higher amount of the transgene. In vivo, this strategy clearly enhanced the antitumor efficacy of an NDV expressing GFP or IL-2. Thus, BLT-1 or another inhibitor could be included as an adjuvant when oncolytic viruses are administered.
In conclusion, we demonstrate in this study that SR-B1 controls IFNα activity through the modulation of clathrin-mediated endocytosis and through the presentation of certain ligands to TLR2 and TLR4. These findings have broad implications for the design of novel treatments for IFNα-mediated autoimmune diseases, to improve oncolytic virotherapy, or to improve IFNα-based therapies.
Experimental procedures
Cells lines
HepaRG cells were donated by Dr. Staels (Université Lille Nord de France, France) and were maintained in William's E medium supplemented with 10% fetal bovine serum, 100 U/mL penicillin, 100 μg/mL streptomycin, 5 μg/mL insulin, and 5 µM hydrocortisone hemisuccinate. MEFs, isolated from wild type and SR-B1-deficient mice were donated by Dr. Widmann (University of Lausanne, Switzerland). MC-38 murine colon adenocarcinoma cells were provided by Dr. Ignacio Melero (CIMA, Spain). The following cells were purchased from the ATCC: L929 (ATCC CCL1), BJ (ATCC CRL2522), 3T3 (ATCC CCL163), RAW 264.7 cells (ATCC TIB-71), MC-38 and CT-26 (ATCC CRL2638). Human monocytes were purified from PBMCs from healthy donors using a Miltenyi monocytes isolation kit and a Miltenyi AutoMACS magnetic cell separator (Miltenyi Biotech, Bergisch, Gladbach, Germany). These cells were cultured in DMEM (Gibco) supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 100 U/mL streptomycin, 100 mg/mL penicillin (DMEM complete).
Reagents
SR-B1 inhibitors BLT-1, ITX-5061 and Glyburide were purchased from Sigma-Aldrich (St Louis, MO), Wuxi Apptech Co (Shanghai, China) and Enzo Life Sciences (Farmingdale, NY), respectively. Mouse interferon α 1, interferon β and interferon gamma were purchased from PBL Biomedical Laboratories (New Brunswick, NJ), Biorbyt (Cambridge, UK) and Immunotools (Friesoythe, Germany). CPZ was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). siRNA was purchased from Ambion, Life Technologies (Austin, TX) and were transfected with TransIT-X2 (Mirus Bio LLC; Madison, WI). Human HDLs were isolated from serum by sequential centrifugation from a healthy volunteer. BSA-FITC, human SAA and human ApoA1 were purchased from Sigma Chemical Co. (St. Louis, MO). L37pA (DWLKAFYDKVAEKLKEAFP), L37pA-FITC, amyloid β (Aβ), LL37 and L37mut (DWLKYKDKLKEKLKEALFP) peptides were purchased from GenScript (Piscataway, NJ). M1 peptide was provided by Dr. Lasarte (CIMA, Spain). For Western blot, anti-phospho STAT1, anti-phospho AKT, anti-AKT, anti-phospho p38, anti-p38 and anti-phospho ERK1/2 were purchased from Cell Signaling Technologies (Beverly, MA). Anti-STAT1 was from Santa Cruz Biotechnology (Santa Cruz, CA) and anti-ERK1/2 was purchased from Millipore (Bedford, MA). Anti-TLR2, anti-TLR4 and anti-IFNAR1 antibodies were purchased from BioLegend (San Diego, CA).
Animal handling
Female BALB/c or C57BL/6 mice were purchased from Harlan laboratories (Barcelona, Spain). The experimental design was approved by the Ethical Committee for Animal Testing of the University of Navarra. Experiments were performed with mice between 5–7 weeks old. For subcutaneous tumors, C57BL/6 or BALB/c mice were subcutaneously inoculated with MC-38 cells or CT-26 cells, respectively (5 × 105) in the right posterior flank in a total volume of 100 µL. Two intratumoral injections of 6.5 × 106NDV, PBS and/or 360 ng of BLT-1 or DMSO as vehicle were administrated at day 7 and 9 after cell inoculations in a total volume of 50 µL. Hepatic tumors were established by direct implantation of 5 × 105 MC38 cells in the left liver lobe of C57BL/6 mice following medial laparotomy in isofluorane-anaesthetized animals. Recombinant AAVs were inoculated via retro-orbital injection with 5 × 1011 viral genomes in a total volume of 200 µL in female C57BL/6 mice. Rag1 knockout mice, Rag2γc knockout mice, SR-B1 knockout mice all in a C57BL/6 background were breed at the University of Navarra.
AAV production
Recombinant AAV vectors were constructed to express the IFNα, L37pA or the reporter gene luciferase under transcriptional control of elongation factor 1α promoter (EF). The signal peptide of the mouse gene for IFNα1 was used to secrete the L37pA. AAV vectors were produced by co-transfection of the plasmids pDP8.ape and pAAV containing IFNα1, L37pA or luciferase into 293T cells. Two days later, AAV was purified from the cell lysates by ultracentrifugation in Optiprep Density Gradient Medium-Iodixanol (Sigma-Aldrich, St Louis MO). To titer the AAV productions, viral DNA was isolated using “The High Pure Viral Nucleic Acid” kit (Roche Applied Science. Mannheim, Germany). The concentration of viral particles was subsequently determined by real-time quantitative PCR using primers specific to the EF promoter (Table S1).
NDV vectors production
To generate recombinant NDV expressing GFP or mIL2, A549 cells and Hep2 cells were plated in a 6-well plate and were infected with MVA-T7 at a multiplicity of infection of 1 and then transfected with the NDV full-length clone together with the following expression plasmids: pTM1-NP (nucleoprotein), pTM1-P (phosphoprotein), and pTM1-L (RNA-dependent RNA polymerase). After overnight incubation, transfected cells were cocultured for an additional 2 d. Cells and supernatants were collected and injected into the allantoic cavities of 9- or 10 d old chicken embryonated eggs. Three days later, allantoic fluids were harvested and virus growth was confirmed by hemagglutination and immunofluorescent assays. Original virus stock was frozen at −80°C. Viruses were amplified by injecting 1000 infection units of the virus stock into 10 d old embryonated eggs. Allantoid fluid was collected after 2 d alliquoted and quick frozen in dry ice. Stocks were kept at −80°C before titration by immunofluoresce in Hep2 cells.
Antiviral assay
Antiviral assay was performed by using a cytopathic-effect reduction assay with EMCV. Cells were plated in 96-well plates (3 × 104 cells/well) and stimulated for 24 h with 1:2 serial dilutions of 125 U/mL murine IFNα. Then, the virus was added for 24 h. When maximum cytopathic effect (CPE) was achieved in control cells, viable IFN-protected cells were measured by luminometry with the ViaLight Plus Kit (Lonza, Rockland, ME).
IFNα cytotoxicity
Mouse L929 cells were plated in 96-well plates (3 × 103 cells per well) and incubated with IFNα (1500 U/mL) in the presence or absence of BLT-1 (15 µM). After 72 h, cell death was determined by the release of adenylate kinase with the ToxiLight BioAssay Kit (Lonza).
Quantitative RT-PCR analysis
Cells were seeded in 24-well plates using DMEM complete medium for 24 h to achieve 80% confluence. Then, the cells were washed twice with PBS and stimulated with mouse (200 U/µL) or human (100 U/mL) IFNα, L37pA or IFNα plus L37pA for 3 h. For experiments with blockers of lipid transport, the cells were pre-incubated for 1 h with 15 µM of BLT-1, 30 µM of ITX-506, 500 µM of glyburide or 10 µM of CPZ in DMEM prior stimulation with mouse (200 U/µL) or human (100 U/mL) IFNα. Total RNA from liver tissue and cell lines was extracted using the automated Maxwell system from Promega (Madison, WI). RNA was treated with DNase I and reverse-retrotranscribed to cDNA with MMLV RT in the presence of RNase OUT (all reagents from Invitrogen) according to the manufacturer´s instructions. Primer sequences are described in Table S1. Real time PCR reaction was performed using SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA, USA). As RPLP0 levels remained constant across different experimental conditions, they were used to standardize gene expression. The amount of each transcript was expressed by the formula 2ΔCt (2ct(RPLP0) – ct(gene)), ct being the point at which the fluorescence rises significantly above background fluorescence.
Western blotting
L929 and MEF cells were seeded in 6-well plates using DMEM at a cell density of 3 × 105 per well and incubated for 24 h. BLT-1 (15 µM) treatment was performed for 1 h. IFNα, L37pA or IFNα plus L37pA stimulations were performed for 30 min and then Western blotting was accomplished using the appropriate antibodies.
Co-immunoprecipitation
Lysates from L929 cells treated or not with L37pA for 30 min were immunoprecipitated with anti-TLR4 beads. Cell samples were lysed for 2 h at 4°C in radioimmune precipitation assay buffer with protease inhibitors. Separately, protein G magnetic beads (Dynabeads Protein G, Novex, San Diego, CA) were washed and resuspended according to the manufacturer's instructions and incubated overnight at 4°C with 2 μg of anti-TLR4 antibody. The antibody-bound beads were washed three times, and samples containing 50 μg of cell lysate were added and incubated for 2 h at 4°C in a volume of 500 µL. After washing, the immunoprecipitated proteins were eluted by heating at 60°C for 15 min in Laemmli sample buffer. Samples of equal total protein content were separated by SDS-PAGE and transferred to PVDF membranes. Membranes were blocked with 5% BSA in TBS/Tween and incubated overnight at 4°C with anti-TLR2 (1:1000) antibody. After washing in TBS, horseradish peroxidase-conjugated secondary antibody was applied, and immunoreactive bands were detected by chemiluminescence.
Determination of IFNα by ELISA analysis
Serum IFNα levels were measured using a VeriKine™ Mouse Interferon Alpha ELISA Kit (PBL Biomedical Laboratories, Piscataway, NJ) following the manufacturer's recommendations.
Hemogram
After 2 or 4 weeks of AAV injection, blood samples were collected in Microvette EDTA-coated tubes (Sarstedt Inc., Newton, NC). Hemograms were analyzed using the Drew Scientific HemaVet Hematology Analyzer according to the manufacturer's recommendations.
Flow cytometry analysis
8 × 104 L929 cells were seeded in 24-well plates using complete DMEM and incubated for 24 h to achieve 80% confluence. Then, cells were washed twice with PBS and incubated with BLT-1 (15 µM) or CPZ (10 µM) in DMEM. For IFNAR1 analyses, cells were incubated for 3 h with BLT-1 or CPZ, detached using citrate buffer and labeled with PE anti-mouse IFNAR1 antibody. For uptake studies, cells were incubated for 1 h with BLT-1 and CPZ. Then, 10 µg/mL of L37pA- or BSA-FITC were added for 3 h, detached using 0.05% trypsin-EDTA, washed twice with PBS and resuspended in 200 µL of PBS containing 0.2 mg/mL of trypan blue. Analyses were performed with a FACSCalibur flow cytometer (Becton Dickinson, San Jose, CA) and data were analyzed using FlowJo software (Tree Star Inc., San Carlos, CA).
Statistical analysis
Prism software (GraphPad Software, Inc.) was employed for statistical analysis. We used the log-rank test to determine the significance of differences in survival curves. Mean differences were compared with t-tests for 2 group comparisons or one-way ANOVA followed by a multiple comparison test for three or more group comparisons. The CPE reduction assay was fitted to a Hill equation and compared with the Extra sum-of-squares F test. p values < 0.05 were considered to be statistically significant.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
KONI_A_1196309_s02.docx
Download MS Word (7.2 MB)Funding
This work was supported by the grant PI13/00207 from Instituto de Salud Carlos III, financed by the FEDER program of the European Union, by a grant from the FAECC and by the EC's H2020 PROCROP project, under grant agreement 635122. Pedro Berraondo was supported by a Miguel Servet and Miguel Servet II (CPII15/00004) contract from Instituto de Salud Carlos III.
References
- Calvo D, Vega MA. Identification, primary structure, and distribution of CLA-1, a novel member of the CD36/LIMPII gene family. J Biol Chem 1993; 268:18929-35; PMID:7689561
- Acton SL, Scherer PE, Lodish HF, Krieger M. Expression cloning of SR-BI, a CD36-related class B scavenger receptor. J Biol Chem 1994; 269:21003-9; PMID:7520436
- Williams DL, de La Llera-Moya M, Thuahnai ST, Lund-Katz S, Connelly MA, Azhar S, Anantharamaiah GM, Phillips MC. Binding and cross-linking studies show that scavenger receptor BI interacts with multiple sites in apolipoprotein A-I and identify the class A amphipathic alpha-helix as a recognition motif. J Biol Chem 2000; 275:18897-904; PMID:10858447; http://dx.doi.org/10.1074/jbc.M002411200
- Calvo D, Gomez-Coronado D, Lasuncion MA, Vega MA. CLA-1 is an 85-kD plasma membrane glycoprotein that acts as a high-affinity receptor for both native (HDL, LDL, and VLDL) and modified (OxLDL and AcLDL) lipoproteins. Arterioscler, Thromb Vasc Biol 1997; 17:2341-9; PMID:9409200; http://dx.doi.org/10.1161/01.ATV.17.11.2341
- Fioravanti J, Medina-Echeverz J, Berraondo P. Scavenger receptor class B, type I: a promising immunotherapy target. Immunotherapy 2011; 3:395-406; PMID:21395381; http://dx.doi.org/10.2217/imt.10.104
- Cai L, Wang Z, Meyer JM, Ji A, van der Westhuyzen DR. Macrophage SR-BI regulates LPS-induced pro-inflammatory signaling in mice and isolated macrophages. J Lipid Res 2012; 53:1472-81; PMID:22589557; http://dx.doi.org/10.1194/jlr.M023234
- Cai L, Ji A, de Beer FC, Tannock LR, van der Westhuyzen DR. SR-BI protects against endotoxemia in mice through its roles in glucocorticoid production and hepatic clearance. J Clin Investigat 2008; 118:364-75; PMID:18064300; http://dx.doi.org/10.1172/JCI31539
- Gowdy KM, Madenspacher JH, Azzam KM, Gabor KA, Janardhan KS, Aloor JJ, Fessler MB. Key role for scavenger receptor B-I in the integrative physiology of host defense during bacterial pneumonia. Mucosal Immunol 2015; 8:559-71; PMID:25336169; http://dx.doi.org/10.1038/mi.2014.88
- Feng H, Guo L, Wang D, Gao H, Hou G, Zheng Z, Ai J, Foreman O, Daugherty A, Li XA. Deficiency of scavenger receptor BI leads to impaired lymphocyte homeostasis and autoimmune disorders in mice. Arterioscler Thromb Vasc Biol 2011; 31:2543-51; PMID:21836069; http://dx.doi.org/10.1161/ATVBAHA.111.234716
- Gomaraschi M, Calabresi L, Rossoni G, Iametti S, Franceschini G, Stonik JA, Remaley AT. Anti-inflammatory and cardioprotective activities of synthetic high-density lipoprotein containing apolipoprotein A-I mimetic peptides. J Pharmacol Exp Therap 2008; 324:776-83; PMID:18042829; http://dx.doi.org/10.1124/jpet.107.129411
- Leelahavanichkul A, Bocharov AV, Kurlander R, Baranova IN, Vishnyakova TG, Souza AC, Hu X, Doi K, Vaisman B, Amar M et al. Class B scavenger receptor types I and II and CD36 targeting improves sepsis survival and acute outcomes in mice. J Immunol 2012; 188:2749-58; PMID:22327076; http://dx.doi.org/10.4049/jimmunol.1003445
- Baranova IN, Vishnyakova TG, Bocharov AV, Kurlander R, Chen Z, Kimelman ML, Remaley AT, Csako G, Thomas F, Eggerman TL et al. Serum amyloid A binding to CLA-1 (CD36 and LIMPII analogous-1) mediates serum amyloid A protein-induced activation of ERK1/2 and p38 mitogen-activated protein kinases. J Biol Chem 2005; 280:8031-40; PMID:15576377; http://dx.doi.org/10.1074/jbc.M405009200
- Nobecourt E, Davies MJ, Brown BE, Curtiss LK, Bonnet DJ, Charlton F, Januszewski AS, Jenkins AJ, Barter PJ, Rye KA. The impact of glycation on apolipoprotein A-I structure and its ability to activate lecithin:cholesterol acyltransferase. Diabetologia 2007; 50:643-53; PMID:17216278; http://dx.doi.org/10.1007/s00125-006-0574-z
- Shao B, Cavigiolio G, Brot N, Oda MN, Heinecke JW. Methionine oxidation impairs reverse cholesterol transport by apolipoprotein A-I. Proc Natl Acad Sci U S A 2008; 105:12224-9; PMID:18719109; http://dx.doi.org/10.1073/pnas.0802025105
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell 2010; 140:805-20; PMID:20303872; http://dx.doi.org/10.1016/j.cell.2010.01.022
- Hervas-Stubbs S, Perez-Gracia JL, Rouzaut A, Sanmamed MF, Le Bon A, Melero I. Direct effects of type I interferons on cells of the immune system. Clin Cancer Res: Off J Am Assoc Cancer Res 2011; 17:2619-27; PMID:21372217; http://dx.doi.org/10.1158/1078-0432.CCR-10-1114
- Sozzani S, Bosisio D, Scarsi M, Tincani A. Type I interferons in systemic autoimmunity. Autoimmunity 2010; 43:196-203; PMID:20298124; http://dx.doi.org/10.3109/08916930903510872
- Pestka S. The interferons: 50 years after their discovery, there is much more to learn. J Biol Chem 2007; 282:20047-51; PMID:17502369; http://dx.doi.org/10.1074/jbc.R700004200
- Motzer RJ, Hutson TE, Tomczak P, Michaelson MD, Bukowski RM, Rixe O, Oudard S, Negrier S, Szczylik C, Kim ST et al. Sunitinib versus interferon alfa in metastatic renal-cell carcinoma. New Engl J Med 2007; 356:115-24; PMID:17215529; http://dx.doi.org/10.1056/NEJMoa065044
- Scheel TK, Rice CM. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat Med 2013; 19:837-49; PMID:23836234; http://dx.doi.org/10.1038/nm.3248
- Escobar-Zarate D, Liu YP, Suksanpaisan L, Russell SJ, Peng KW. Overcoming cancer cell resistance to VSV oncolysis with JAK1/2 inhibitors. Cancer Gene Ther 2013; 20:582-9; PMID:24030211; http://dx.doi.org/10.1038/cgt.2013.55
- Nguyen TL, Abdelbary H, Arguello M, Breitbach C, Leveille S, Diallo JS, Yasmeen A, Bismar TA, Kirn D, Falls T et al. Chemical targeting of the innate antiviral response by histone deacetylase inhibitors renders refractory cancers sensitive to viral oncolysis. Proc Natl Acad Sci U S A 2008; 105:14981-6; PMID:18815361; http://dx.doi.org/10.1073/pnas.0803988105
- Vigil A, Park MS, Martinez O, Chua MA, Xiao S, Cros JF, Martinez-Sobrido L, Woo SL, Garcia-Sastre A. Use of reverse genetics to enhance the oncolytic properties of Newcastle disease virus. Cancer Res 2007; 67:8285-92; PMID:17804743; http://dx.doi.org/10.1158/0008-5472.CAN-07-1025
- Bocharov AV, Baranova IN, Vishnyakova TG, Remaley AT, Csako G, Thomas F, Patterson AP, Eggerman TL. Targeting of scavenger receptor class B type I by synthetic amphipathic alpha-helical-containing peptides blocks lipopolysaccharide (LPS) uptake and LPS-induced pro-inflammatory cytokine responses in THP-1 monocyte cells. J Biol Chem 2004; 279:36072-82; PMID:15199068; http://dx.doi.org/10.1074/jbc.M314264200
- Fernandez-Lizarbe S, Montesinos J, Guerri C. Ethanol induces TLR4/TLR2 association, triggering an inflammatory response in microglial cells. J Neurochem 2013; 126:261-73; PMID:23600947; http://dx.doi.org/10.1111/jnc.12276
- Wang YC, Zhou Y, Fang H, Lin S, Wang PF, Xiong RP, Chen J, Xiong XY, Lv FL, Liang QL et al. Toll-like receptor 2/4 heterodimer mediates inflammatory injury in intracerebral hemorrhage. Ann Neurol 2014; 75:876-89; PMID:24752976; http://dx.doi.org/10.1002/ana.24159
- Barton GM, Medzhitov R. Toll-like receptor signaling pathways. Science 2003; 300:1524-5; PMID:12791976; http://dx.doi.org/10.1126/science.1085536
- Nieland TJ, Penman M, Dori L, Krieger M, Kirchhausen T. Discovery of chemical inhibitors of the selective transfer of lipids mediated by the HDL receptor SR-BI. Proc Natl Acad Sci U S A 2002; 99:15422-7; PMID:12438696; http://dx.doi.org/10.1073/pnas.222421399
- Masson D, Koseki M, Ishibashi M, Larson CJ, Miller SG, King BD, Tall AR. Increased HDL cholesterol and apoA-I in humans and mice treated with a novel SR-BI inhibitor. Arterioscler Thromb Vasc Biol 2009; 29:2054-60; PMID:19815817; http://dx.doi.org/10.1161/ATVBAHA.109.191320
- Nieland TJ, Chroni A, Fitzgerald ML, Maliga Z, Zannis VI, Kirchhausen T, Krieger M. Cross-inhibition of SR-BI- and ABCA1-mediated cholesterol transport by the small molecules BLT-4 and glyburide. J Lipid Res 2004; 45:1256-65; PMID:15102890; http://dx.doi.org/10.1194/jlr.M300358-JLR200
- Zoon KC, Arnheiter H, Zur Nedden D, Fitzgerald DJ, Willingham MC. Human interferon alpha enters cells by receptor-mediated endocytosis. Virology 1983; 130:195-203; PMID:6314645; http://dx.doi.org/10.1016/0042-6822(83)90127-7
- Kumar KG, Barriere H, Carbone CJ, Liu J, Swaminathan G, Xu P, Li Y, Baker DP, Peng J, Lukacs GL et al. Site-specific ubiquitination expose s a linear motif to promote interferon-alpha receptor endocytosis. J Cell Biol 2007; 179:935-50; PMID:18056411; http://dx.doi.org/10.1083/jcb.200706034
- Marchetti M, Monier MN, Fradagrada A, Mitchell K, Baychelier F, Eid P, Johannes L, Lamaze C. Stat-mediated signaling induced by type I and type II interferons (IFNs) is differentially controlled through lipid microdomain association and clathrin-dependent endocytosis of IFN receptors. Mol Biol Cell 2006; 17:2896-909; PMID:16624862; http://dx.doi.org/10.1091/mbc.E06-01-0076
- Nestle FO, Di Meglio P, Qin JZ, Nickoloff BJ. Skin immune sentinels in health and disease. Nat Rev Immunol 2009; 9:679-91; PMID:19763149
- Guo L, Song Z, Li M, Wu Q, Wang D, Feng H, Bernard P, Daugherty A, Huang B, Li XA. Scavenger receptor BI protects against septic death through its role in modulating inflammatory response. J Biol Chem 2009; 284:19826-34; PMID:19491399; http://dx.doi.org/10.1074/jbc.M109.020933
- Zhu P, Liu X, Treml LS, Cancro MP, Freedman BD. Mechanism and regulatory function of CpG signaling via scavenger receptor B1 in primary B cells. J Biol Chem 2009; 284:22878-87; PMID:19542230; http://dx.doi.org/10.1074/jbc.M109.018580
- Anantharamaiah GM, Jones JL, Brouillette CG, Schmidt CF, Chung BH, Hughes TA, Bhown AS, Segrest JP. Studies of synthetic peptide analogs of the amphipathic helix. Structure of complexes with dimyristoyl phosphatidylcholine. J Biol Chem 1985; 260:10248-55; PMID:4019510
- Barbalat R, Lau L, Locksley RM, Barton GM. Toll-like receptor 2 on inflammatory monocytes induces type I interferon in response to viral but not bacterial ligands. Nat Immunol 2009; 10:1200-7; PMID:19801985; http://dx.doi.org/10.1038/ni.1792
- Brandt KJ, Fickentscher C, Kruithof EK, de Moerloose P. TLR2 ligands induce NF-kappaB activation from endosomal compartments of human monocytes. PloS One 2013; 8:e80743; PMID:24349012; http://dx.doi.org/10.1371/journal.pone.0080743
- Zhao LJ, Wang W, Wang WB, Ren H, Qi ZT. Involvement of ERK pathway in interferon alpha-mediated antiviral activity against hepatitis C virus. Cytokine 2015; 72:17-24; PMID:25544181; http://dx.doi.org/10.1016/j.cyto.2014.11.031
- Bieniasz PD. Intrinsic immunity: a front-line defense against viral attack. Nat Immunol 2004; 5:1109-15; PMID:15496950; http://dx.doi.org/10.1038/ni1125
- Nieland TJ, Ehrlich M, Krieger M, Kirchhausen T. Endocytosis is not required for the selective lipid uptake mediated by murine SR-BI. Biochimica et Biophysica Acta 2005; 1734:44-51; PMID:15866482; http://dx.doi.org/10.1016/j.bbalip.2005.02.007
- Liu B, Krieger M. Highly purified scavenger receptor class B, type I reconstituted into phosphatidylcholine/cholesterol liposomes mediates high affinity high density lipoprotein binding and selective lipid uptake. J Biol Chem 2002; 277:34125-35; PMID:12110672; http://dx.doi.org/10.1074/jbc.M204265200
- Faloon PW, Dockendorff C, Youngsaye W, Yu M, Nag PP, Lewis TA, Bennion M, Paterson C, Lam G, Dandapani S et al. A Small Molecule Inhibitor of Scavenger Receptor BI-mediated Lipid Uptake—Probe 1. 2011 Dec 15 [Updated 2014 Sep 18]. In: Probe Reports from the NIH Molecular Libraries Program [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2010; PMID:23658945
- Eckhardt ER, Cai L, Shetty S, Zhao Z, Szanto A, Webb NR, Van der Westhuyzen DR. High density lipoprotein endocytosis by scavenger receptor SR-BII is clathrin-dependent and requires a carboxyl-terminal dileucine motif. J Biol Chem 2006; 281:4348-53; PMID:16368683; http://dx.doi.org/10.1074/jbc.M513154200
- Eckhardt ER, Cai L, Sun B, Webb NR, van der Westhuyzen DR. High density lipoprotein uptake by scavenger receptor SR-BII. J Biol Chem 2004; 279:14372-81; PMID:14726519; http://dx.doi.org/10.1074/jbc.M313793200
- Thilakawardhana S, Everett DM, Murdock PR, Dingwall C, Owen JS. Quantification of apolipoprotein E receptors in human brain-derived cell lines by real-time polymerase chain reaction. Neurobiol Aging 2005; 26:813-23; PMID:15718039; http://dx.doi.org/10.1016/j.neurobiolaging.2004.08.004
- Azhar S, Reaven E. Scavenger receptor class BI and selective cholesteryl ester uptake: partners in the regulation of steroidogenesis. Mol Cell Endocrinol 2002; 195:1-26; PMID:12354669; http://dx.doi.org/10.1016/S0303-7207(02)00222-8
- Uze G, Schreiber G, Piehler J, Pellegrini S. The receptor of the type I interferon family. Curr Topics Microbiol Immunol 2007; 316:71-95; PMID:17969444
- Crouse J, Kalinke U, Oxenius A. Regulation of antiviral T cell responses by type I interferons. Nat Rev Immunol 2015; 15:231-42; PMID:25790790; http://dx.doi.org/10.1038/nri3806
- Melero I, Quetglas JI, Reboredo M, Dubrot J, Rodriguez-Madoz JR, Mancheno U, Casales E, Riezu-Boj JI, Ruiz-Guillen M, Ochoa MC et al. Strict requirement for vector-induced type I interferon in efficacious antitumor responses to virally encoded IL12. Cancer Res 2015; 75:497-507; PMID:25527611; http://dx.doi.org/10.1158/0008-5472.CAN-13-3356
- Quetglas JI, Fioravanti J, Ardaiz N, Medina-Echeverz J, Baraibar I, Prieto J, Smerdou C, Berraondo P. A Semliki forest virus vector engineered to express IFNalpha induces efficient elimination of established tumors. Gene Ther 2012; 19:271-8; PMID:21734727; http://dx.doi.org/10.1038/gt.2011.99
