ABSTRACT
Tumor-derived exosomes (TEX) are ubiquitously present in the tumor microenvironment and plasma of cancer patients. TEX carry a cargo of multiple stimulatory and inhibitory molecules and deliver them to recipient cells, serving as a communication network for the tumor. The mechanisms TEX use for delivering messages to recipient cells were evaluated using PKH26-labeled TEX produced by cultured human tumor cells, exosomes produced by dendritic cells-derived exosomes (DEX), or exosomes isolated from plasma of cancer patients (EXO). Human T-cell subsets, B cells, NK cells, and monocytes were co-incubated with TEX, DEX, or EXO and binding or internalization of labeled vesicles was evaluated by confocal microscopy and/or Amnis-based flow cytometry. Vesicle-induced Ca2+ influx in recipient T cells was monitored, and TEX-induced inosine production in Treg was determined by mass spectrometry. In contrast to B cells, NK cells or monocytes, conventional T cells did not internalize labeled vesicles. Minimal exosome uptake was only evident in Treg following prolonged co-incubation with TEX. All exosomes induced Ca2+ influx in T cells, with TEX and EXO isolated from cancer patients' plasma delivering the strongest, sustained signaling to Treg. Such sustained signaling resulted in the significant upregulation of the conversion of extracellular ATP to inosine (adenosine metabolite) by Treg, suggesting that TEX signaling could have functional consequences in these recipient cells. Thus, modulation of Treg suppressor functions by TEX is mediated by mechanisms dependent on cell surface signaling and does not require TEX internalization by recipient cells.
Introduction
Exosomes are the smallest subset of extracellular vesicles, measuring 30–150 nm in diameter.Citation1,2 Exosomes are produced by all cells and can be found in all body fluids.Citation3 Their origin from the endocytic compartment of the parent cell sets them apart from other vesicles, which are either formed by “blebbing” of the surface membrane or represent remnants of apoptotic cells.Citation4 The exosome biogenesis involves the fusion of multivesicular bodies enclosing pools of endosomal vesicles with the cell plasma membrane and the subsequent release of the vesicles into intercellular space.Citation5 The exosome cargo of proteins, lipids, and glycans characteristically contains endocytic components, such as ALIX or TSG101 as well as tetraspanins (e.g., CD63 and CD81) commonly used as exosome “markers.”Citation6
Exosomes are considered to be vehicles for information transfer between the parent and recipient cells. As such, exosomes carry and deliver messages from the parent to the recipient cell. While considerable knowledge has accumulated about the biogenesis of exosomes,Citation7,8 relatively little is known about their uptake by recipient cells. It is known, however, that exosomes can interact with target cells by several different mechanisms, ranging from receptor/ligand interactions to endocytosis [reviewed in Citationref. 9]. The mechanism used for the delivery of the exosome cargo and its uptake by recipient cells depends primarily on the nature of the recipient cell. Thus, cells equipped with the ability to phagocytose, such as macrophages, readily internalize exosomes, un-coat them and process their content, which results in alterations in the transcriptome and ultimately cellular functions.Citation10 Non-phagocytic cells, such as subsets of T lymphocytes, which express receptors for ligands carried by exosomes, depend on specific receptor-ligand recognition for the implementation of exosome-delivered signals.Citation11 In this case, the molecular content of exosome membrane-bound cargo could serve as an “address,” and might play a role in directing exosomes to the appropriate recipient cells.
Cellular responses to the exosome-mediated information transfer involve phenotypic, functional, and genetic modifications in responding cells. We have previously reported that tumor-derived exosomes (called TEX) suppress functions of immune cells either directly by delivering suppressive signals/molecules to activated lymphocytesCitation12-15 or indirectly by promoting expansion and activity of regulatory T cells (Treg).Citation16 However, mechanistic aspects of direct or indirect T cell interactions with TEX remain poorly understood. Specifically, it is not clear whether exosomes have to be internalized by T cells before inducing cellular changes or whether signals received upon exosome binding to the cell surface are sufficient for of T cell responses. Based on our preliminary (i.e., receptor-ligand signaling) data reported by Muller et al.,Citation17 we hypothesized that negative signaling by TEX interacting with conventional CD8+ and CD4+ T cells depends on the receptor-ligand contact on the cell surface. In contrast, the upregulation of Treg functions might require TEX uptake and re-programming of cellular activities in Treg. To gain insights into exosome-T cell crosstalk, we co-incubated dye-labeled TEX with human T cells or other mononuclear immune cells and evaluated signal delivery via Ca2+ signaling and exosome uptake by the recipient cells. We report that T lymphocytes, unlike other mononuclear cells, do not internalize exosomes but rather respond to exosome signals delivered to the cell surface by Ca2+ influx followed by down-stream functional responses. Thus, functions of Treg as well as conventional T cells are regulated by TEX via receptor-ligand signaling on the cell surface rather than uptake mechanisms.
Results
Co-incubation with TEX induces apoptosis of activated CD8± T cells
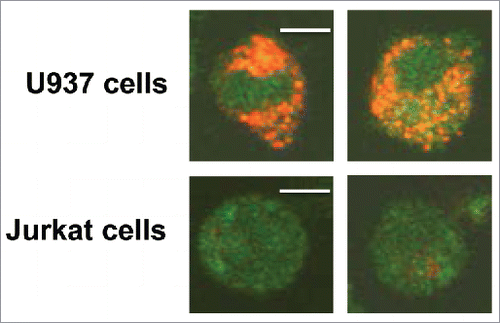
Our previously reported and current data consistently show that activated human CD8+T cell co-incubated with TEX bind AnnexinV and undergo apoptosis (Fig. S1). Interestingly, when we co-incubated CD8+ Jurkat cells (as surrogates for primary T cells) with the PKH26 dye-labeled TEX isolated from supernatants of tumor cell lines or plasma of patients with cancer for up to 24 h, we did not observe TEX uptake by these T cells (). No TEX were internalized even after prolonged co-incubation (up to 72 h). In contrast, a tumor cell (U937) strongly internalized the same TEX (). Staining intensity of tumor cells treated with trypsin after co-incubation with TEX was comparable to that of untreated cells, suggesting the TEX uptake and not just their binding to the cell surface. These studies suggested that TEX can induce functional changes (i.e., apoptosis) in CD8+ Jurkat cells without internalization.
Figure 1. Differential uptake of PKH26-labeled TEX by U937tumor cells and by Jurkat T cells. Representative images showing strong internalization of PKH26-labeled TEX by U937 cells (upper row) but no TEX uptake in CD8+ Jurkat cells (lower row). TEX were isolated from supernatants of PCI-13 tumor cells and labeled with the PKH26 dye as described in Methods. Cells co-incubated with 10 µg of TEX for 24 h in a 35 mm MatTek plate were examined in a confocal microscope. The bar = 5 μm.
Binding or internalization of TEX by human mononuclear cell subsets (MNC)
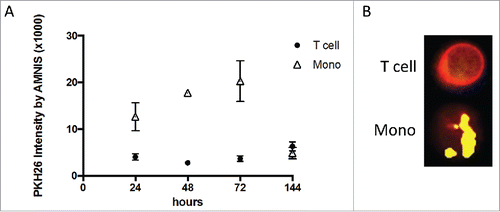
To further evaluate exosome interactions with the MNC subsets, we used the AMNIS image analysis system. Binding to or internalization of TEX, DEX, or EXO by immune cells was found to be time dependent, dose dependent, and recipient cell type dependent. As shown in , the co-incubation of PKH26-labeled TEX with T cells did not lead to the uptake of TEX even after >72 h co-incubation. In contrast, monocytes readily internalized labeled TEX, as indicated by the cell images taken at 48 h ().
Figure 2. Differences in uptake of PKH26-labeled TEX cells and monocytes. (A) Time-dependent uptake of TEX by monocytes contrasts with the slower and lower TEX uptake by T cells. Amnis measurements of PKH26 intensity in cells were acquired after increasing co-incubation times. (B) Representative Amnis images show TEX uptake by monocytes vs. no uptake by T cells after 48 h of co-incubation.
In addition to monocytes, B cells and NK cells internalized TEX, as illustrated in .
Figure 3. Amnis-generated data showing differences in TEX uptake by various human MNC subsets. (A) Various MNC subsets isolated from different ND were co-incubated with TEX for 24–48 h. T cells T show significantly lower TEX uptake compared with other MNC subsets. (B) Comparisons of TEX uptake by resting or activated CD8+ T cells, CD4+ T cells, or CD4+CD39+ Treg show a lack of TEX internalization by CD8+ T cells relative to low but significantly increased TEX uptake by Treg. T cell subsets were isolated from the peripheral blood of four different ND. In (A) and (B), the data are presented as mean levels of PKH26 intensity in recipient cells ± SD. The p values denote significant differences.
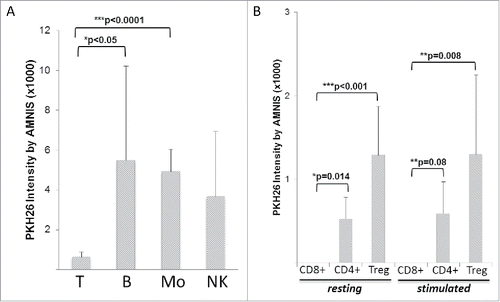
Differences in the exosome uptake at 24 h between T cells and the other MNC subsets were highly significant (). Clearly, the uptake of exosomes by immune cells depended on the type of recipient immune cells: T cells did not internalize exosomes, while the other MNCs did.
To determine whether pre-activation of the recipient cells influences exosome uptake, we co-incubated resting or activated T cells with PKH26-labeled TEX or DEX. As shown in Fig. S2A, the activation of the recipient T cells had no effect on the uptake of either TEX or DEX, which was equally low and not significantly different for these two exosome types. In contrast to T cells, resting or activated B cells, effectively internalized TEX or DEX, and the uptake by activated B cells was greater (p = 0.03) than that by resting B cells (Fig. S2B). Also, activated NK cells and monocytes internalized TEX or DEX with significantly greater efficiency (p < 0.0001) than activated recipient T cells (Fig. S2C). In aggregate, the Amnis-generated results showed that TEX and DEX are equally well internalized by MNC, except for T cells that did not internalize either. Pre-activation of recipient cells appears to improve the uptake of TEX and DEX by monocytes and NK cells as well as B cells.
Exosome interactions with Treg
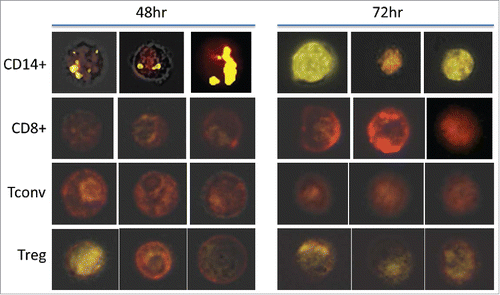
We have previously reported that the co-incubation of CD4+CD25hiCD39+ Treg with TEX or DEX induced changes in the transcriptome of the recipient cells.Citation17 Therefore, it was of interest to determine whether Treg internalized any TEX or DEX relative to CD8+ or CD4+Tconv cells. As shown in , pre-activated or resting CD8+ T cells did not take up labeled exosomes during 24 h co-incubation, with CD4+ Tconv cells demonstrating only weak positivity by Amnis, and Treg showing significantly better but still very low uptake at 24 h (compare with the uptake by other MNC in ). The activation of Treg via the T cell receptor (TcR) did not improve exosome uptake, and the uptake of EXO obtained from cancer patients' or normal control's (NC's) plasma was equivalent to that of TEX or DEX (data not shown). presents representative Amnis images of the TEX uptake by monocytes and various T cell subsets following 48 h and 72 h co-incubation with labeled exosomes. The images clearly show that in comparison to negative CD8+T cells and CD4+Tconv, weak but detectable remnants of PKH26+ exosomes can be encountered in some but not all Treg. Thus, interactions of TEX, DEX, or EXO with T lymphocytes did not involve their internalization, except in the case of Treg, where the binding of exosomes to the cell surface was followed by weak and reluctant internalization.
Figure 4. Amnis-generated representative images of recipient MNC co-incubated with PKH26-labeled TEX for 48 or 72 h. Immune cell subsets were isolated from healthy donors' plasma and analyzed by Amnis Image Stream as described in Methods. The presented images are representative results of four experiments performed with MNC of different donors and show results obtained by a triple overlay (PKH26-stain in yellow, surface stain in red, and a brightfield image) as described in Methods.
Exosomes induce Ca2+ influx in T cells
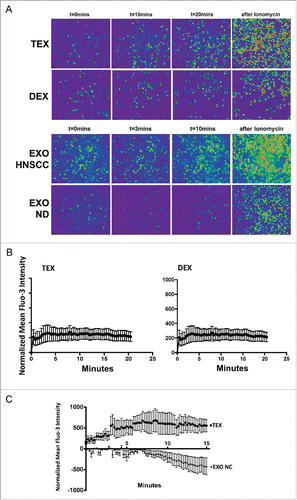
The data we previously reported showed that TEX induced significant changes in the phenotype and functions of T lymphocytes, including Treg.Citation15 The Amnis uptake data for TEX described above suggest that these phenotypic and functional changes in T cells are not accompanied by TEX internalization. Therefore, we considered the possibility that signals delivered by TEX to the T cell surface are responsible for the observed effects. Given that Ca2+ signaling is essential for T cell activation and T-cell-dependent immune responses,Citation18 we next sought to determine whether co-incubation of T cells with exosomes induced Ca2+ influx in recipient cells. We measured the strength of Ca2+ flux induced by TEX or EXO in various T cell subsets by confocal microscopy after labeling T cells with the Fluo-3 dye. As expected, Ca2+ flux in T cells was time dependent. illustrates results of confocal microscopy for T cells co-incubated with TEX or DEX for various time periods. Ca2+ influx induced by DEX appears weaker and is already extinguished by 10 min relative to TEX-induced flux. EXO isolated from the plasma of a cancer patient induced stronger and longer lasting flux than TEX, while EXO isolated from NC's plasma were negative. Among T cells, CD4+CD39+ Treg were preferentially targeted by TEX, showing the strongest and sustained Ca2+ influx within the first 20 min of co-incubation with TEX (). DEX co-incubated with Treg induced only a very weak signal that remained flat during 20 min co-incubation (). Furthermore, we compared Ca2+ flux in conventional CD4+T cells co-incubated with EXO obtained from the plasma of head and neck squamous cell carcinoma (HNSCC) patients (n = 3) with active disease (AD) vs. EXO obtained from the plasma of NDs (n = 3). Representative data () indicate that the EXO of patients with cancer induced Ca2+ flux and signaling in T cells, while the EXO of ND did not, similar to what we saw with EXO isolated from the plasma of NC in . These data are consistent with our previous reports on effects of exosomes on activated immune cells, with a caveat that TEX-induced signaling leads to activation of inhibitory pathways in these T cells, while EXO from NC's plasma do not induce immunosuppression.Citation12
Figure 5. Ca2+ flux in human T cells co-incubated with TEX, DEX, or EXO. In (A), T cells co-incubated with exosomes were labeled with Fluo-3 and examined in a Nikon A1 microscope as indicated in Methods. Ionomycin, which induces strong Ca2+ flux, was used as a positive control. T cells were imaged at different times after Fura-2 addition. The upper panel compares effects of TEX and DEX. The lower panel compares effects of EXO isolated from plasma of a representative HNSCC patient vs. EXO isolated from ND's plasma. In (B), Fluo-3 intensity levels in T cells co-incubated with TEX or DEX. The data are from a representative experiment of 3 performed with T cells of different donors. Note the strong and prolonged signal in T cells co-incubated with TEX and the absence of signaling in T cells co-incubated with DEX. In C, Fluo-3 intensity levels in CD4+T conv cells co-incubated with EXO from the plasma of HNSCC patient with active disease (AD) or with EXO from ND's plasma. Note the strong, prolonged signal induced by patients' EXO vs. an absence of signal with ND's EXO. Shown are representative Ca2+ flux data obtained from four to six experiments performed with T cells of different subjects.
TEX induce functional alterations in T cells
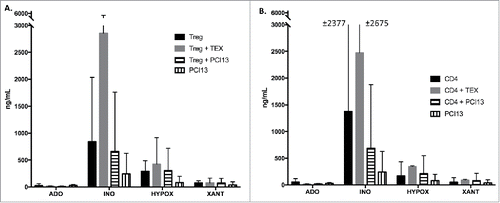
In experiments performed in parallel with Ca2+ flux measurements, we co-incubated CD4+CD39+ Treg or CD4+ T cells with TEX in the presence of exogenous ATP for 16 h. Production of purines by the cells ± TEX was measured by mass spectrometry (). In addition, PCI-13 cells (the TEX parent cells) were co-incubated with T cells (). The data showed that after 16 h of co-incubation, adenosine is consumed and inosine is the main product of enzymatic ATP hydrolysis. Co-incubation with TEX induced significant (p < 0.0074) upregulation of inosine production in Treg but not in CD4+ T cells, probably because of large differences among the three cell donors (). These data are consistent with the results of Ca2+ experiments. Interestingly, PCI-13 cells did not significantly upregulate inosine production, suggesting that TEX deliver a stronger signal to recipient T cells than to the parent tumor cells. Overall, the data indicate that in T cells, signaling by TEX leads to detectable, biologically relevant changes in functions of recipient cells.
Figure 6. Mass spectrometry for purine derivatives of adenosine in co-cultures of TEX with Treg or CD4+ T cells. TEX were co-incubated with T cells or PCI-13 tumor cells in the presence of exogenous ATP for 16 h. T cells were isolated from three different donors. Cells and supernatants were collected and prepared for measurements of purines by mass spectrometry as described in Methods. In (A), TEX but not the parent PCI-13 tumor cells induced a large increase in inosine production by CD4+CD39+ Treg. In (B), TEX co-incubated with CD4+T cells also induced the upregulation of inosine production, although there were large differences among cells of different donors. The data are means ± SEM from experiments performed with T cells of three different donors.
Discussion
Exosomes are considered to be information-transfer vehicles that deliver messages from the parent to recipient cells located within or outside of the microenvironment. The mechanisms used for directing exosome trafficking and message delivery and the role of exosomes in inducing phenotypic and functional changes in recipient cells are not yet understood. Evidence has emerged that TEX exert inhibitory effects on immune cells.Citation15,19,20 TEX, which are ubiquitous in the tumor microenvironment, carry a cargo of suppressive and stimulatory moleculesCitation15 and, upon contacting the surface of immune cells, might suppress functions of some cells, while upregulating functions of others. Specifically, based on our previous data,Citation14,16 we assumed that Treg would respond to stimulatory signals, while CD8+ effector T cells would be suppressed by interactions with TEX. Furthermore, it is likely that TEX and exosomes produced by normal cells, e.g., DEX, which carry molecularly distinct cargos, as we previously reported,Citation17 will exert different effects upon co-incubation with human T cell subsets.
Previously, we and others have shown that TEX-induced immunosuppression involves signaling of FasL+ exosomes via CD95 (Fas) receptor on activated CD8+ T cells.Citation13,21 Other receptor-ligand interactions (e.g., TRAIL/TRAILR, TGF-β/TGF-βR, or ADO/A2AR) are also implicated in TEX-mediated suppression in the tumor microenvironment.Citation17,22,23 The data suggest that TEX convey inhibitory signals to T cells by delivering ligands tethered to the exosome membrane to the cognate receptors on immune cells. We and others have observed that phenotypic and functional changes brought about by TEX interacting with T cells result in transcriptional alterations of multiple immunoregulatory genes.Citation17 As exosome cargos contain mRNAs and miRNAs,Citation24,25 transfer of genetic materials from the parent to recipient cells could be the mechanism responsible for the observed effects. Indeed, evidence for transfer of genes by exosomes has been forthcoming from numerous studies involving different cell types.Citation26-28 Exosomes transferring their contents to recipient cells must bind, fuse with the surface membrane, and unload their cargo inside the recipient cell. Several mechanisms have been proposed for this type of information transfer, largely based on the use of inhibitors able to partially (but almost never completely) block the uptake.Citation9 While endocytosis, a mechanistically complex and varied process of internalization, appears to be a major method of exosome entry into recipient cells, both the origin of exosomes and the cell type receiving them might be of critical importance for productive information transfer.Citation29
The mechanisms of information transfer and delivery by TEX to immune cells are critical for understanding the crosstalk between the tumor and the host immune system. Here, we show that signals delivered by TEX to the cell surface are sufficient for eliciting changes in the phenotypic or functional profile of recipient T cells and do not require TEX internalization by these cells. Among MNC, T cells alone did not internalize TEX, DEX, or EXO even following activation via the TcR. Their responses largely depended on surface signaling as indicated by Ca2+ influx, which was especially strong with TEX, weaker with DEX and EXO of cancer patients and negative with EXO isolated from NC's plasma. In contrast to CD8+ and CD4+ conv T cells, Treg showed the evidence of weak but positive uptake of TEX after prolonged (48 h) exposure to TEX. Importantly, TEX also induced strong and sustained Ca2+ signal in Treg, while in Treg co-incubated with DEX, the Ca2+ signal was weak and flat. This observation suggests that the exosomal cargo, most likely the presence of relevant ligands in the exosomal membrane, determines the strength of signals that translate downstream into functional alterations in T cells. Several examples of ligands carried by exosomes (or produced by enzymatic cleavage of exosome membrane proteins) interacting with receptors and inducing downstream signaling effects have been reported. For example, stimulation by exosomes of the MAPK pathway leading to altered activity in recipient cells has been reported.Citation30 Importantly, in our hands, only TEX induced sustained Ca2+ influx in Treg, while EXO from NC's plasma did not. This confirms that in cancer patients, TEX represent a highly efficient signaling system programmed by the parent tumor cell to crosstalk with Treg that regulate antitumor immune responses.
To demonstrate that TEX-delivered signals alter the functional repertoire of the immune cells expressing receptors for the cognate signals, we focused on the adenosine pathway. T cells are known to express the A2AR and respond to adenosine by cAMP upregulation, which results in alterations in their functions, as we and others have shown.Citation31 More recently, it has been reported that inosine, a metabolite of adenosine, also binds to and signals via A2AR.Citation32 Furthermore, inosine sustains prolonged A2AR signaling long after its more potent predecessor, adenosine, is degraded, thus maintaining the immunomodulatory role of the A2AR.Citation32 Our previous observations that CD39+CD73+ TEX signaling via the A2AR rapidly induce adenosine production by TregCitation22 are now extended to illustrate that prolonged exposure to TEX upregulates the production of inosine in Treg. This may be an example of TEX-mediated “A to I editing,” which is now thought to play an important role in a variety of physiologically relevant cell functions.Citation33 It also appears that A2AR-mediated signals delivered to Treg by TEX were significantly more effective in stimulating inosine production than were the TEX parent cells, emphasizing the importance of exosomes in mediating intercellular communication. The ability of TEX to regulate suppressive activities of Treg at a distance, using receptors present on the recipient cell surface, represents an efficient and so far underestimated mechanism of tumor-induced immune suppression operating in cancer.
Materials and methods
Peripheral blood specimens
Buffy coats obtained from healthy volunteers and used for isolations of human leukocyte subsets were purchased from the Central Blood Bank of Pittsburgh. Fresh specimens of venous peripheral blood were obtained from patients with HNSCC or NC, all of whom signed the informed consent approved by the institutional IRB (IRB #991206). Peripheral blood mononuclear cells (PBMC) were isolated by centrifugation on Ficoll-Hypaque, washed in the RPMI medium and immediately used for experiments.
Exosome isolation
Exosomes were isolated from: (a) supernatants of the HNSCC cell line, PCI-13, maintained in a long-term culture.Citation34 This cell line served as a source of pure TEX; (b) the plasma of patients with cancer [HNSCC or acute myelogenous leukemia (AML)] and the plasma of NC as described below. These plasma-derived exosomes are referred to as EXO; (c) supernatants of human dendritic cells (DC) cultured from monocytes isolated from PBMC by adherence to plastic and incubated in the presence of IL-4 and GM-CSFCitation35 for 4 d. These DC supernatants were used as a source of DC-derived exosomes (DEX). The DC cultured from plastic-adherent monocytes were >90% CD40+CD83+CD86+DR+ by flow cytometry. The media used for cell cultures contained FCS that was ultracentrifuged at 100,000×g for 3 h to deplete it of bovine exosomes.
The method for exosome isolation was described previously by us.Citation36 Briefly, differential centrifugation (1,000×g for 10 min and 2,000×g for 10 min at 4°C and then 10,000×g for 30 min at 4°C) was followed by ultrafiltration (0.22 μm filter; Millipore, Billerica, MA, USA) and then by size-exclusion chromatography (SEC) on mini-columns packed with Sepharose 2B (Sigma-Aldrich, St. Louis, MO, USA). The exclusion-volume fractions were collected and concentrated by Vivaspin 500 centrifugal concentrator (Sartorius Corp., New York, NY). The exosome fractions isolated from plasma contained a mix of vesicles originating from different parent cells, although in patients with cancer, these fractions were variably enriched in TEX.
Characterization of isolated exosomes
Protein concentrations of exosome fractions were determined using a BCA Protein Assay kit as recommended by the manufacturer (Pierce, Thermo Scientific, Rockford, IL 61105, USA). The isolated exosomes were evaluated for morphology by transmission electron microscopy (TEM), particle distribution and size in a NanoSight instrument and biologic activity by flow cytometry to demonstrate their ability to downregulate NKG2D expression in isolated human NK cells as described previously.Citation36 TEM of isolated exosomes was performed at the Center for Biologic Imaging at the University of Pittsburgh. Briefly, freshly isolated exosomes were put on a copper grid coated with 0.125% Formvar in chloroform. The grids were stained with 1% (v/v) uranyl acetate in ddH2O, and the exosome samples were examined immediately. A JEM 1011 transmission electron microscope was used for imaging.
Exosome labeling with the PKH-26 dye
Isolated exosomes (up to 200μg protein) were added to wells containing 2 mL of PKH26 (10−6 M in diluent buffer) and after 60 sec the staining reaction was stopped by adding 7 mL of PBS. Exosomes were pelleted by ultracentrifugation at 100,000×g for 2 h. They were then re-suspended in PBS at the concentration of ∼1 μg protein/μL and used for co-incubation with immune cells.
Isolation of human mononuclear cell subsets
T cell subsets were isolated from human PBMC via an immunoaffinity-based capture procedure, using Miltenyi beads as described previously.Citation37 Briefly, before immunocapture, mononuclear cells were plated on Ab-coated plates to collect CD14+ monocytes. Negative selection was used to recover CD19+ B cells, CD4+, or CD8+ T cells or CD3-CD56+CD16+ NK cells as needed. To isolate Treg, negatively selected CD4+ T cells were separated by positive selection using anti-CD39 Ab-coated Miltenyi beads into CD4+CD39+ Treg and CD4+CD39neg T effector cells. The purity of the isolated cell subsets was determined by flow cytometry. The isolated T cell subsets were either directly used for experiments (resting T cells) or activated by incubation in the presence of anti-CD3/anti-CD28 antibody (Ab)-coated beads and IL-2 (150 U/mL) for 4 h or overnight, depending on the experiment. To confirm activation, cells were harvested, stained for CD69, and the frequency of CD69+ T cells as well as CD69 expression (MFI) on the cell surface were determined by flow cytometry.Citation36 The MFI values were converted into MESF units, based on fluorescent intensity curves generated with calibration beads.
Flow cytometry of PBMC
The phenotype of freshly-isolated primary mononuclear cell subsets was evaluated by multiparameter flow cytometry. Cells were incubated in the dark for 20 min at room temperature (RT) with a panel of labeled monoclonal antibodies (mAbs): anti-CD4-eFluor 450 (48–0048–42, eBioscience, CA, USA), CD8-ECD (6604728, Beckman Coulter, Brea, CA, USA), CD39-PC7 (25–0399–42, eBioscience); CD20-PE (IM1451U, Beckman Coulter); CD56-PE (IM2073U, Beckman Coulter); CD14-FITC (IM0645U, Beckman Coulter); CD16-FITC (IM0814U, Beckman Coulter); CD25-APC/Cy7 (302613, BioLegend, San Diego, CA, USA) and Foxp3-APC (17–4776–41, eBioscience). Isotype controls were used for each experiment. After incubation, cells were again washed, re-suspended in flow buffer and analyzed using a Gallios flow cytometer (Beckman Coulter). At least 5 × 104 events were collected, and the data were analyzed using Kaluza software (Beckman Coulter). For fluorescence quantitation standardized fluorochrome microspheres were used as recommended by the company (Quantum™ FITC-5 MESF, 555, Bangs Laboratories, Fishers, Indiana, USA).
Co-incubation of resting or activated PBMC subsets with PKH26-labeled exosomes
Isolated subsets of PBMC (lymphocyte subsets or monocytes) were used either without or with in vitro activation. T lymphocytes were incubated in a medium containing 150I U/mL of IL-2 (Miltenyi Biotech Inc.,) and 150 µL of microbeads coated with anti-CD3/CD28 Abs (Miltenyi). B cells were activated using CD40L and IL-4 as described previously.Citation38 Monocytes were activated with 10 ng/mL of LPS (Sigma Aldrich, St. Louis). All cells were activated for 24 h. Pre-activated or resting cells were seeded in wells of a 96-well plate at the cell concentration of 1 × 106 per mL in triplicate. The PKH26-labeled exosomes (50–100 µg protein) were added to each well and the plate was incubated for various time periods from 4 to 72 h at 37°C. Control wells contained cells but no exosomes. Following extensive washing in PBS to remove exosomes, cells were examined by confocal microscopy or stained with labeled Abs for Amnis image cytometry.
AMNIS image cytometry
Following co-incubation with PKH26-labeled exosomes cells was harvested, washed, and stained for surface markers with antibodies specific for CD4-BY421, CD8-ALEXA488, CD39-APC, CD19-BY421 or CD14-APC as described above. Image analysis was performed using an Amnis cytometer. Cells were first visualized in a bright field, identified as T cells (CD4+, CD8+, CD39+), B cells (CD19+), or monocytes (CD14+) based on surface staining and observed for the presence of intracytoplasmic PKH26. The cell images were merged to confirm uptake of PKH26 by individual cells. Exosome binding to immune cells and their uptake by the cells were monitored over time. The IDEAS software was used to evaluate exosome uptake by cells based on the number of PKH-26+ cells and overall PKH-26 fluorescence intensity.
Confocal microscopy
Human cell lines U937 (purchased from ATCC) and CD8+Jurkat cells (a gift from Dr. H. Rabinowich, U. of Pittsburgh) were maintained in cultures of RPMI medium supplemented with 10% (v/v) fetal bovine serum that was pre-centrifuged at 100,000× g for 3 h to remove nanovesicles. Cells were harvested while in the log phase of growth and labeled with CFSE as described previously.Citation35 Exosomes were isolated from plasma of patients with AML as described by usCitation36 and labeled with PKH26 (see above). Exosomes (10 μg protein) were added to 50 × 104 cells in a 35 mm culture dish, and co-cultures were incubated at 37°C in the atmosphere of 5% CO2 in air for 24 h. Confocal microscopy was performed using a Nikon A1 microscope at the Center of Bioimaging, University of Pittsburgh.
To demonstrate that exosomes are internalized and not just bound to the cell surface, labeled cells were treated with trypsin before microscopy and images of untreated vs. trypsin-treated cells were compared.
Image analysis for Ca2+ flux
Freshly isolated T cells (200 × 103 cells/50 μL HBSS (with calcium and magnesium, no phenol red) medium; Thermo Fisher Scientific, Waltham, MA 02451) were stained with Fluo-3 AM (diluted 1: 200; Sigma-Aldrich, St. Louis, MO 63178) for 20 min. Cells were centrifuged at 300×g for 5 min, resuspended in 2 mL medium, placed in a Matek dish and incubated at 37°C in an atmosphere of 5% CO2 in air for 15 min. The dish then was placed on the microscope table (preheated at 37°C) and after establishing the settings, continuous readings were taken for 30 min using a Nikon A1 microscope. While the first cell images were taken TEX, DEX, or EXO were added. The acquired image data were extracted processed using the Imaris Image software (Bitplane, Concord, MA). Cells were defined as spots (min. n = 500) and cell movements were tracked over time (max. move = 2 cm). The mean intensity was measured for each cell over each time point. The data were normalized, averaged and the results plotted in Prism (GraphPad Software, Inc., La Jolla, CA 92037).
Co-incubation of T cells with TEX and mass spectrometry
Aliquots of CD4+ T cells or CD4+CD39+ Treg isolated from PBMC of ND as described above were co-incubated with TEX (10 μg protein added to 105 cells/well in 96-well plates) or PCI-13 cells in the presence of exogenous ATP (20 μM) for 16 h. Control wells contained T cells alone or tumor cells alone. All experiments were performed in triplicate. Cells and supernatants were collected, centrifuged twice, and boiled for 5 min to inactivate ADO-degrading enzymes, and stored at −80°C for subsequent analyses. Purine nucleosides were measured using liquid chromatography-tandem mass spectrometry as described previously by us.Citation2,22
Statistics
Results are shown as mean values. Significance (p-value <0.05) was determined using Wilcoxon's signed rank test. Prism (GraphPad Software, Inc., La Jolla, CA 92037 USA) was used to perform the statistical analysis.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
KONI_A_1261243_s02.pptx
Download MS Power Point (173.8 KB)Funding
Supported in part by NIH grants R01 CA168628 and R21 CA205644 to TLW and NIH grants DKO79307 and DK068575 to EKJ. LM was a fellow of the Swiss National Foundation. This project used the Flow Cytometry Facility and the Center for Biological Imaging at UPCI, supported in part by award P30 CA047904.
References
- Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 2013; 200:373-83; PMID:23420871; https://doi.org/10.1083/jcb.201211138
- Figueiro F, Muller L, Funk S, Jackson EK, Battastini AM, Whiteside TL. Phenotypic and functional characteristics of CD39high human regulatory B cells (Breg). Oncoimmunology 2016; 5:e1082703; PMID:27057473; https://doi.org/10.1080/2162402X.2015.1082703
- Keller S, Ridinger J, Rupp AK, Janssen JW, Altevogt P. Body fluid derived exosomes as a novel template for clinical diagnostics. J Transl Med 2011; 9:86; PMID:21651777; https://doi.org/10.1186/1479-5876-9-86
- Brinton LT, Sloane HS, Kester M, Kelly KA. Formation and role of exosomes in cancer. Cell Mol Life Sci 2015; 72:659-71; PMID:25336151; https://doi.org/10.1007/s00018-014-1764-3
- Kowal J, Tkach M, Thery C. Biogenesis and secretion of exosomes. Curr Opin Cell Biol 2014; 29:116-25; PMID:24959705; https://doi.org/10.1016/j.ceb.2014.05.004
- Andreu Z, Yanez-Mo M. Tetraspanins in extracellular vesicle formation and function. Front Immunol 2014; 5:442; PMID:25278937; https://doi.org/10.3389/fimmu.2014.00442
- Colombo M, Moita C, van Niel G, Kowal J, Vigneron J, Benaroch P, Manel N, Moita LF, Thery C, Raposo G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J Cell Sci 2013; 126:5553-65; PMID:24105262; https://doi.org/10.1242/jcs.128868
- Cocucci E, Meldolesi J. Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol 2015; 25:364-72; PMID:25683921; https://doi.org/10.1016/j.tcb.2015.01.004
- Mulcahy LA, Pink RC, Carter DR. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles 2014; 3:24641; PMID:25143819; https://doi.org/10.3402
- Feng D, Zhao WL, Ye YY, Bai XC, Liu RQ, Chang LF, Zhou Q, Sui SF. Cellular internalization of exosomes occurs through phagocytosis. Traffic 2010; 11:675-87; PMID:20136776; https://doi.org/10.1111/j.1600-0854.2010.01041.x
- Mincheva-Nilsson L, Baranov V. Cancer exosomes and NKG2D receptor-ligand interactions: impairing NKG2D-mediated cytotoxicity and anti-tumour immune surveillance. Semin Cancer Biol 2014; 28:24-30; PMID:24602822; https://doi.org/10.1016/j.semcancer.2014.02.010
- Whiteside TL. Immune modulation of T-cell and NK (natural killer) cell activities by TEXs (tumour-derived exosomes). Biochem Soc Trans 2013; 41:245-51; PMID:23356291; https://doi.org/10.1042/BST20120265
- Kim JW, Wieckowski E, Taylor DD, Reichert TE, Watkins S, Whiteside TL. Fas ligand-positive membranous vesicles isolated from sera of patients with oral cancer induce apoptosis of activated T lymphocytes. Clin Cancer Res 2005; 11:1010-20; PMID:15709166
- Wieckowski EU, Visus C, Szajnik M, Szczepanski MJ, Storkus WJ, Whiteside TL. Tumor-derived microvesicles promote regulatory T cell expansion and induce apoptosis in tumor-reactive activated CD8+ T lymphocytes. J Immunol 2009; 183:3720-30; PMID:19692638; https://doi.org/10.4049/jimmunol.0900970
- Whiteside TL. Exosomes and tumor-mediated immune suppression. J Clin Invest 2016; 126:1216-23; PMID:26927673; https://doi.org/10.1172/JCI81136
- Szajnik M, Czystowska M, Szczepanski MJ, Mandapathil M, Whiteside TL. Tumor-derived microvesicles induce, expand and up-regulate biological activities of human regulatory T cells (Treg). PLoS One 2010; 5:e11469; PMID:20661468; https://doi.org/10.1371/journal.pone.0011469
- Muller L, Mitsuhashi M, Simms P, Gooding WE, Whiteside TL. Tumor-derived exosomes regulate expression of immune function-related genes in human T cell subsets. Sci Rep 2016; 6:20254; PMID:26842680; https://doi.org/10.1038/srep20254
- Joseph N, Reicher B, Barda-Saad M. The calcium feedback loop and T cell activation: how cytoskeleton networks control intracellular calcium flux. Biochim Biophys Acta 2014; 1838:557-68; PMID:23860253; https://doi.org/10.1016/j.bbamem.2013.07.009
- Taylor DD, Gercel-Taylor C. Exosomes/microvesicles: mediators of cancer-associated immunosuppressive microenvironments. Semin Immunopathol 2011; 33:441-54; PMID:21688197; https://doi.org/10.1007/s00281-010-0234-8
- Valenti R, Huber V, Iero M, Filipazzi P, Parmiani G, Rivoltini L. Tumor-released microvesicles as vehicles of immunosuppression. Cancer Res 2007; 67:2912-5; PMID:17409393; https://doi.org/10.1158/0008-5472.CAN-07-0520
- Andreola G, Rivoltini L, Castelli C, Huber V, Perego P, Deho P, Squarcina P, Accornero P, Lozupone F, Lugini L et al. Induction of lymphocyte apoptosis by tumor cell secretion of FasL-bearing microvesicles. J Exp Med 2002; 195:1303-16; PMID:12021310; https://doi.org/10.1084/jem.20011624
- Schuler PJ, Saze Z, Hong CS, Muller L, Gillespie DG, Cheng D, Harasymczuk M, Mandapathil M, Lang S, Jackson EK et al. Human CD4(+) CD39(+) regulatory T cells produce adenosine upon co-expression of surface CD73 or contact with CD73(+) exosomes or CD73(+) cells. Clin Exp Immunol 2014; 177:531-43; PMID:24749746; https://doi.org/10.1111/cei.12354
- Szczepanski MJ, Szajnik M, Welsh A, Whiteside TL, Boyiadzis M. Blast-derived microvesicles in sera from patients with acute myeloid leukemia suppress natural killer cell function via membrane-associated transforming growth factor-beta1. Haematologica 2011; 96:1302-9; PMID:21606166; https://doi.org/10.3324/haematol.2010.039743
- Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Jr., Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 2008; 10:1470-6; PMID:; PMID:19011622; https://doi.org/10.1038/ncb1800
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007; 9:654-9; PMID:17486113; https://doi.org/10.1038/ncb1596
- Balaj L, Lessard R, Dai L, Cho YJ, Pomeroy SL, Breakefield XO, Skog J. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun 2011; 2:180; PMID:21285958; https://doi.org/10.1038/ncomms1180
- Waldenstrom A, Genneback N, Hellman U, Ronquist G. Cardiomyocyte microvesicles contain DNA/RNA and convey biological messages to target cells. PLoS One 2012; 7:e34653; PMID:22506041; https://doi.org/10.1371/journal.pone.0034653
- Xiao D, Ohlendorf J, Chen Y, Taylor DD, Rai SN, Waigel S, Zacharias W, Hao H, McMasters KM. Identifying mRNA, microRNA and protein profiles of melanoma exosomes. PLoS One 2012; 7:e46874; PMID:23056502; https://doi.org/10.1371/journal.pone.0046874
- Abels ER, Breakefield XO. Introduction to extracellular vesicles: biogenesis, RNA cargo selection, content, release, and uptake. Cell Mol Neurobiol 2016; 36:301-12; PMID:27053351; https://doi.org/10.1007/s10571-016-0366-z
- Li C, Liu DR, Li GG, Wang HH, Li XW, Zhang W, Wu YL, Chen L. CD97 promotes gastric cancer cell proliferation and invasion through exosome-mediated MAPK signaling pathway. World J Gastroenterol 2015; 21:6215-28; PMID:26034356; https://doi.org/10.3748/wjg.v21.i20.6215
- Muller-Haegele S, Muller L, Whiteside TL. Immunoregulatory activity of adenosine and its role in human cancer progression. Expert Rev Clin Immunol 2014; 10:897-914; PMID:24871693; https://doi.org/10.1586/1744666X.2014.915739
- Welihinda AA, Kaur M, Greene K, Zhai Y, Amento EP. The adenosine metabolite inosine is a functional agonist of the adenosine A2A receptor with a unique signaling bias. Cell Signal 2016; 28:552-60; PMID:26903141; https://doi.org/10.1016/j.cellsig.2016.02.010
- Hasko G, Sitkovsky MV, Szabo C. Immunomodulatory and neuroprotective effects of inosine. Trends Pharmacol Sci 2004; 25:152-7; PMID:15019271; https://doi.org/10.1016/j.tips.2004.01.006
- Heo DS, Snyderman C, Gollin SM, Pan S, Walker E, Deka R, Barnes EL, Johnson JT, Herberman RB, Whiteside TL. Biology, cytogenetics, and sensitivity to immunological effector cells of new head and neck squamous cell carcinoma lines. Cancer Res 1989; 49:5167-75; PMID:2766286
- Bergmann C, Strauss L, Zeidler R, Lang S, Whiteside TL. Expansion and characteristics of human T regulatory type 1 cells in co-cultures simulating tumor microenvironment. Cancer Immunol Immunother 2007; 56:1429-42; PMID:17265021; https://doi.org/10.1007/s00262-007-0280-9
- Hong CS, Funk S, Muller L, Boyiadzis M, Whiteside TL. Isolation of biologically active and morphologically intact exosomes from plasma of patients with cancer. J Extracell Vesicles 2016; 5:29289; PMID:27018366; https://doi.org/10.3402/jev.v5.29289
- Skriner K, Adolph K, Jungblut PR, Burmester GR. Association of citrullinated proteins with synovial exosomes. Arthritis Rheum 2006; 54:3809-14; PMID:17133577; https://doi.org/10.1002/art.22276
- Saze Z, Schuler PJ, Hong CS, Cheng D, Jackson EK, Whiteside TL. Adenosine production by human B cells and B cell-mediated suppression of activated T cells. Blood 2013; 122:9-18; PMID:23678003; https://doi.org/10.1182/blood-2013-02-482406
