ABSTRACT
Neuromedin U (NmU) -a neuropeptide belonging to the neuromedin family– plays a substantial role in HER2-positive breast cancer, correlating with increased aggressiveness, resistance to HER2-targeted therapies and overall significantly poorer outcome for patients. However, the mechanism through which it exerts these effects remains unclear. To elucidate this, initially we used HER2-positive breast cancer cells stably over-expressing NmU. These cells and their released extracellular vesicles (EVs) had increased amounts of the immunosuppressive cytokine TGFβ1 and the lymphocyte activation inhibitor PD-L1. Furthermore, these cells also showed enhanced resistance to antibody-dependent cell cytotoxicity (ADCC) mediated by trastuzumab, indicating a role of NmU in enhancing immune evasion. All these features were also found in HER2-targeted drug-resistant cells which we previously found to express higher levels of NmU than their drug-sensitive counterparts. Interestingly, EVs from drug-resistant cells were able to increase levels of TGFβ1 in drug-sensitive cells. In our neo-adjuvant clinical trial, TGFβ1 levels were significantly higher in EVs isolated from the serum of patients with HER2-overexpressing breast cancers who went on to not respond to HER2-targeted drug treatment, compared with those who experienced complete or partial response. Taken together, our results report a new mechanism-of-action for NmU in HER2-overexpressing breast cancer that enhances resistance to the anti-tumor immune response. Furthermore, EV levels of TGFβ1 correlating with patients' response versus resistance to HER2-targeted drugs suggests a potential use of EV-TGFβ1 as a minimally-invasive companion diagnostic for such treatment in breast cancer.
Introduction
Exosomes and/or ectosomes, collectively termed extracellular vesicles (EVs), are vesicles released from cells and are typically considered to be miniature maps of their cell of origin.Citation1 Accumulating evidence from our own research and that of others suggests that EVs are useful cargos of biomarkers representing the cells from which they were derived. They also appear to play important roles in cell-to-cell communication and are implicated in tumor growth and transferring drug-resistance.Citation2,3
HER2 is overexpressed in ∼25% of breast cancers and is associated with poorest prognosis.Citation4 Therapies targeting HER2 (including monoclonal antibodies such as trastuzumab/Herceptin and small molecules lapatinib and neratinib) have shown encouraging results in the clinic. Unfortunately, not all HER2-overexpressing tumors respond to these therapies and many that initially respond later acquire resistance.Citation5,6 Predictive companion biomarkers for HER2-targeting drugs are thus urgently needed for improved patient selection and enhanced patient outcome. Ideally, such biomarkers should be extracellular so as to be easily obtained through minimally invasive-techniques from blood specimens. Of course, novel therapeutic strategies to circumvent resistance are also badly needed.
It has been established that mechanisms that enhance drug-resistance may also result in protection against cancer cell destruction by the immune system.Citation7 In a study representing mesothelioma, leukemia and prostate cancers, tumor cell-derived EVs were found to possess immunosuppressive properties and are proposed to participate in the transfer of these traits to recipient cells.Citation8 TGFβ1 present in EVs was found to be at least partly responsible for the immunosuppressive effects in various cancer types.Citation9,10 Whether EVs from HER2-targeted drug resistant breast cancer cells have different immunosuppressive properties than those from drug sensitive cells has not been investigated to date.
Neuromedin U (NmU) is a neuropeptide for which several different functions have been ascribed. These include role(s) in feeding, energy balance, smooth muscle contraction, stress response, immune system activation and pain sensing.Citation11,12 A limited number of studies to date have associated NmU with cancer, with conflicting observations depending on the cancer type. In breast cancer, we have shown that NmU expression is increased in cells with innate- or acquired-resistance to HER2-targeted therapies; furthermore, NmU overexpression was also associated with increased migration, invasion and resistance to anoikis.Citation13 Assessing data from approximate 3,500 breast tumor specimens, our work identified association between NmU expression and poor survival outcome for breast cancer patients; particularly those with HER2-overexpressing tumors. Furthermore, our work also found an association between NmU levels and the cell population with cancer stem cell (CSC) phenotype (Martinez et al, unpublished results). Our in vitro and in vivo NmU knock-down experiments pointed to a role for NmU as a new therapeutic target to help circumvent innate- and acquired- drug resistance, although the precise mechanisms of action remained unexplained.
Here we show that HER2-targeted drug resistance in HER2-positive breast cancer cells correlates with increased levels of the immunosuppressive molecules TGFβ1 and PD-L1 and resistance to the anti-tumor immune response. Furthermore, these molecules are carried by EVs, which are able to transfer the traits of their cell of origin to drug-sensitive cells. EV-associated TGFβ1 levels also correlate with response to HER2-targeted treatment in HER2-overexpressing breast cancer patients, suggesting it could be used as a biomarker of response to therapy. We have therefore revealed druggable targets – TGFβ1 and PD-L1 – to enhance the efficacy of currently used HER2-targeted therapies. Furthermore, we have also shown that circulating levels of EV-associated TGFβ1 have potential as a predictive biomarker of patients' treatment response.
Results
NmU overexpression increases TGFβ1 levels
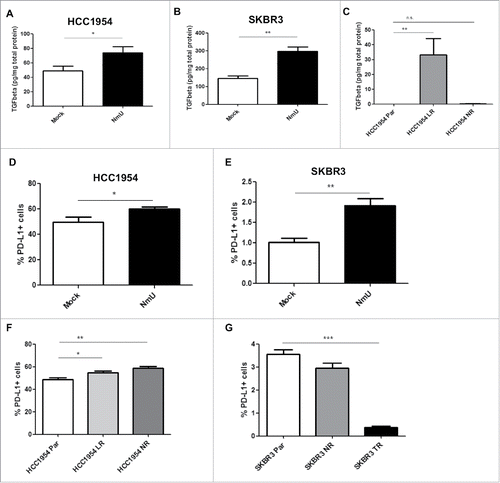
Our previous studies showed that increased expression of NmU in tumor tissue is associated with poor prognosis in HER2-overexpressing breast cancer patientsCitation13 and with expansion of the CSC population (Martinez et al, unpublished results). As has been shown previously,Citation14 breast cancer cells with CSC phenotype secrete high levels of TGFβ1. We then set out to determine whether NmU-overexpressing cells also showed increased levels of TGFβ1. As shown in and , TGFβ1 levels were increased in conditioned media from NmU-overexpressing HCC1954 and SKBR3 cells. As expected, TGFβ1 levels were also increased in lapatinib-resistant HCC1954 cells compared with their sensitive counterparts; the increase in TGFβ1 levels was not significant for neratinib-resistant HCC1954 cells (). TGFβ1 levels secreted by SKBR3 parent and resistant cell variants were below the level of detection of the ELISA (data not shown). Short-term treatment with NmU did not result in release of TGFβ1.
Figure 1. Overexpression of NmU correlates with increased TGFβ1 and PD-L1 levels. (A), (B), and (C), TGFβ1 levels were determined by ELISA in serum-free conditioned medium of HCC1954 and SKBR3 cell variants and expressed as pg TGFβ1 per mg total cell protein. (D) and F, HCC1954 cell variants were stained with APC-labeled anti-PD-L1, followed by FACS analysis in a FACSCanto II flow cytometer. (E) and (G), Same as in (D) for SKBR3 cell variants. Results represent averaged replicates from at least 3 independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001.
NmU overexpression increases cancer cell expression of immunosuppressive mediators
We have previously reported that NmU overexpression in HER2-positive breast cancer cells causes an enhancement in drug resistance and a more aggressive phenotype, which is commonly associated with resistance to the anti-tumor immune response.Citation15-17 Here our results show that NmU-overexpressing cells secrete increased levels of TGFβ1, which is a well-known immunosuppressive cytokine. To evaluate the possibility that NmU confers cells the ability to evade destruction by the immune system, expression of cell surface proteins known to inhibit the immune response were assessed by flow cytometry in different cell variants. As shown in and , overexpression of NmU in HER2-positive breast cancer cells also increased the expression of the PD-L1 ligand, which has been widely shown to suppress the anti-tumor immune response; the levels of the immunosuppressive receptor CTLA-4, however, were unchanged in both cell line variants (data not shown). HER2-targeted drug-resistant HCC1954 cells also showed increased levels of PD-L1 (): however, no differences in PD-L1 expression were observed in neratinib-resistant SKBR3 cells, while PD-L1 expression was actually decreased in SKBR3 TR cells (). Overall, our results show that overexpression of NmU increases the levels of immunosuppressive molecules TGFβ1 and PD-L1 in HER2-positive breast cancer cells; while the PD-L1 results differed in cells with acquired resistance, depending on the drug in question and the cell line.
NmU-overexpressing cells are more resistant to ADCC
To analyze the functional consequences of NmU-overexpression, we performed antibody-dependent cell cytotoxicity (ADCC) assays in different cell variants. NmU-overexpressing HCC1954 cells were significantly more resistant to ADCC mediated by trastuzumab than control transfected counterparts (); similar results were obtained with NmU-overexpressing SKBR3 cells, although the differences in this case were not significant (). On the other hand, lapatinib- and neratinib-resistant HCC1954 cell variants were also more resistant to ADCC than their drug-sensitive counterparts (): this was not observed in SKBR3 drug-resistant cell variants. These results suggest that, as well as enhancing resistance to HER2-targeted drugs, overexpression of NmU increases the ability of HER2-positive breast cancer cells to evade ADCC, which is one of the main mechanisms of cancer cell death mediated by trastuzumab.
Figure 2. Overexpression of NmU confers resistance to ADCC. Cells (A) and (C), HCC1954 cells, (B) and (D), SKBR3 cells) were seeded in a round bottom 96-well plate and incubated with trastuzumab (100µg/ml). PBMCs were isolated from healthy donor blood by Ficoll gradient centrifugation and added on top of cells at the appropriate ratios. Cells were incubated at 37°C for a further 6h and cell lysis was then assessed by LDH release. (B) and (D), Same as in (A), but for SKBR3 cell variants. Averaged results from technical triplicates from at least 2 independent experiments with PBMCs from different donors are shown. *p < 0.05, **p < 0.01, ***p < 0.001. [Note The 100% of cell lysis is calculated based on cancer cells alone and so sometimes the percentage of cell lysis following addition of PBMCs can be over 100% as some PBMCs also die and release their LDH content into the medium].
![Figure 2. Overexpression of NmU confers resistance to ADCC. Cells (A) and (C), HCC1954 cells, (B) and (D), SKBR3 cells) were seeded in a round bottom 96-well plate and incubated with trastuzumab (100µg/ml). PBMCs were isolated from healthy donor blood by Ficoll gradient centrifugation and added on top of cells at the appropriate ratios. Cells were incubated at 37°C for a further 6h and cell lysis was then assessed by LDH release. (B) and (D), Same as in (A), but for SKBR3 cell variants. Averaged results from technical triplicates from at least 2 independent experiments with PBMCs from different donors are shown. *p < 0.05, **p < 0.01, ***p < 0.001. [Note The 100% of cell lysis is calculated based on cancer cells alone and so sometimes the percentage of cell lysis following addition of PBMCs can be over 100% as some PBMCs also die and release their LDH content into the medium].](/cms/asset/94b6e65b-07bb-493b-9c97-4c699706281a/koni_a_1362530_f0002_b.gif)
EVs from NmU-overexpressing cells have more TGFβ1 and PD-L1
EVs are known to transport cargo that is representative of their cell of origin. Since NmU-overexpressing and HER2-targeted drug-resistant cells displayed higher levels of TGFβ1 and PD-L1, we investigated whether this was also true of their derived EVs. EVs were isolated from cell culture conditioned media by filtration and ultracentrifugation and characterized by specific marker expression by immunoblotting and particle size analysis (Suppl. Fig. 1) and also by transmission electron microscopy (Suppl. Fig. 2). As expected, NmU levels were increased in EVs derived from NmU-overexpressing and drug-resistant HCC1954 cells, particularly in the case of neratinib-resistant cells (Supp. Fig 3A and B). NmU levels were also increased in EVs from neratinib-resistant EFM192A cells (Supp. Fig. 3C), but not in any of the drug-resistant SKBR3 cell variants (Suppl. Fig. 3D).
Figure 3. EVs from NmU-overexpressing and HER2-targeted drug-resistant variants show altered content of TGFβ1 and PD-L1. (A), (C), and €, TGFβ1 levels were determined by ELISA in intact (unlysed) EVs from HCC1954 (A), EFM192A (C) and SKBR3 (E) cell variants and expressed as pg TGFβ1 per mg total EV protein. B, D, and F, PD-L1 levels were determined by ELISA in EVs from HCC1954 (B), EFM192A (D) and SKBR3 (F) cell variants and expressed as pg PD-L1 per mg total EV protein. Results represent averaged replicates of at least 3 independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001.
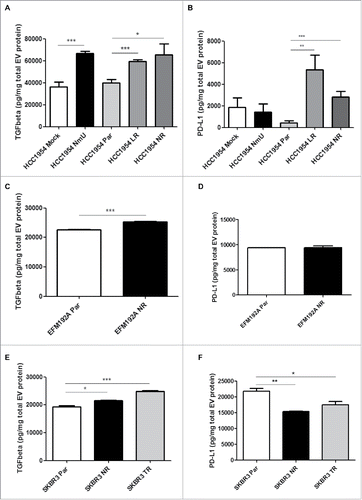
Interestingly, TGFβ1 levels were increased in intact (unlysed) EVs from lapatinib, neratinib and trastuzumab-resistant HCC1954, EFM192A and SKBR3 cells compared with their sensitive counterparts (, and ); this effect appears to be at least partly mediated by NmU, as EVs from NmU-overexpressing HCC1954 cells also showed significantly increased TGFβ1 levels compared with EVs from control-transfected cells ().
Furthermore, EVs from drug-resistant HCC1954 variants also displayed increased levels of PD-L1 (). This was not observed for EVs from EFM192A and SKBR3 cell variants ( and ); in fact, EVs from drug-resistant SKBR3 cell variants displayed significantly lower levels of PD-L1 compared with EVs from their drug-sensitive counterparts, in agreement with observations on their cells of origin. The increase in EV-bound PD-L1 in HCC1954 cells did not appear to be mediated by NmU, as EVs derived from NmU-overexpressing cells showed similar levels of PD-L1 as those EVs derived from control transfected cells (). Our results show that EVs from HER2-targeted drug-resistant cells have higher content of TGFβ1, while changes in PD-L1 content are dependent on the particular cell line.
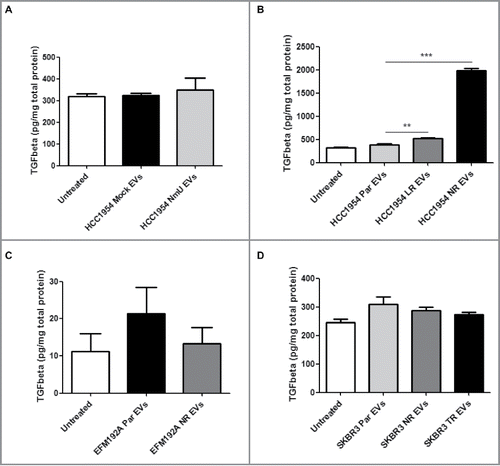
EVs from drug-resistant cell variants can enhance TGFβ1 secretion and PD-L1 expression in drug-sensitive cells
EVs have been shown to transfer traits from their cell of origin to other, recipient cells. We then treated drug-sensitive HCC1954, EFM192A and SKBR3 cells with EVs from their drug-resistant variants and measured TGFβ1 secretion and PD-L1 expression. As shown in , EVs from drug-resistant HCC1954 cell variants induced an increase in TGFβ1 secretion by HCC1954 Par cells. This increase did not appear to be mediated by NmU (). On the other hand, EVs from drug-resistant EFM192A and SKBR3 cell variants did not show the same effect ( and ). It should be noted that TGFβ1 secretion levels were much lower in these cells than in HCC1954 cells.
Figure 4. Treatment with EVs from NmU-overexpressing and HER2-targeted drug-resistant variants enhances TGFβ1 secretion. (A) and (B), HCC1954 Par cells were treated with EVs from different HCC1954 cell variants (20 µg EV/1 × 105 cells) for 48 hr. Cell conditioned medium was then analyzed for TGFβ1 content by ELISA, expressed as pg TGFβ1 per mg total cell protein. (C), Same as in (A) for EFM192A cells. (D), Same as in (A) for SKBR3 cells. Results represent averaged replicates of at least 3 independent experiments. **p < 0.01, ***p < 0.001.
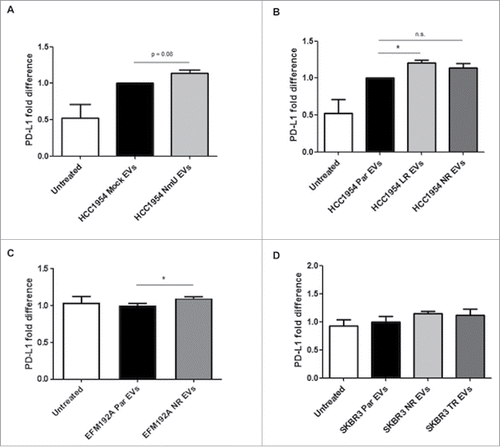
Expression of PD-L1 was however increased by treatment with EVs from drug-resistant variants from HCC1954 and EFM192A cells (, and ), even when the EVs themselves did not appear to contain higher levels of PD-L1. In the case of EVs from HCC1954 NR and SKBR3 cell variants the increase was not significant, but an upwards trend was observed. This increase in PD-L1 expression could have been at least partly mediated by NmU, as treatment with EVs from NmU-overexpressing HCC1954 cells increased expression of PD-L1 in HCC1954 Par cells (). Overall, our results so far show that EVs from HER2-targeted drug-resistant cell variants can affect expression levels of TGFβ1 and PD-L1 in recipient, drug-sensitive cells.
Figure 5. Treatment with EVs from NmU-overexpressing and HER2-targeted drug-resistant variants increases PD-L1 expression. (A) and (B), HCC1954 Par cells were treated with EVs from different HCC1954 cell variants (20 µg EV/1 × 105 cells) for 48 hr. PD-L1 surface staining was then analyzed by flow cytometry. (C), Same as in A for EFM192A cells. (D), Same as in (A) for SKBR3 cells. Results represent averaged replicates of at least 3 independent experiments and are expressed as fold difference with respect to cells treated with EVs from parent cell variants, which is considered to be equal to 1. *p < 0.05.
TGFβ1 is a potential marker of response for HER2-positive breast cancer treatment
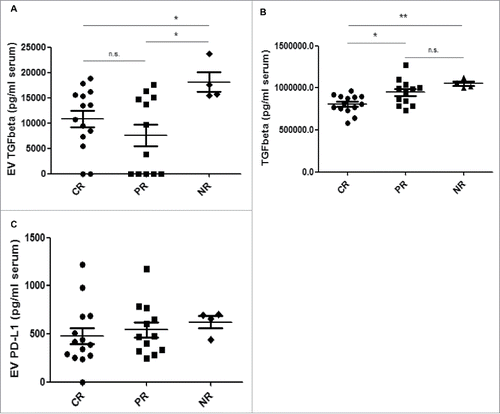
Analysis of EVs from drug-resistant cell variants pointed to a correlation between TGFβ1 and resistance to HER2-targeted drugs; this suggested a possible use of this molecule as a companion diagnostics/predictive biomarker of response in breast cancer patients with HER2-overexpressing tumors. To evaluate this possibility, EVs were isolated from serum obtained from a blinded study of patients before neo-adjuvant treatment including trastuzumab with or without lapatinib. Isolated EVs were characterized as described by immunoblotting and particle size analysis (Suppl. Fig. 4). TGFβ1 and PD-L1 levels were determined on intact EVs by ELISA. Patients were then followed up and classified as complete responders (CR, n = 14), partial responders (PR, n = 12) and non-responders (NR, n = 4). As shown in , EVs from NR patients showed significantly higher levels of TGFβ1 than those of CR or PR patients. Of note, no differences in EV quantities were observed from specimens in the CR, PR or NR cohorts, indicating that the higher amount of EV TGFβ1 is not just a consequence of increased numbers of EVs in patient serum. These results were different from those obtained when measuring TGFβ1 levels directly in raw serum (): while CR patients displayed significantly lower levels than either PR or NR patients, there were no significant differences between PR and NR patients, suggesting that serum TGFβ1 levels cannot distinguish between these 2 groups. No significant differences were observed between CR, PR and NR patient EVs regarding PD-L1 levels, although increased quantities of EV-carried PD-L1 were found for those who were classified as NR vs. PR vs. CR (). These findings suggest that EV-carried TGFβ1 –but not raw serum TGFβ1–may be able to identify those patients that would benefit from HER2-targeted treatment.
Figure 6. EV TGFβ1 levels correlate with response to HER2-targeted therapy in HER2-overexpressing breast cancer patients. EVs were isolated from 250 µl patients' sera by serial ultracentrifugation. (A), TGFβ1 was detected on intact EVs isolated from patient serum by ELISA. Values are expressed as pg TGFβ1 per ml of serum. (B), TGFβ1 was detected in patients' sera by ELISA. Values are expressed as pg TGFβ1 per ml of serum. C, PD-L1 was detected on intact EVs by ELISA. Values are expressed as pg PD-L1 per mg total EV protein. *p ≤ 0.05, **p < 0.01.
Discussion
In this study we show that NmU in HER2-positive breast cancer cells promotes evasion of the immune response, increasing the expression of immunosuppressive molecules TGFβ1 and PD-L1 and functionally affecting ADCC, a key mechanism of action of monoclonal antibodies used in the treatment of breast cancer. Interestingly, TGFβ1 and PD-L1 are also detectable in EVs from HER2-targeted drug-resistant cell variants, which have been found to express high levels of NmU. EVs from these cells can induce an increase in TGFβ1 secretion in drug-sensitive cells, promoting further immune evasion. Finally, EV-associated TGFβ1 levels also correlate with response to HER2-targeted treatment in HER2-overexpressing breast cancer patients, suggesting it could be used as a marker of response to therapy.
EVs from cancer cells have been shown to carry immunosuppressive molecules such as FasL, TRAIL, TGFβ1 and PD-L1,Citation9,18-19 and also to be involved in immunosuppression at several different stages of the immune response, including T cell proliferation, activation and apoptosis,Citation9,20-21 regulatory T cell abundance and function,Citation23-25 NK cell function,Citation22,26-27 dendritic cell maturation,Citation28 induction of suppressive mesenchymal stem cells,Citation29-30 and macrophage polarization.Citation31 TGFβ1 in EVs has been proposed to mediate at least some of the immunosuppressive effects.Citation9,10
Our previous work had shown an association between overexpression of NmU and an expansion of cells with CSC phenotype.Citation32 The CSC phenotype is characterized by increased drug resistance and also by enhanced immune evasion.Citation33-35 In accordance with this, as well as driving HER2-targeted drug resistance, NmU overexpression was also associated with increased expression of TGFβ1 and PD-L1, known inhibitors of the immune response. Moreover, HER2-targeted drug-resistant cells were also shown to be more resistant to ADCC mediated by trastuzumab, and this effect was at least partly mediated by NmU. These results show that NmU-overexpressing cells are indeed more capable of avoiding destruction by the immune system.
EVs have gathered a lot of interest in recent times due to the understanding that they carry valuable information about their cell of origin. Indeed, we found increased levels of TGFβ1 in EVs from drug-resistant cell variants, and this increase appeared to be at least partly mediated by NmU. On the other hand, PD-L1 levels were not consistently increased in EVs from drug-resistant cell variants, similarly to what we observed on their cells of origin. To our knowledge, this is the first time that EVs from drug-resistant cells have been shown to possess enhanced immunosuppressive abilities compared with EVs from drug-sensitive cells. Future studies where high levels of NmU in resistant cells are knocked-down with siRNA or shRNA to establish if this would reverse the increased vesicular release of TGFβ1/PD-L1 are now warranted. Their success may lead to pre-clinical in vivo studies to establish the potential therapeutic relevance of this approach.
Our previous work and that of others has shown that EVs are able to transfer trends from their cell of origin to recipient cells.Citation1-3 Indeed, EVs from most of the drug-resistant cells tested were able to increase the levels of secreted TGFβ1 and cell surface PD-L1 in parent, drug-sensitive cells.
It is noteworthy that HCC1954, SKBR3 and EFM192A -all HER2-positive cell lines- were found to sometimes behave in different ways under the same conditions. Possible contributing reasons for this may include the fact that HCC1954 cells were originally isolated from a primary breast tumor, while EFM192A and SKBR3 cells were isolated from metastatic sites. Also, their tumor type of origin is different i.e. HCC1954 cells represent a ductal carcinoma, while EFM192A and SKBR3 cells represent adenocarcinoma cells. The 3 cell lines also carry different driver mutations due to the diverse origin of their transformation. The commonality of substantial importance here is that they all represent HER2-overexpressing breast tumors. Crucially, the fact that all 3 cell lines –despite tumor/patient-to-patient differences– display increased TGFβ1 levels on EVs further highlights the relevance of this finding and how it potentially could be applied to assessing patients whose HER2-overexpressing breast tumors are different.
TGFβ1 levels in EVs correlated particularly well with HER2-targeted drug resistance displayed by their cells of origin. A study in a small group of patients undergoing neo-adjuvant treatment showed encouraging results, suggesting that high levels of circulating EV TGFβ1 may indicate that the tumor is resistant to HER2-targeted therapy. Interestingly, the levels of raw serum TGFβ1 did not correlate as well with response to treatment, indicating that there is an advantage in analyzing EVs. If confirmed, this would point to EV TGFβ1 as a novel biomarker of response to HER2-targeted therapy in HER2-overexpressing breast cancer. However, these results should be validated in a larger cohort.
It may also be interesting, in future studies, to consider the multiple forms of TGFβ1. Our results represent analysis, by ELISA, of the mature form of TGFβ1. However, using an immunoblotting approach, Graner et al.Citation36 reported that EVs from glioma patient sera have multiple forms of TGFβ1 i.e., forms of pro-peptide, LAP, dimer -presumably the active form- and monomer. More recently and also using immunoblotting, multiple TGFβ1 forms have been shown on EVs from AML patient plasma (albeit in very small numbers of specimens) and the EVs with more of the “active/mature” form of TGFβ1 correlated with greater capacity to suppress NK cell activity.Citation37 For our studies reported here, we selected the approach (ELISA) that we took because we are not confident that immunoblotting techniques would easily translate into routine clinical utility for a blood-based biomarker (e.g. to predict response to therapy for patients enrolling in a clinical trial), while ELISAs are routinely used in clinics and hospital laboratories. However, undoubtedly using the less quantitative immunoblotting approach could help generate new scientific knowledge -which is also fundamentally important- that could complement the quantitative ELISA data.
NmU is an attractive target for HER2-overexpressing breast cancer, but the multiple physiologic roles it plays would make it difficult to alter its function without side-effects. The identification of downstream druggable mediators of its effects such as TGFβ1 and PD-L1 suggest novel ways in which NmU function can be targeted for the treatment of HER2-overexpressing breast cancer. Combination with therapies that block these downstream effectors of NmU could then add value to currently approved HER2-targeted therapies.
Materials and methods
Cell culture and treatments
HCC1954, SKBR3 and EFM192A cells, obtained from ATCC, were cultured in RPMI-1640 (Sigma-Aldrich, Ireland) with 10% FCS (PAA Laboratories, UK) and 1% L-glutamine (Sigma-Aldrich, Ireland). Our HCC1954 and SKBR3 NmU and mock-transfected cells have been described previously,Citation3 as have lapatinib-resistant (HCC1954 LR), neratinib-resistant (HCC1954 NR, SKBR3 NR and EFM192A NR) and trastuzumab-resistant (SKBR3 TR) variants.Citation13,38 For short-term treatment with NmU peptide, cells (1 × 105 cells) were seeded in a 24-well plate, allowed to attach overnight, fed with serum-free medium and subsequently treated with 1–10 μmol/L of NmU-25 (Bachem AG, Switzerland) for 1 hour. Cell lines are authenticated by short tandem repeat profiling (at least every 2 years) and tested for Mycoplasma using qPCR methods (quarterly). All experiments are performed within 10 passages of thawing and biologic replicate experiments are performed on consecutive passages numbers of cells. Lapatinib and neratinib were purchased from Sequoia Chemicals Ltd (UK); trastuzumab (Herceptin) was obtained from St. James' Hospital Pharmacy, TCD.
Immunoblotting
Total cellular proteins (10µg for extracellular vesicles, 30–40 μg for cell lysates, depending on the specific protein abundance but constant for any given protein) were resolved on 10% SDS-PAGE and transferred to polyvinylidene difluoride membranes (Millipore, Ireland). Primary antibodies used included Grp94 (1:1000, Abcam, UK), PDC6I/Alix (1:1000, Abcam, UK) and CD81 (1:200, Abcam, UK). Membranes were incubated with appropriate horseradish peroxidase-conjugated secondary antibodies (Cell Signaling Technology, Netherlands) and proteins were visualized by chemiluminescence (Millipore, Ireland). Detection was performed with a Chemidoc exposure system (Bio-Rad Laboratories, UK).
Nanoparticle tracking analysis
Analysis was performed on all EVs with the NTA NS500 system (NanoSight, Amesbury, UK) to determine particle size based on Brownian motion. EV samples were diluted 1:100 in PBS and injected into the NTA system, which obtained 6 60 second videos of the particles in motion. Videos were then analyzed with the NTA software to determine particle size.
ELISA
ELISA kits for human TGFβ1, PD-L1 (R&D Systems, UK) and NmU (Peninsula Laboratories, CA) were used according to the manufacturer's instructions. For analysis of conditioned media, HCC1954, SKBR3 and EFM192A cell variants were seeded at 1 × 105 cells/well in a 24-well plate and left to attach overnight. The following day, cells were washed and fed with serum-free media and incubated for a further 72hr. Supernatant was then collected and analyzed immediately or stored at −80°C until analysis. Results were normalized by total cell protein contained in the well. 10µg of protein from lysed or intact EVs, as stated, were tested per well.
Flow cytometry
HCC1954 and SKBR3 cell variants (1 × 105/well) were seeded in a 6-well plate and allowed to attach overnight. They were then trypsinised, blocked with 10% FCS in PBS and stained with APC-conjugated anti-CD24 (1:75 dilution, eBiosciences, UK) and FITC-conjugated anti-CD44 (1:800 for HCC1954, 1:400 for SKBR3, eBiosciences, UK) or APC-conjugated anti-PD-L1 antibody (1:800 for HCC1954, 1:400 for SKBR3, eBiosciences, UK) for 30 min at 4°C. For EV treatment, cells were seeded at 0.5 × 105 (HCC1954) or 1 × 105 (SKBR3 and EFM192A) cells/well in a 24-well plate and allowed to attach overnight. The following day, cells were washed with serum-free media, fed with media supplemented with extracellular vesicle (EV)-depleted-FBS and treated with EVs for a further 48 hr before staining as above. Staining was assessed in a FACSCanto II flow cytometer, followed by analysis with BD FACSDiva software.
Antibody-dependent cell cytotoxicity (ADCC) assay
HCC1954 cell variants were seeded at 1 × 104 cells/well in a round bottom 96-well plate and incubated with trastuzumab (100µg/ml) for 30 min at 37°C. Whole blood was obtained from healthy donors in accordance with a protocol approved by the Research Ethics Committee at Trinity College Dublin. PBMCs were isolated from blood by Ficoll gradient centrifugation (Histopaque, Sigma-Aldrich, UK) and added on top of cells at the appropriate ratios. Cells were incubated at 37°C for a further 6 h and cell lysis was then assessed by LDH release (CytoTox-ONE™ Homogeneous Membrane Integrity Assay, Promega, UK).
Extracellular vesicle isolation from conditioned media
HCC1954, SKBR3 and EFM192A cell variants were seeded (1.7 × 107 cells/flask in a 15 ml volume) in CELLine Adhere 1000 flasks (Integra Biosciences AG, Switzerland) in complete medium containing 10% extracellular vesicle (EV)-depleted-FBS, 2 mM L-Glutamine, 1% penicillin/streptomycin (Sigma–Aldrich, Ireland), and grown as described previously.Citation39 Conditioned media (CM) was collected every 7 d and processed for EV isolation. In brief, CM was centrifuged at 400 g for 10 min and 2000 g for 15 min to eliminate cells and cell debris, then filtered through a 0.45 μm pore filter and subsequently ultracentrifuged at 110,000 g for 75 min at 4°C, using an SW32.Ti swing bucket rotor. EV pellets were washed in PBS and ultracentrifugation repeated. EVs were then assessed using the BCA assay (Bio-Rad Laboratories, UK) and further characterized by nanoparticle tracking analysis (NanoSight) and immunoblotting for positive and negative markers.Citation40
Extracellular vesicle isolation from patients' specimens
Serum specimens were obtained from patients accrued to the All Cooperative Oncology Research Group (ICORG/Cancer Trials Ireland) translational trial NCT01485926. The procurement and analysis of these specimens was approved by the Ethics Committees of the associated hospitals and in accordance with the Helsinki Declaration of 1975. Specimens were taken before neo-adjuvant treatment that included trastuzumab with or without lapatinib. Following 6 cycles of treatment, response was assessed in accordance with ICORG guidelines, and defined as pathological complete response (CR) by the absence of invasive carcinoma in the breast and lymph nodes. Pathological partial response (PR) was defined as a decrease in either or both the tumor or lymph nodes compared with the pre-treatment tumor or lymph nodes, while having no increase in either the tumor or lymph nodes, while no pathological response (NR) was defined as no apparent change in either the tumor or lymph nodes compared with the clinical (pre-treatment) assignment or an increase in the tumor or lymph nodes at the time of post-treatment pathologic evaluation. Patient information (age, tumor score, lymph node status, metastasis status and overall staging) can be found in Supplementary Table 1. EVs were isolated from 250µl serum diluted in 8 ml PBS, filtered through 0.2µm syringe filter and ultracentrifuged at 110,000 g for 75 min at 4°C, using a 70.1 Ti fixed angle rotor. EV pellets were washed in PBS and ultracentrifugation repeated. EVs were then assessed using the BCA assay (Bio-Rad Laboratories, UK), nanoparticle tracking analysis (NanoSight), immunoblotting for positive and negative markers and transmission electron microscopy (TEM) as described previously.Citation11,12 EV pellets re-suspended in 200µl PBS were stored at −80°C until analysis.
Statistical analysis
One-way ANOVA followed by Tukey's post-test was used for analysis of multiple comparison groups. Two-tailed Student's unpaired t-test was used to compare 2 groups. Statistical analyses were performed using GraphPad Prism 5 (GraphPad Software, Inc., CA). Results are expressed as mean of a minimum of 3 independent experiments (except when noted otherwise) ± SEM. Statistical significance was set at *p < 0.05; **p < 0.01; ***p < 0.001.
Funding details
This work was supported by HRB's Health Research Award under grant HRA-POR-2014–658; Breast Cancer Now under grant 2015NovSP68; Irish Cancer Society's Breast-Predict under grant CCRC13GAL; H2020 COST Action ME-HaD under grant BM1202; and Science Foundation Ireland's support, under grant 12/RI/2340(7) of the FACS equipment used.
supp.zip
Download Zip (1.8 MB)Acknowledgments
The authors wish to acknowledge the All Ireland Cooperative Oncology Research Group (ICORG/Cancer Trials Ireland) for facilitating the neo-adjuvant clinical trial (ICORG: 10–05; NCT01485926) and Ms. Laura Ivers for help with specimen collection. We wish to thank Mr. Neal Leddy for his transmission electron microscopy expertise.
Disclosure statement
JC has acted in a consultancy role for GSK and has received honoraria and other remuneration from Roche and GSK. None of these monies were used for any of these studies and no drugs used were received as a gift from any company. There is no other disclosure to report.
References
- O'Driscoll L. Expanding on exosomes and ectosomes in cancer. N Engl J Med. 2015;372(24):2359-62. doi:10.1056/NEJMcibr1503100. PMID:26061842
- Corcoran C, Rani S, O'Brien K, O'Neill A, Prencipe M, Sheikh R, Webb G, McDermott R, Watson W, Crown J, O'Driscoll L. Docetaxel-resistance in prostate cancer: evaluating associated phenotypic changes and potential for resistance transfer via exosomes. PLoS One. 2012;7(12):e50999. doi:10.1371/journal.pone.0050999. PMID:23251413
- O'Brien K, Lowry MC, Corcoran C, Martinez VG, Daly M, Rani S, Gallagher WM, Radomski MW, MacLeod RA, O'Driscoll L. Exosomes from triple-negative breast cancer cells can transfer phenotypic traits representing their cells of origin to secondary cells. Eur J Cancer. 2013;49(8):1845-59. doi:10.1016/j.ejca.2013.01.017
- Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, Levin WJ, Stuart SG, Udove J, Ullrich A, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244(4905):707-12. doi:10.1126/science.2470152. PMID:2470152
- Nahta R, Esteva FJ. Trastuzumab: triumphs and tribulations. Oncogene. 2007;26(25):3637-43. doi:10.1038/sj.onc.1210379. PMID:17530017
- Arribas J, Baselga J, Pedersen K, Parra-Palau JL. p95HER2 and breast cancer. Cancer Res. 2011;71(5):1515-9. doi:10.1158/0008-5472.CAN-10-3795. PMID:21343397
- Bruttel VS, Wischhusen J. Cancer stem cell immunology: key to understanding tumorigenesis and tumor immune escape? Front Immunol. 2014;5:360. doi:10.3389/fimmu.2014.00360. PMID:25120546
- Altevogt P, Bretz NP, Ridinger J, Utikal J, Umansky V. Novel insights into exosome-induced, tumor-associated inflammation and immunomodulation. Semin Cancer Biol. 2014;28:51-7. doi:10.1016/j.semcancer.2014.04.008
- Clayton A, Mitchell JP, Court J, Mason MD, Tabi Z. Human tumor-derived exosomes selectively impair lymphocyte responses to interleukin-2. Cancer Res. 2007;67(15):7458-66. doi:10.1158/0008-5472.CAN-06-3456. PMID:17671216
- Rong L, Li R, Li S, Luo R. Immunosuppression of breast cancer cells mediated by transforming growth factor-beta in exosomes from cancer cells. Oncol Lett. 2016;11(1):500-4. PMID:26870240
- Mitchell JD, Maguire JJ, Davenport AP. Emerging pharmacology and physiology of neuromedin U and the structurally related peptide neuromedin S. Br J Pharmacol. 2009;158(1):87-103. doi:10.1111/j.1476-5381.2009.00252.x. PMID:19519756
- Martinez VG, O'Driscoll L. Neuromedin U: a multifunctional neuropeptide with pleiotropic roles. Clin Chem. 2015;61(3):471-82. doi:10.1373/clinchem.2014.231753. PMID:25605682
- Rani S, Corcoran C, Shiels L, Germano S, Breslin S, Madden S, McDermott MS, Browne BC, O'Donovan N, Crown J, et al. Neuromedin U: A Candidate Biomarker and Therapeutic Target to Predict and Overcome Resistance to HER-Tyrosine Kinase Inhibitors. Cancer Res. 2014;74(14):3821-33. doi:10.1158/0008-5472.CAN-13-2053. PMID:24876102
- Korkaya H, Kim GI, Davis A, Malik F, Henry NL, Ithimakin S, Quraishi AA, Tawakkol N, D'Angelo R, Paulson AK, et al. Activation of an IL6 inflammatory loop mediates trastuzumab resistance in HER2+ breast cancer by expanding the cancer stem cell population. Mol Cell. 2012;47(4):570-84. doi:10.1016/j.molcel.2012.06.014
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871-90. doi:10.1016/j.cell.2009.11.007
- Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147(2):275-92. doi:10.1016/j.cell.2011.09.024
- Brabletz T. EMT and MET in metastasis: where are the cancer stem cells? Cancer Cell. 2012;22(6):699-701. doi:10.1016/j.ccr.2012.11.009
- A Andreola G, Rivoltini L, Castelli C, Huber V, Perego P, Deho P, Squarcina P, Accornero P, Lozupone F, Lugini L, et al. Induction of lymphocyte apoptosis by tumor cell secretion of FasL-bearing microvesicles. J Exp Med. 2002;195(10):1303-16. doi:10.1084/jem.20011624. PMID:12021310
- Abusamra AJ, Zhong Z, Zheng X, Li M, Ichim TE, Chin JL, Min WP. Tumor exosomes expressing Fas ligand mediate CD8+ T-cell apoptosis. Blood Cells Mol Dis. 2005;35(2):169-73. doi:10.1016/j.bcmd.2005.07.001. PMID:16081306
- Kim JW, Wieckowski E, Taylor DD, Reichert TE, Watkins S, Whiteside TL. Fas ligand-positive membranous vesicles isolated from sera of patients with oral cancer induce apoptosis of activated T lymphocytes. Clin Cancer Res. 2005;11(3):1010-20. PMID:15709166
- Taylor DD, Gercel-Taylor C, Lyons KS, Stanson J, Whiteside TL. T-cell apoptosis and suppression of T-cell receptor/CD3-zeta by Fas ligand-containing membrane vesicles shed from ovarian tumors. Clin Cancer Res. 2003;9(14):5113-9. PMID:14613988
- Wen SW, Sceneay J, Lima LG, Wong CS, Becker M, Krumeich S, et al. The Biodistribution and Immune Suppressive Effects of Breast Cancer-Derived Exosomes. Cancer Res. 2016;76(23):6816-27. doi:10.1158/0008-5472.CAN-16-0868. PMID:27760789
- Klibi J, Niki T, Riedel A, Pioche-Durieu C, Souquere S, Rubinstein E, Le Moulec S, Guigay J, Hirashima M, Guemira F, et al. Blood diffusion and Th1-suppressive effects of galectin-9-containing exosomes released by Epstein-Barr virus-infected nasopharyngeal carcinoma cells. Blood. 2009;113(9):1957-66. doi:10.1182/blood-2008-02-142596. PMID:19005181
- Wieckowski EU, Visus C, Szajnik M, Szczepanski MJ, Storkus WJ, Whiteside TL. Tumor-derived microvesicles promote regulatory T cell expansion and induce apoptosis in tumor-reactive activated CD8+ T lymphocytes. J Immunol. 2009;183(6):3720-30. doi:10.4049/jimmunol.0900970. PMID:19692638
- Szajnik M, Czystowska M, Szczepanski MJ, Mandapathil M, Whiteside TL. Tumor-derived microvesicles induce, expand and up-regulate biological activities of human regulatory T cells (Treg). PLoS One. 2010;5(7):e11469. doi:10.1371/journal.pone.0011469. PMID:20661468
- Clayton A, Tabi Z. Exosomes and the MICA-NKG2D system in cancer. Blood Cells Mol Dis. 2005;34(3):206-13. doi:10.1016/j.bcmd.2005.03.003. PMID:15885603.
- Liu C, Yu S, Zinn K, Wang J, Zhang L, Jia Y, Kappes JC, Barnes S, Kimberly RP, Grizzle WE, et al. Murine mammary carcinoma exosomes promote tumor growth by suppression of NK cell function. J Immunol. 2006;176(3):1375-85. doi:10.4049/jimmunol.176.3.1375. PMID:16424164
- Valenti R, Huber V, Filipazzi P, Pilla L, Sovena G, Villa A, Corbelli A, Fais S, Parmiani G, Rivoltini L.. Human tumor-released microvesicles promote the differentiation of myeloid cells with transforming growth factor-beta-mediated suppressive activity on T lymphocytes. Cancer Res. 2006;66(18):9290-8. doi:10.1158/0008-5472.CAN-06-1819. PMID:16982774
- Liu Y, Xiang X, Zhuang X, Zhang S, Liu C, Cheng Z, Michalek S, Grizzle W, Zhang HG. Contribution of MyD88 to the tumor exosome-mediated induction of myeloid derived suppressor cells. Am J Pathol. 2010;176(5):2490-9. doi:10.2353/ajpath.2010.090777. PMID:20348242
- Xiang X, Poliakov A, Liu C, Liu Y, Deng ZB, Wang J, Cheng Z, Shah SV, Wang GJ, Zhang L, et al. Induction of myeloid-derived suppressor cells by tumor exosomes. Int J Cancer. 2009;124(11):2621-33. doi:10.1002/ijc.24249. PMID:19235923
- Marton A, Vizler C, Kusz E, Temesfoi V, Szathmary Z, Nagy K, Szegletes Z, Varo G, Siklos L, Katona RL, et al. Melanoma cell-derived exosomes alter macrophage and dendritic cell functions in vitro. Immunol Lett. 2012;148(1):34-8. doi:10.1016/j.imlet.2012.07.006
- Martinez VG, Crown J, Porter RK, O'Driscoll L. Neuromedin U alters bioenergetics and expands the cancer stem cell phenotype in HER2-positive breast cancer. Int J Cancer. 2017;140(12):2771-2784. doi:10.1002/ijc.30705
- Abdullah LN, Chow EK. Mechanisms of chemoresistance in cancer stem cells. Clin Transl Med. 2013;2(1):3. doi:10.1186/2001-1326-2-3. PMID:23369605
- Schatton T, Frank MH. Antitumor immunity and cancer stem cells. Ann N Y Acad Sci. 2009;1176:154-69. doi:10.1111/j.1749-6632.2009.04568.x. PMID:19796244
- Codony-Servat J, Rosell R. Cancer stem cells and immunoresistance: clinical implications and solutions. Transl Lung Cancer Res. 2015;4(6):689-703. doi:10.3978/j.issn.2218-6751.2015.12.11
- Graner MW, Alzate O, Dechkovskaia AM, Keene JD, Sampson JH, Mitchell DA, Bigner DD. Proteomic and immunologic analyses of brain tumor exosomes. FASEB J. 2009;23(5):1541-1557. doi:10.1096/fj.08-122184. PMID:19109410
- Hong CS, Muller L, Whiteside TL, Boyiadzis M. Plasma exosomes as markers of therapeutic response in patients with acute myeloid leukemia. Front Immunol. 2014;5:160. eCollection 2014. doi:10.3389/fimmu.2014.00160. PMID:24782865
- Breslin S, Lowry MC, O'Driscoll L. Neratinib resistance and cross-resistance to other HER2-targeted drugs due to increased activity of metabolism enzyme cytochrome P4503A4. Br J Cancer. 2017;116(5):620-625. doi:10.1038/bjc.2016.445. PMID:28152547.
- Mitchell JP, Court J, Mason MD, Tabi Z, Clayton A. Increased exosome production from tumour cell cultures using the Integra CELLine Culture System. J Immunol Methods. 2008;335(1-2):98-105. doi:10.1016/j.jim.2008.03.001
- Lötvall J, Hill AF, Hochberg F, Buzás EI, Di Vizio D, Gardiner C, Gho YS, Kurochkin IV, Mathivanan S, Quesenberry P, et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J Extracell Vesicles. 2014;3:26913. doi:10.3402/jev.v3.26913. PMID:25536934
