ABSTRACT
Both in vivo data in preclinical cancer models and in vitro data with T cells from patients with advanced cancer support a role for Tim-3 blockade in promoting effective anti-tumor immunity. Consequently, there is considerable interest in the clinical development of antibody-based therapeutics that target Tim-3 for cancer immunotherapy. A challenge to this clinical development is the fact that several ligands for Tim-3 have been identified: galectin-9, phosphatidylserine, HMGB1, and most recently, CEACAM1. These observations raise the important question of which of these multiple receptor:ligand relationships must be blocked by an anti-Tim-3 antibody in order to achieve therapeutic efficacy. Here, we have examined the properties of anti-murine and anti-human Tim-3 antibodies that have shown functional efficacy and find that all antibodies bind to Tim-3 in a manner that interferes with Tim-3 binding to both phosphatidylserine and CEACAM1. Our data have implications for the understanding of Tim-3 biology and for the screening of anti-Tim-3 antibody candidates that will have functional properties in vivo.
Introduction
Tim-3, a member of the T cell immunoglobulin and mucin domain (TIM) family of molecules, has garnered much attention as a co-inhibitory/checkpoint receptor target for cancer immunotherapy. Studies in multiple pre-clinical cancer models have demonstrated that Tim-3 antibody treatment can promote anti-tumor immunity. These studies have employed three different anti-murine Tim-3 antibody clones. Anti-Tim-3 RMT3-23 has been shown to have monotherapeutic efficacy in WT3 sarcoma and MC38 colon carcinoma,Citation1 and anti-Tim-3 5D12 has been shown to have monotherapeutic efficacy in EL4 lymphoma.Citation2 These clones, along with anti-Tim-3 B8.2C12, have been shown to have remarkable efficacy when combined with anti-PD-1/L1 in multiple tumor types.Citation1,3 Similarly, an anti-human Tim-3 antibody has shown single agent efficacy in improving the functional responses of NY-ESO-1-reactive CD8+ T cells from patients with advanced melanoma with more potent efficacy observed when anti-Tim-3 was combined with anti-PD-1.Citation4 Moreover, two recent studies implicate Tim-3 expression as a mechanism of resistance to anti-PD-1 therapy.Citation5,6 Collectively, these data support the use of anti-Tim-3 antibodies for cancer immunotherapy. However, these antibodies are diverse in both species and Ig subclass, and their binding epitopes on Tim-3 have not been defined, thus raising the issue of what shared features might underlie the functional activity of these antibodies. Such information is critical for the development of anti-human Tim-3 antibodies for clinical translation.
Tim-3 has been reported to have multiple ligands. The soluble C-type lectin, galectin-9, was the first reported ligand for Tim-3.Citation7 Subsequently, elucidation of the crystal structure of Tim-3 led to the discovery of a unique binding cleft that is shared among members of the TIM family and is responsible for the binding of TIM family molecules to phosphatidylserine (PtdSer).Citation8-10 In this regard, the binding affinity of Tim-3 for PtdSer is at least five times lower than that of either Tim-1 or Tim-4 for PtdSer.Citation11 More recently, high mobility group box 1 (HMGB1)Citation12 and carcinoembryonic antigen cell adhesion molecule 1 (CEACAM1)Citation13 have also been identified as Tim-3 ligands.
The unique binding cleft that mediates PtdSer binding to TIM family members is framed by the CC' and FG loops.Citation8 PtdSer binding to Tim-3 depends on hydrophobic residues in the FG loop.Citation11 Interestingly, CEACAM1 binding is predicted to involve residues in both the CC' and FG loops, based upon analyses of the mouse and human orthologues, respectively.Citation11,13 Lastly, HMGB1 binding to Tim-3 has also been reported to involve the FG loop.Citation12 In contrast, the galectin-9 binding site is distinct and predicted to involve N-linked sugars on the opposite side of the FGCC' face of Tim-3.Citation7 Thus, the CEACAM1/PtdSer/HMGB1 binding sites and the galectin-9 binding sites comprise distinct and opposed ligand-binding surfaces.
The existence of multiple ligands for Tim-3 raises the important issue of which ligand-binding interaction is responsible for driving the functional effects of Tim-3. Another outstanding question is whether antibodies that have shown functional efficacy do so because they block a specific ligand interaction or because they deplete Tim-3-expessing cells. Here, we have addressed the functional and ligand blocking properties of both anti-mouse and anti-human Tim-3 antibodies that have demonstrated immunomodulatory properties.
Results
Anti-murine Tim-3 antibodies bind non-overlapping epitopes
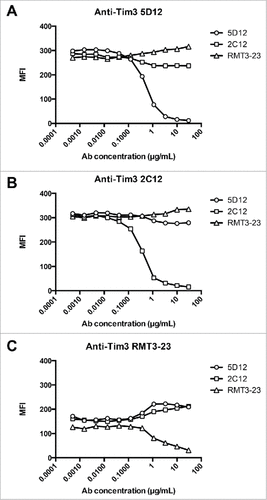
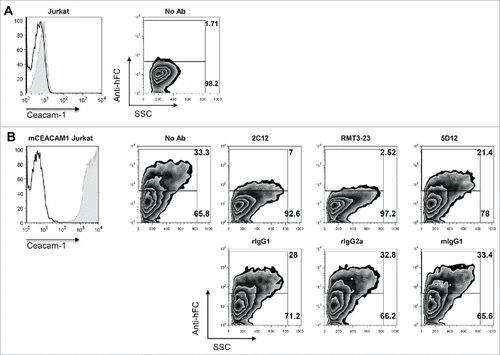
Three different anti-murine Tim-3 antibodies have been reported to have immunomodulatory properties in vivo in multiple pre-clinical models of cancer.Citation1–3 These antibody clones – B8.2C12, RMT3-23, and 5D12 – are of different species origin and isotype. B8.2C12 is a rat IgG1, RMT3-23 is a rat IgG2a, and 5D12 is a mouse IgG1 monoclonal antibody (mAb). To understand the epitope recognition of these antibodies, we performed antibody cross-blocking experiments to address whether these antibodies bind to similar or distinct epitopes on Tim-3. We found that the three antibody clones do not interfere with each other's binding, indicating that they recognize non-overlapping epitopes on Tim-3 (). The fact that these antibodies do not compete for binding as assessed on murine Tim-3 provides a means by which the fate of Tim-3-expressing cells can be reliably tracked in the context of treatment in vivo with each of these antibody clones.
Functional Tim-3 antibodies do not deplete Tim-3+ cells
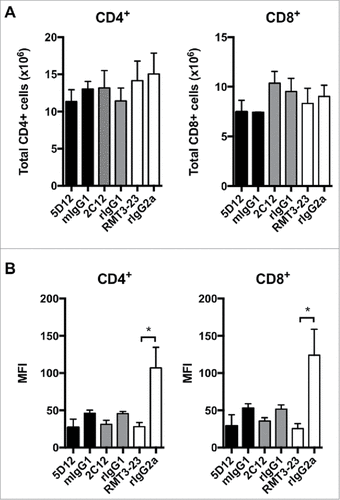
We next addressed whether any of these antibody clones can deplete Tim-3+ T cells in vivo. For this we took advantage of Tim-3 transgenic (Tg) mice that constitutively express high levels of Tim-3 on approximately 40% of peripheral CD4+ and CD8+ T cells.Citation2 We administered B8.2C12, RMT3-23, 5D12 or isotype control antibodies to Tim-3 Tg mice and assessed both Tim-3 expression and the number of CD4+ and CD8+ T cells. We found that administration of B8.2C12, RMT3-23, or 5D12 had no effect on the total number of CD4+ or CD8+ T cells compared to mice administered isotype control antibodies (). We further investigated the ability of the three antibody clones to deplete cells in the tumor microenvironment (TME). Our previous studies showed that Tim-3 is highly expressed on intra-tumoral CD4±FoxP3± regulatory T (Treg) cells.Citation14 We therefore examined the frequency of Treg in tumor-bearing mice after treatment with B8.2C12, RMT3-23, or 5D12 and found no difference in Treg frequencies compared to mice treated with isotype control antibodies (Figure S1). Interestingly, we noted that the Tim-3 Tg mice that received RMT3-23 showed significant down-regulation of Tim-3 as determined by staining with a non-competing anti-Tim-3 antibody clone (). This was not true of clones B8.2C12 and 5D12, which showed a non-statistically significant trend towards receptor down modulation. Further investigation of RMT3-23-mediated Tim-3 down modulation showed that it requires antibody cross-linking (Figure S2). Collectively, these data show that the anti-murine Tim-3 antibodies that have demonstrated efficacy in vivo may function in part by down-modulating Tim-3 surface expression but not by depleting Tim-3+ cells.
Figure 2. Anti-murine Tim-3 do not deplete Tim-3+ cells in vivo. B8.2C12 (n = 3), 5D12 (n = 3), RMT3-23 (n = 3) or matched isotype control antibodies were administered i.p. to Tim-3 Tg mice. After 48 h, the frequency of CD4+ and CD8+ T cells was determined by flow cytometry. A, Total number of CD4+ and CD8+ cells in treated mice. Error bars represent s.e.m. B, Down-modulation of Tim-3 expression was examined by staining with a non-competing Tim-3 antibody clone. Error bars represent s.e.m. #p = 0.03, t-test.
Ligand-blocking properties of murine anti-Tim-3 antibodies
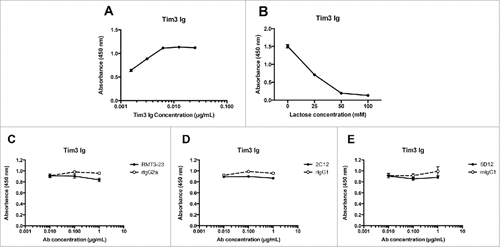
Given that anti-murine Tim-3 antibodies do not deplete Tim-3+ T cells, we next addressed the ligand-blocking properties of the different antibody clones. We first examined the ability of anti-Tim-3 antibodies to interfere with Tim-3 binding to galectin-9. For this, we established an ELISA-based assay wherein we could detect binding of murine Tim-3-Ig to galectin-9 over a dose titration (). Galectins bind their ligands via recognition of oligosaccharide side chains by their carbohydrate recognition domain (CRD). We confirmed that the binding of Tim-3-Ig to galectin-9 in our ELISA assay was indeed dependent on this mechanism by performing our binding assay in the presence of increasing concentrations of lactose (). Having confirmed the specificity of Tim-3:galectin-9 binding in our assay, we tested the ability of the different anti-murine Tim-3 antibody clones to block this binding. None of the three mAbs tested – B8.2C12, 5D12, or RMT3-23 – were able to block the Tim-3:galectin-9 interaction ().
Figure 3. Anti-murine Tim-3 antibodies do not block galectin-9 binding. A, mTim-3-Ig was added at the indicated concentrations to ELISA plates coated with galectin-9. Similar results were obtained in an independent experiment. B, mTim-3-Ig (0.1 μg/ml) was added to ELISA plates coated with galectin-9 in the presence of lactose at the indicated concentrations. mTim-3-Ig was incubated with RMT3-23 (C), B8.2C12 (D), 5D12 (E) or matched isotype control antibody prior to addition to ELISA plates coated with galectin-9. Similar results were obtained in an independent experiment.
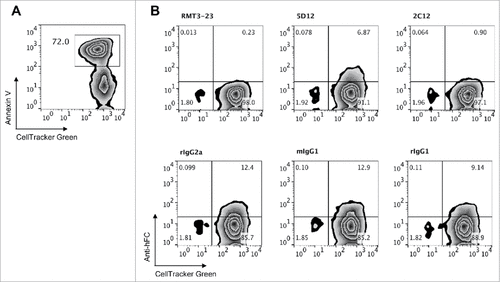
Next, we examined the ability of anti-murine Tim-3 antibodies to interfere with Tim-3 binding to phosphatidylserine (PtdSer) and CEACAM1. The binding sites for these two ligands are located on the opposite side of the Tim-3 IgV domain from the galectin-9 binding site and are in close spatial proximity to each other. Tim-3 binds to PtdSer via the unique binding cleft formed by the presence of four non-canonical cysteine residues that are characteristic of all members of the TIM family.Citation8–10 CEACAM1 is predicted to bind within the vicinity of the CC' and FG loops that frame the PtdSer binding cleft.Citation13 Because of the proximity of these binding sites, it is possible that a given anti-Tim-3 antibody could interfere with binding to either of these ligands. To test whether the anti-murine Tim-3 antibodies affect Tim-3 binding to PtdSer, we determined the ability of each antibody to block the binding of Tim-3-Ig to dexamethasone-treated apoptotic thymocytes, which express PtdSer as determined by staining with Annexin V (). We found that each antibody clone interfered with Tim-3 Ig binding, with RMT3-23 being the most potent blocker, followed by B8.2C12, and then 5D12. Collectively, our data indicate that although these antibodies bind to non-overlapping epitopes on Tim-3, they each bind in such a way that disrupts the Tim-3:PtdSer interaction.
Figure 4. Effect of anti-murine Tim-3 antibodies on binding to phosphatidylserine. A, Dexamethasone-treated thymocytes were labeled with CellTracker Green and stained with Annexin V to detect phosphatidylserine expression. Representative flow cytometry data are shown. B, mTim-3-Ig was incubated with RMT3-23, B8.2C12, 5D12 or matched isotype control antibody prior to addition to dexamethasone treated thymocytes. Data are representative of 4 independent experiments.
To examine whether the antibody clones affect Tim-3 binding to CEACAM1, we transiently expressed full-length murine CEACAM1 in Jurkat T cells and determined the ability of each antibody clone to block Tim-3-Ig binding to CEACAM1-over-expressing cells relative to untransfected parent Jurkat T cells. We found that Tim-3-Ig specifically bound to CEACAM1 transduced, but not un-transduced parent, Jurkat T cells ( and ). We further found that RMT3-23 was the most potent blocker of Tim-3 Ig binding to Jurkat-mCEACAM1 cells, followed closely by B8.2C12, and to a lesser extent, 5D12 (). Lastly, we sought to examine whether anti-murine Tim-3 antibodies could interfere with the binding of HMGB1 to Tim-3; however, we were unable to confirm the previously reported binding interaction between these two proteins.Citation12 Together our data indicate that the anti-murine Tim-3 antibody clones that show efficacy in vivo bind to Tim-3 in such a way that they interfere with both PtdSer and CEACAM1 binding. Moreover, these antibodies can similarly be ranked in a hierarchy for PtdSer and CEACAM1 blockade with RMT3-23 being best, followed by B8.2C12, and lastly by 5D12.
Figure 5. Effect of anti-murine Tim-3 antibodies on binding to CEACAM1. A, Expression of murine CEACAM1 on transduced Jurkat T cells. B, Untransduced or murine CEACAM1-transduced Jurkat T cells were stained with mTim-3-Ig that was pre-incubated with no antibody, RMT3-23, 2C12, 5D12 or matched isotype control antibody. Data are representative of 3 independent experiments.
Ligand-blocking properties of a functional anti-human Tim-3 antibody
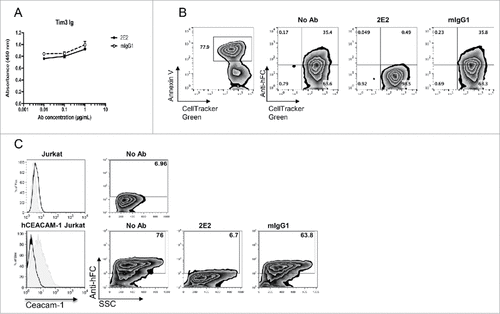
Given our data showing that functional anti-murine Tim-3 antibodies interfere with PtdSer and CEACAM1 binding but not galectin-9 binding, we addressed whether an anti-human TIM-3 antibody clone (clone F38.2E2) that has shown functional properties in vitroCitation4 has similar ligand blocking properties. In line with our murine anti-Tim-3 antibody data, we found that anti-human TIM-3 F38.2E2 does not interfere with galectin-9 binding to human TIM-3 (). However, F38.2E2 results in near complete blockade of Tim-3-Ig binding to both PtdSer and CEACAM1 ( and ). These data indicate that the ligand-blocking properties of functional anti-human and anti-murine Tim-3 antibodies phenocopy each other.
Figure 6. Ligand-blocking properties of an anti-human TIM-3 antibody. A, hTIM-3-Ig was incubated with F38.2E2 or matched isotype control antibody prior to addition to ELISA plates coated with galectin-9. B, hTim-3-Ig was incubated with F38.2E2 or matched isotype control antibody prior to addition to dexamethasone treated thymocytes. C, Untransduced or human CEACAM1-transduced Jurkat T cells were stained with hTim-3-Ig that was pre-incubated with no antibody, F38.2E2 or matched isotype control antibody. Data are representative of 6 independent experiments.
Epitope mapping by hydrogen deuterium exchange mass spectrometry (HDxMS)
We next undertook epitope mapping by HDxMS in order to elucidate the physical basis for these ligand-blocking properties. For this purpose, we used the human and mouse TIM-3 IgV domains and anti-human TIM-3 antibody (F38.2E2) and anti-murine TIM-3 antibodies (B8.2C12, RMT3-23, 5D12), respectively, and performed Hydrogen-deuterium exchange (HDx) in combination with mass spectrometry (MS).Citation15 In HDxMS, backbone amide hydrogens of proteins are replaced by deuterium. This process is sensitive to protein structure/dynamics and solvent accessibility and, therefore, able to report on locations that are directly affected by ligand binding as well as locations that undergo structural modulations such as allosteric effects upon ligand binding.
shows the change in hydrogen exchange observed between free hTIM-3 and the hTIM-3/F38.2E2 complex. The horizontal bars represent individual peptide observations with the sequence aligned along the hTIM-3 sequence (horizontal axis). This provides an impression of the obtained sequence coverage and sequence resolution of the experiments. The shading of the bars represents the average deuteration difference or protection observed for a fragment and provides information about the consistency of observations between fragments. To compensate for differences resulting from size differences of the observed peptides, observed deuteration differences have been normalized by the number of observable amides in each fragment under experimental conditions. The data clearly show that the dominant protection occurs on a limited number of residues in the N-terminal region and a second region protected to a lesser extent can be identified in the C-terminal part of the protein. Detailed inspection of the relative protection observed for overlapping peptides in those regions further allows narrowing of the dominantly protected amides to the sequences of the CC' and FG loops.
Figure 7. Hydrogen-deuterium exchange mass spectrometry mapping. A, Coverage map for hTim-3 as determined by HDxMS analysis. Green shading shows observable amide normalized protection (white 0.02Da per amide of deprotection, darkest green corresponds to −0.19Da per amide of protection). White vertical lines mark the position of proline, which has no exchangeable amide. B, Mapping of the dominantly protected sequence by F38.2E2 (blue) and lesser protected sequence (red) onto the crystal structure of hTIM-3 (PDB 5F71). Orange residues denote N-linked glycosylation sites and pink residues denote polymorphic residues in mTim-3. C-E, coverage map of mTim-3 as determined by HDxMS analysis as shown in A for hTim-3. Dominantly protected sequences by RMT3-23, 5D12, and B8.2C12 are shown in red, purple, and yellow, respectively. Shared protected sequence is shown in blue. F, Mapping of the protected sequences onto the crystal structure of mTim-3 (PDB3 KAA).
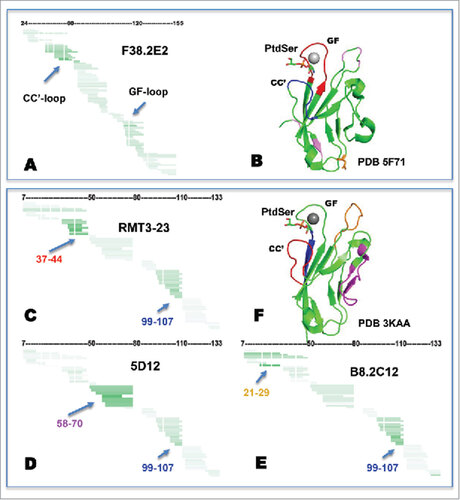
shows the protected sequences color-coded onto a hTIM-3 structure (PDB 5F71) with calcium and phosphatidylserine modeled in, based on the mouse Tim-3 co-structure PDB 3 KAA, for reference. The observed dominantly protected region (blue) and secondary protected region (red) clearly suggest that at least some of the residues of the CC' loop participate in the epitope that is potentially discontinuous and also includes residues in the FG loop. Thus, the observed protections provide strong corroborative evidence for F38.2E2 binding to the FGCC' face of the protein, which is consistent with our data showing loss of PtdSer binding and loss of CEACAM1 binding in the presence of F38.2E2.
Examination of the protection profiles of the three anti-murine clones shows different areas of dominant protection for each clone (), which is in agreement with their non-competitive nature (). However, all three clones additionally showed protection of residues 99–107 (, residues marked blue). This region of allosteric protection is in close proximity to the PtdSer binding site, thus providing a basis for how 5D12 and B8.2C12 are able to block CEACAM1/PtdSer interactions despite having epitopes remote of these ligand-binding sites. Further, it is noteworthy that the epitope of B8.2C12 covers the residues of the BC-loop that are polymorphic across the C57BL/6 and Balb/c mouse strains,Citation11 which is consistent with the differential binding of B8.2C12 to the BALB/c and C57BL/6 mTim-3 alleles (data not shown).
Discussion
The existence of multiple ligands and ligand-binding surfaces for Tim-3 has posed a challenge for understanding Tim-3 biology. Given the established role of Tim-3 as a checkpoint receptor target for cancer immunotherapy, it is important to understand which structural features of Tim-3 are important in mediating its inhibitory function. This is critically important for the development of agents that target Tim-3 for clinical translation. We have addressed this issue by examining the properties of both anti-murine Tim-3 and anti-human TIM-3 antibodies that have shown functional efficacy in vivo and in vitro, respectively. Our data indicate that anti-murine and anti-human Tim-3 antibodies closely phenocopy each other in that they do not interfere with binding to galectin-9 but do interfere with binding to both PtdSer and CEACAM1.
That binding of a given anti-Tim-3 antibody can interfere with binding of PtdSer and CEACAM1 is not surprising given that CEACAM1 is predicted to bind in the vicinity of the CC' and FG loops that frame the PtdSer binding cleft. Some of the critical residues involved include Glu62 in the CC' loop and Asp120 in the FG loop of human TIM-3 which when mutated both abrogate biochemical interactions with human CEACAM1Citation13 and are part of the 2E2 epitope which inhibits CEACAM1-TIM-3 interactions as shown here. Thus, an antibody that binds anywhere on the FGCC' face could interfere directly with CEACAM1 binding while also inducing allosteric changes that either alter the PtdSer binding cleft such that PtdSer can no longer bind or bind in such a way that sterically hinders access of PtdSer to the binding cleft. It is important to note that while our ligand-blocking data implicate interactions with PtdSer and/or CEACAM1 as important for Tim-3 inhibitory function, we cannot discern whether PtdSer and/or CEACAM1 is the physiologically relevant ligand for binding Tim-3 and triggering inhibitory function. Further, our data do not inform as to whether Tim-3 can simultaneously bind to both CEACAM1 and PtdSer.
While our data show that none of the functional anti-Tim-3 antibodies interfere with galectin-9 binding, we cannot discount a role for galectin-9 in Tim-3 function. Galectin-9 has been shown to trigger phosphorylation of the Tim-3 tail and release of the intracellular adaptor Bat-3.Citation16 Loss of Bat-3 allows for binding of the src kinase Fyn, which binds to the same region on the Tim-3 tail as Bat-3Citation16,17 and has been associated with inhibition of T cell responses.Citation18 Moreover, galectin-9 has been shown to promote recruitment of Tim-3 to the immunological synapse, where Tim-3 can co-localize with phosphatases (CD45 and CD148) that antagonize proximal TCR signaling.Citation19 Galectin-9 is a tandem galectin, containing two identical CRD domains linked by a hinge region. Thus, galectin-9 could act as a bridging molecule that promotes the aggregation of adjacent Tim-3:ligand complexes at the cell surface via its tandem CRDs. In such a model, galectin-9 would play an important, although accessory, role in Tim-3 signaling.
Our data examining murine surrogate anti-Tim-3 antibodies has some additional implications for anti-human TIM-3 antibody development. Our data show that murine surrogate anti-Tim-3 antibodies do not deplete Tim-3+ cells. This observation would support the generation of anti-human TIM-3 antibodies that do not mediate ADCC. Further, we found that among murine surrogate anti-Tim-3 antibodies, clone RMT3-23 seemed to be the most potent at blocking both PtdSer and CEACAM1 binding to Tim-3. Interestingly, this antibody also induced significant down-modulation of surface Tim-3 expression. Whether Tim-3 was internalized or shed from the cell surface was not determined. In this regard, some recent studies have demonstrated that ADAM 10/17 metalloproteases can drive Tim-3 shedding from the surface of both monocytes and T cells upon activation.Citation20,21 While the biological impact of shed Tim-3 has not been elucidated, it is interesting to speculate that shed Tim-3 could block Tim-3:ligand interactions and interfere with Tim-3 inhibitory function as has been shown previously for soluble Tim-3-Ig.Citation22,23 Whether the ability of Tim-3 antibodies to promote Tim-3 shedding correlates with functional efficacy will require further investigation.
Our study addresses an outstanding question in Tim-3 biology by showing that blockade of PtdSer and CEACAM1 is a shared property of anti-Tim-3 antibodies with demonstrated functional efficacy. Our findings have important implications not only for understanding how Tim-3 achieves its inhibitory effects but also for the clinical development of antibodies that target Tim-3 for clinical translation.
Material and methods
Animals
Wild type Balb/c mice and C57 BL/6 mice were obtained from the Jackson Laboratory (Bar Harbor, ME). Tim-3 transgenic (Tim-3 Tg) mice were previously described.Citation2 In some experiments, Balb/c mice bred with FoxP3-GFP knock-in mice were used. All experiments were performed in accordance to the guidelines outlined by the Harvard Medical Area Standing Committee on Animals (Boston, MA).
Flow cytometry
Single cell suspensions were stained with antibodies against CD4 (RM4-5), CD8 (53–6.7), and Tim-3 (B8.2C12, RMT3-23, 5D12). All antibodies were purchased from BioLegend or eBioscience except for anti-murine Tim-3 5D12 which was generated in-house. All flow cytometry data was collected on a FACSCalibur (BD Biosciences) and analyzed using FlowJo software (Tree Star).
Antibody-mediated depletion studies
Tim-3 Tg mice were injected i.p. with 200 µg of anti-murine Tim-3 antibody or isotype control. After 48 hours, spleens were harvested and stained with antibodies against CD4, CD8, and non-competing antibodies against Tim-3 prior to analysis by flow cytometry. In some experiments, C57Bl/6 mice were implanted with MC38 colon carcinoma (1 × 106/mouse) and Balb/c FoxP3-GFP KI mice were implanted with CT26 (1 × 106/mouse). Mice bearing established tumors (40–60 mm2) were treated with 200 μg of B8.2C12 (for CT26) or RMT3-23 or 5D12 (for MC38) or isotype control i.p. every third day for a total of 2 shots. Tumors were harvested and analyzed 48–72 hrs after the last injection.
Antibody cross-blocking assay
Jurkat T-cells (ATCC) expressing the Balb/c isoform of murine Tim-3 were incubated with unlabeled B8.2C12, 5D12, or RMT3-23 and then stained individually with either PE-labeled B8.2C12, 5D12, or RMT3-23 and analyzed by flow cytometry.
Antibody-mediated TIM-3 down-modulation
Raw 264.7 cells were treated with rat IgG2a or RMT3-23 at 10 ug/mL, in the presence or absence of anti-rat IgG Fc secondary antibody (ThermoFisher) at 30 ug/mL for the indicated time, and then individually stained with labeled 5D12 or RMT3-23 and analyzed by flow cytometry.
CEACAM1 blocking assay
Jurkat T-cells (ATCC) were transduced with lentiviruses engineered to express either mouse or human full-length CEACAM1. To detect interaction between mouse CEACAM1 and Tim-3, 10 μg/ml recombinant mouse Tim-3-Ig fusion protein harboring a human IgG1 tail (Chimerigen) was incubated with 10 μg/ml of either anti-murine Tim-3 antibody or isotype control antibody for 30 minutes at room temperature prior to addition to the transfected cells. For detection of the interaction between human CEACAM1 and human Tim-3, 10 μg/ml recombinant human Tim-3-Ig harboring a human IgG1 tail (Sino Biological) was incubated with 10 μg/ml of either anti-human Tim-3 (F38.2E2) or isotype control. Tim-3-Ig binding was detected with goat anti-human IgG secondary antibody (Jackson ImmunoResearch) for 15 minutes at 4°C. Cells were analyzed by flow cytometry.
ELISA
To detect the interaction between galectin-9 and Tim-3, Immulon 4HBX Microtiter plates (Thermo Scientific) were coated with 5 µg/mL galectin-9 protein (US Biological) at 4°C overnight. 0.1 µg/mL murine Tim-3-Ig (Chimerigen) or human Tim-3-Ig (Sino Biological) was incubated with either anti-Tim-3 antibody or isotype control for 30 minutes at room temperature prior to addition to the galectin-9 coated plate. Tim-3-Ig binding was detected with 0.5 µg/mL goat anti-human IgG1-biotin (Jackson ImmunoResearch) and 1 µg/mL avidin-peroxidase (Sigma) and quantified using an ELISA Microplate reader (Promega). In some assays, lactose was used to competitively block the binding of Tim-3-Ig to galectin-9.
Phosphatidylserine binding assay
Thymocytes were harvested from Balb/c mice and labeled using 0.1 µM CellTracker Green (ThermoFisher) according to the manufacturer's instructions. Cells were then treated with 1 µM dexamethasone (Sigma) in DMEM with 10% FCS at 37°C for 6 hours. Apoptosis was confirmed by staining with Annexin V (BD Pharmingen). To detect interaction between phosphatidylserine and Tim-3, 10 μg/ml recombinant mouse Tim-3-Ig (Chimerigen) or human Tim-3-Ig (Sino Biological) was incubated with either 10 μg/ml anti-Tim-3 antibody or isotype control for 30 minutes at room temperature prior to addition to the apoptotic cells. Tim-3 binding was detected with goat anti-human-IgG secondary antibody (Jackson ImmunoResearch). Cells were analyzed by flow cytometry.
Fragmentation hydrogen deuterium-exchange mass spectroscopy
HDx-MS experiments were performed as described.Citation15,24 In-house produced hTIM-3 protein expressed in E. coli as a C-terminal cmyc-His6 fusion (Uniprot ID Q8TDQ0[Ser22-Thr135]-EQKLISEEDLNGAAHHHHHH) was used for F38.2E2 mapping. Similarly, Balb/c mTIM-3 (GenBank AAL65156.1 [Asp24-Lys131]-HHHHHHGLNDIFEAQKIEWHE, deglycosylated using PNGase F) produced by mammalian expression was used for mapping of B8.2C12, RMT3-23, and 5D12 antibodies. The method describing the mapping of the human antibody that follows was also applied to murine antibody mapping.
Room temperature on-exchange experiments were performed by manual addition of 50 μl of in-exchange buffer (50 mM phosphate, 150 mM NaCl, pHread 7.6, in D20) to 10μl of 0.25 mg/ml hTIM-3 protein (50 mM phosphate, pH7.6 in H20) or a similar amount of target combined with an equimolar amount of anti-human Tim-3 (F38.2E2, nominally 2 fold excess of Fab-domains). Samples were quenched after 60 s of in-exchange by addition of 250μL quench buffer (4 M guanidinium hydrochloride, 0.5 M TCEP-HCl, 0.2 M phosphate, pH 2.5) and rapidly further diluted with 300μl of storage buffer (20% glycerol, 0.25% formic acid in water) before flash freezing with liquid nitrogen and storage at −80°C until use or transfer into a −70°C drawer attached to the rail of a liquid handler (PAL HTS, LEAP Technologies, Carrboro, NC) located inside 0°C enclosure. Samples were thawed for 120 s by flowing N2 gas over the vial (facilitated by the liquid handler in conjunction with a thaw fixture) and loaded into the sample loop of the injection valve (Valco, Houston, TX). Loading of samples (load buffer 0.05% TFA, 500 µl/min) onto the pre-column of the chromatographic system and online pepsin digestion (immobilized pepsin on Poros AL20, 2.1 × 150 mm) was performed with a dual pump system (2x, Accela 1250, Thermo Scientific, Waltham MA) that allowed admixing of another flow of 550μL/min (0.05% TFA, 500 µl/min) of load buffer post digestion through a mixing tee. The combined flow was directed into the chromatographic system maintained at 0°C for desalting and gradient LC separation at a flow rate of 15 µL/min (Dionex UltiMate 3000, Thermo Scientific, Waltham, MA), which was followed by MS analysis (QExactive, Thermo Scientific, Waltham, MA). The chromatographic system consisted of a valve (15 kPSI Valco, Houston, TX), a 4 µL EXP Halo C18 reversed-phase trap cartridge (Optimize Technologies Inc., Oregon City, OR), and an analytical column (2.1 × 10 mm ID, Prozap 1.5 µm C18, Grace). Gradient separation was from 0% to 40% B over 20 min followed by 40% to 75% B over 5 min using buffer compositions A: 99.75:0.25%v/v (H20: formic acid) and B: 99.75:0.25%v/v (acetonitrile:formic acid). MS scans were acquired at a resolution of 70,000 over the m/z range of 350–2000 for MS, and 35,000 for MS/MS. The instrument parameters used for all experiments including spray voltage of 2.5 kV, a maximum injection time of 120 ms, AGC target for MS of 500,000 ions were maintained the same for all runs. Samples were analyzed in triplicate. Peptide identification was performed by converting raw data to .mgf format using Proteome Discoverer 1.4, searched against the construct sequence using MASCOT 2.4 (Matrix Science, London, UK), and filtered using Scaffold 1.4. Filtered results were imported into HDExaminer (v1.3, Sierra Analytics, Modesto, CA) together with raw data files for quantitation of deuteration. Deuteration values were exported into Microsoft Excel for back exchange correction, calculation of deuteration differences, and normalization of deuterium incorporation by the number of observable amides (number of residues less 2, less the number of prolines in the sequence excluding the N-terminus and penultimate residues) in a peptide. Water, deuterium oxide, guanidine hyrdochloride, sodium chloride, glycerol, formic acid, trifluoroacetic acid (TFA), trifluoroethanol (TFE), acetonitrile (ACN) were from Sigma Chemical Company (St. Louis, MO). Tris (2-carboxyethyl)phosphine) TCEP was from Gold Biotechnology Inc (St. Louis, MO).
Disclosure of interest
A.C. Anderson serves on the scientific advisory boards for Potenza Therapeutics, Tizona Therapeutics, and Idera Pharmaceuticals, which have interests in cancer immunotherapy. V.K. Kuchroo has an ownership interest and is a member of the SAB for Potenza Therapeutics and Tizona Therapeutics. R.S. Blumberg is a consultant to Syntalogic that is developing immune-oncology therapeutics. R.S. Blumberg's, V.K. Kuchroo's, and A.C. Anderson's interests were reviewed and managed by the Brigham and Women's Hospital and Partners Healthcare in accordance with their conflict of interest policies. A.C. Anderson, C.A. Sabatos-Peyton, and V.K. Kuchroo are inventors on patents related to Tim-3. C. Sabatos-Peyton, A. Brock, J. Venable, F. Xu, J. Taraszka, and Xiaomo Jiang are employees of Novartis.
Funding details
This work was supported by grants from the National Institutes of Health (P01AI073748 to VKK, RSB, and ACA; R01NS045937 to VKK; R01CA187975 to ACA; R01DK51362 to RSB), the Melanoma Research Alliance (ACA), and a Drug Discovery Program grant from Novartis (ACA).
supp_file.zip
Download Zip (579.5 KB)Acknowledgments
The authors would like to thank the NIBR Biologics Center (NBC) Basel for providing human Tim-3-Ig used in the HDx-MS experiments. The authors would like to thank Tiancen Hu and Pushpa Jayaraman for helpful discussion.
References
- Ngiow SF, von Scheidt B, Akiba H, Yagita H, Teng MW, Smyth MJ. Anti-TIM3 antibody promotes T cell IFN-{gamma}-mediated anti-tumor immunity and suppresses established tumors. Cancer Res. 2011;71:3540–51. doi:10.1158/0008-5472.CAN-11-0096. PMID:21430066
- Dardalhon V, Anderson AC, Karman J, Apetoh L, Chandwaskar R, Lee DH, Cornejo M, Nishi N, Yamauchi A, Quintana FJ, et al. Tim-3/galectin-9 pathway: regulation of Th1 immunity through promotion of CD11b+Ly-6G+ myeloid cells. J Immunol. 2010;185:1383–92. doi:10.4049/jimmunol.0903275. PMID:20574007
- Sakuishi K, Apetoh L, Sullivan JM, Blazar BR, Kuchroo VK, Anderson AC. Targeting Tim-3 and PD-1 pathways to reverse T cell exhaustion and restore anti-tumor immunity. J Exp Med. 2010;207:2187–94. doi:10.1084/jem.20100643. PMID:20819927
- Fourcade J, Sun Z, Benallaoua M, Guillaume P, Luescher IF, Sander C, Kirkwood JM, Kuchroo V, Zarour HM. Upregulation of Tim-3 and PD-1 expression is associated with tumor antigen-specific CD8+ T cell dysfunction in melanoma patients. J Exp Med. 2010;207:2175–86. doi:10.1084/jem.20100637. PMID:20819923
- Koyama S, Akbay EA, Li YY, Herter-Sprie GS, Buczkowski KA, Richards WG, Gandhi L, Redig AJ, Rodig SJ, Asahina H, et al. Adaptive resistance to therapeutic PD-1 blockade is associated with upregulation of alternative immune checkpoints. Nat Commun. 2016;7:10501. doi:10.1038/ncomms10501. PMID:26883990
- Shayan G, Srivastava R, Li J, Schmitt N, Kane LP, Ferris RL. Adaptive resistance to anti-PD1 therapy by Tim-3 upregulation is mediated by the PI3 K-Akt pathway in head and neck cancer. Oncoimmunology. 2017;6:e1261779. doi:10.1080/2162402X.2016.1261779. PMID:28197389
- Zhu C, Anderson AC, Schubart A, Xiong H, Imitola J, Khoury SJ, Zheng XX, Strom TB, Kuchroo VK. The Tim-3 ligand galectin-9 negatively regulates T helper type 1 immunity. Nat Immunol. 2005;6:1245–52. doi:10.1038/ni1271. PMID:16286920
- Cao E, Zang X, Ramagopal UA, Mukhopadhaya A, Federov A, Federov E, Zencheck WD, Lary JW, Cole JL, Deng H, et al. T cell immunoglobulin mucin-3 crystal structure reveals a novel ligand binding surface. Immunity. 2007;26:311–21. doi:10.1016/j.immuni.2007.01.016. PMID:17363302
- Santiago C, Ballestros A, Tami C, Martinez-Munoz L, Kaplan GG, Casanovas JM. Structures of T Cell immunoglobulin mucin receptors 1 and 2 reveal mechanisms for regulation of immune responses by the TIM receptor family. Immunity. 2007;26:299–310. doi:10.1016/j.immuni.2007.01.014. PMID:17363299
- Santiago C, Ballestros A, Martinez-Munoz L, Mellado M, Kaplan GG, Freeman GJ, Casanovas JM. Structures of T Cell Immunoglobulin Mucin Protein 4 show a metal-ion-dependent ligand binding site where phosphatidylserine binds. Immunity. 2007;27:941–5. doi:10.1016/j.immuni.2007.11.008. PMID:18083575
- DeKruyff RH, Bu X, Ballesteros A, Santiago C, Chim YL, Lee HH, Karisola P, Pichavant M, Kaplan GG, Umetsu DT, et al. T cell/transmembrane, Ig, and mucin-3 allelic variants differentially recognize phosphatidylserine and mediate phagocytosis of apoptotic cells. J Immunol. 2010;184:1918–30. doi:10.4049/jimmunol.0903059. PMID:20083673
- Chiba S, Baghdadi M, Akiba H, Yoshiyama H, Kinoshita I, Dosaka-Akita H, Fujioka Y, Ohba Y, Gorman JV, Colgan JD, et al. Tumor-infiltrating DCs suppress nucleic acid-mediated innate immune responses through interactions between the receptor TIM-3 and the alarmin HMGB1. Nat Immunol. 2012;13:832–42.
- Huang YH, Zhu C, Kondo Y, Anderson AC, Gandhi A, Russell A, Dougan SK, Petersen BS, Melum E, Pertel T, et al. CEACAM1 regulates TIM-3-mediated tolerance and exhaustion. Nature. 2015;517:386–90. doi:10.1038/nature13848. PMID:25363763
- Sakuishi K, Ngiow SF, Sullivan JM, Teng MWL, Kuchroo VK, Smyth MJ, Anderson AC. TIM3+FOXP3+ regulatory Tcells are tissue-specific promoters of T-cell dysfunction in cancer. OncoImmunology. 2013;2:e23849. doi:10.4161/onci.23849. PMID:23734331
- Burns-Hamuro LL, Hamuro Y, Kim JS, Sigala P, Fayos R, Stranz DD, Jennings PA, Taylor SS, Woods VL, Jr. Distinct interaction modes of an AKAP bound to two regulatory subunit isoforms of protein kinase A revealed by amide hydrogen/deuterium exchange. Protein Sci. 2005;14:2982–92. doi:10.1110/ps.051687305. PMID:16260760
- Rangachari M, Zhu C, Sakuishi K, Xiao S, Karman J, Chen A, Angin M, Wakeham A, Greenfield EA, Sobel RA, et al. Bat3 protects T cell responses by repressing Tim-3-mediated exhaustion and death. Nat Med. 2012;18:1394–400. doi:10.1038/nm.2871. PMID:22863785
- Lee J, Su EW, Zhu C, Hainline S, Phuah J, Moroco JA, Smithgall TE, Kuchroo VK, Kane LP. Phosphotyrosine-dependent coupling of Tim-3 to T-cell receptor signaling pathways. Mol Cell Biol. 2011;31:3963–74. doi:10.1128/MCB.05297-11. PMID:21807895
- Davidson D, Schraven B, Veillette A. PAG-associated FynT regulates calcium signaling and promotes anergy in T lymphocytes. Mol Cell Biol. 2007;27:1960–73. doi:10.1128/MCB.01983-06. PMID:17210649
- Clayton KL, Haaland MS, Douglas-Vail MB, Mujib S, Chew GM, Ndhlovu LC, Ostrowski MA. T cell Ig and mucin domain-containing protein 3 is recruited to the immune synapse, disrupts stable synapse formation, and associates with receptor phosphatases. J Immunol. 2014;192:782–91. doi:10.4049/jimmunol.1302663. PMID:24337741
- Clayton KL, Douglas-Vail MB, Nur-ur Rahman AK, Medcalf KE, Xie IY, Chew GM, Tandon R, Lanteri MC, Norris PJ, Deeks SG, et al. Soluble T cell immunoglobulin mucin domain 3 is shed from CD8+ T cells by the sheddase ADAM10, is increased in plasma during untreated HIV infection, and correlates with HIV disease progression. J Virol. 2015;89:3723–36. doi:10.1128/JVI.00006-15. PMID:25609823
- Moller-Hackbarth K, Dewitz C, Schweigert O, Trad A, Garbers C, Rose-John S, Scheller J. A disintegrin and metalloprotease (ADAM) 10 and ADAM17 are major sheddases of T cell immunoglobulin and mucin domain 3 (Tim-3). J Biol Chem. 2013;288:34529–44.
- Sabatos CA, Chakravarti S, Cha E, Schubart A, Sanchez-Fueyo A, Zheng XX, Coyle AJ, Strom TB, Freeman GJ, Kuchroo VK. Interaction of Tim-3 and Tim-3 ligand regulates T helper type 1 responses and induction of peripheral tolerance. Nat Immunol. 2003;4:1102–10. doi:10.1038/ni988. PMID:14556006
- Sanchez-Fueyo A, Tian J, Picarella D, Domenig C, Zheng XX, Sabatos CA, Manlongat N, Bender O, Kamradt T, Kuchroo VK, Gutierrez-Ramos JC, Coyle AJ, Strom TB. TIM-3 inhibits T helper type 1-mediated auto- and alloimmune responses and promotes immunological tolerance. Nat. Immunol. 2003;4:1093–101. doi:10.1038/ni987. PMID:14556005
- Park IH, Venable JD, Steckler C, Cellitti SE, Lesley SA, Spraggon G, Brock A. Estimation of Hydrogen-Exchange Protection Factors from MD Simulation Based on Amide Hydrogen Bonding Analysis. J Chem Inf Model. 2015;55:1914–25. doi:10.1021/acs.jcim.5b00185. PMID:26241692