ABSTRACT
Cancer immunotherapy with antibodies targeting immune checkpoints such as the PD-1/PD-L1 pathway have emerged as breakthrough treatment for multiple solid tumors with high response rates and durable remissions. Despite the benefit for patients and encouraging safety profile, severe inflammatory reactions are observed in some patients. Such immune-related adverse events (irAEs) frequently lead to temporary or permanent cessation of the treatment and require systemic immunosuppression yet underlying mechanisms of irAEs are not known in detail. Here, we describe the T cell-mediated immune reaction in irAE lesions of four patients that developed pneumonitis during therapy with a PD-1 blocking antibody. Immunohistochemical analysis was performed to map the environment of the inflammatory lesions. Tumor infiltrating T cell clones were identified by sequencing the T cell receptor, and comparison with clones from peripheral blood or secondary lymphoid organs. A significant overlap of clones infiltrating irAE lesions and tumors was found. The most prevalent clones were also expanded in peripheral blood, but only a minor fraction of clonal overlap was found. Our findings suggest that irAE lesions in patients under PD-1 blockade are infiltrated by T cells with similar specificity as tumor-infiltrating T cells. These results raise the possibility that the immune response is elicited in these patients against antigens shared by the tumor and distant organs affected by irAEs.
Introduction
Treatment of cancer patients with blocking antibodies that target immune checkpoints including CTLA-4 or PD-1 and PD-L1 lead to impressive response rates and, most notably, may lead to durable remission in patients across different types of cancer.Citation1–3 In particular, long-term benefits have been observed in patients with carcinogen-induced cancers and an increased mutational load such as melanoma or tobacco-induced lung cancer.Citation4 Positive results of randomized phase III trials have led to the approval of antibodies blocking CTLA-4 and PD-1 or PD-L1 for the treatment of melanoma, non-small cell lung cancer, bladder cancer and squamous cell carcinoma of the head and neck.Citation5–9 Various other indications are being tested in ongoing trials. Moreover, the combination of PD-1 and CTLA-4 inhibitors has led to an increase in response rates in patients with melanoma and is currently studied in different trials for other cancer types.Citation5 In addition, many ongoing trials combine checkpoint blockade with other immunotherapies or conventional anti-cancer treatment.Citation2,10
Yet, while these data established immunotherapy as one of the most promising clinical approaches to cancer treatment, marked clinical response is observed in only a fraction of patients, and is limited to cancers with multiple mutations and high levels of expression of checkpoint molecules. Furthermore, checkpoint blockade may entail severe inflammatory off-site toxicity, so called immune-related adverse events (irAEs).Citation11–14 These irAEs can be even life threatening and may involve various organ systems including the skin, the gastrointestinal tract, endocrine organs and the lung.Citation11–14 Severe grade 3–4 irAEs are found in about 22–24% of patients treated with the CTLA-4 inhibitor ipilimumab and in about 5–10% with PD-1 blocking antibodies.Citation11–14 Combination immunotherapy with PD-1 and CTLA-4 inhibitors significantly increases the rate of severe inflammatory side effects to about 50 percent.Citation5
The mechanisms that lead to loss of immune tolerance to autoantigens and irAEs during checkpoint blockade are poorly understood. In particular, it is unclear which patient will develop clinically significant side effects. Here, we report the analysis of the T-cell repertoire diversity and clonal expansions in four patients that developed irAEs during PD-1 blockade.
Results
Development of pneumonitis in patients during PD-1 blockade. Adverse events leading to pulmonary lesions are seen in around 1–2% of patients treated with PD-1 blocking agents.Citation11,12 To further analyze the T cell response in patients that developed pulmonary toxicities, we identified four patients that underwent PD-1 blockade and developed a histologically or cytologically confirmed inflammatory pneumonitis (). Three patients had metastatic melanoma and one patient non-small cell lung cancer (). All patients were treated with a PD-1 inhibitor (either nivolumab or pembrolizumab). Two patients with melanoma had an objective response with one complete and one good partial remission. One patient with melanoma had no response to the treatment with disease progression. The patient with an adenocarcinoma of the lung had a partial response to PD-1 blockade. The time to the development of the irAEs varied between three months and two years. Three patients had affection of a single organ with the development of anti-PD1 related pneumonitis that radiologically and histologically presented as an organizing pneumonia (). In one patient with melanoma and fatal tumor progression, immune toxicities were detected only during necropsy and different organs including the lung, the heart and the meninges were affectedCitation15 (). Molecular analysis of the tumors was performed by a focused next generation sequencing (NGS, OncoMine AmpliSeq Cancer Hotspot Panel v2). Patient 1, 2 and 3 had a BRAF V600E mutation. Also, the adenocarcinoma of patient 4 was studied by focused NGS analysis and no mutation was detected in panel tested. In all 3 patients with an objective response to PD-1 inhibition, pneumonitis was controlled by treatment with corticosteroids and steroids were tapered over 4–6 weeks period.
Table 1. Characteristics of patients examined for the T cell response in tumor lesions and irAE lesions.
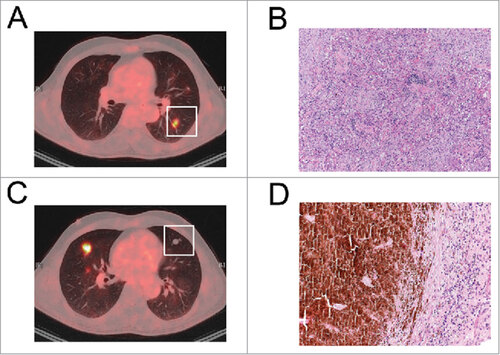
Figure 1. Development of pneumonitis during PD-1 blockade. (A, B) An FDG-positive pulmonary lesion of patient 1 (A) was resected and presented as organizing pnmeumonia in the histological examination (B). (C, D) An FDG-negative lesion in the same patient was resected (C) and a necrotic melanoma metastasis was found by histological analysis (D).
Immunohistochemical analysis of T cell infiltrates. To compare the immune infiltrates in irAE lesions to the tumor or metastasis, we subjected the tissues of patient 1 and 2 to immunohistochemical analysis with a special focus on markers of T cell infiltration and T cell activation ( and ). We had not enough tissue from patient 3 and 4 to perform the extensive immunohistochemical analysis as for patient 1 and 2. Quantification and subtyping of T-cell infiltrates was carried out by digital pathology methods (Figure S1). We observed a similar infiltration of CD3 positive cells and higher CD4/CD8 ratios in the tumoral tissue compared to the corresponding irAE lesions (). Both CD4 and CD8 T cell infiltration was increased in patient 1 in the tumor lesion compared to the pulmonary irAE lesion, while in patient 2 more CD8 T cells was seen in pneumonitis tissue (). Also, activated CD8 T cells were increased in the irAEs (, TIA1+ cells), which points towards an increased cytotoxic T cell response in these lesions. We also stained for PD-1, PD-L1 and FoxP3 positive cells in the tumor and in the irAE lesion (, ). In particularly in patient 1, there was a strong presence of PD-L1 positive cells within the the pulmonary irAE tissue (, ). However, the limited availability of tissue from all 4 patients for these markers prevents the inference of clear conclusions from this analysis and requires a larger number of samples to be analyzed.
Figure 2. Histological analysis of tumor and irAE-infiltrating T cells. (A) Representative images of IHC analysis of the pulmonary irAE lesion from patient 1 for the indicated markers. (B) Quantification of CD3, CD4, and CD8 positive cells, ratios of CD4+ vs CD8+ and TIA1+ cells in tissue sections from the tumor and in the irAE lesions from patients 1 and 2. Numbers were calculated on.
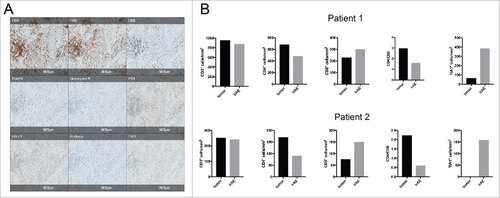
Table 2. Quantification of infiltrating immune cells in tumor and irAE lesions from patient 1 and 2.
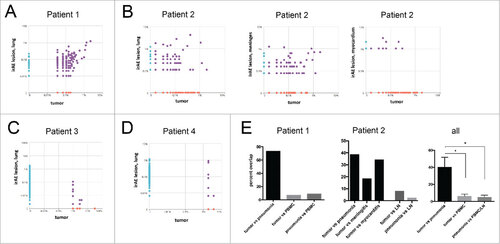
Overlap of T cell clonotypes in the tumor and irAE lesions. To identify unique T cell clones we sequenced the complementary determining region 3 (CDR) of the TCRβ chain of irAE lesions and compared it to tumor infiltrating lymphocytes (TILs). We found a significant overlap of T cell clones present in the irAEs and corresponding tumor lesions in all the four patients (). The most frequent T cell clones were identified in the first two patients in both the tumor and the irAE lesions and represented the most prevalent clonotype (). The overlap of sequences in patient 1 between the lung metastasis and irAE lesion was significantly higher when compared to overlaps with T cell clones in the peripheral blood of this patient ( and ). We had several irAE lesions available from patient 2 (). There was a variable proportion of shared T cell clonotypes between the primary tumor and the lung, meninges and myocardium. The tumor tissue of patient 3 and 4 was limited and the analysis produced fewer sequences ( and ). Nevertheless, there was a clear overlap of clones found in the pulmonary irAEs and in tumor tissue. Overall, the overlap of the T cell repertoire in all patients was significantly higher between tumors and irAE lesions when compared to T cell clones in the periphery or lymph node (, Supplementary Table S1).
Figure 3. Analysis of TCR repertoire in tumors and irAE lesions. (A) Next generation sequencing of the complementary determining region 3 (CDR3) of the beta chain of the TCR was performed. The prevalence (frequency of reads compared to all reads in percent) is compared between inflammatory lung tissue (irAE, y-axis) and the tumor from patient 1. (B) Post-mortem frequency of T cell clones found in different tissues from patient 2. The frequency in the tumor was compared to irAE lesions of the lung (left panel), meninges (middle panel), and the myocardium (right panel). (C, D) Frequency of T cell clones in patient 3 (C) and patient 4 (D). (E) Analysis of overlap of sequences between tumor lesions and pneumonitis lesions or PBMCs/lymph node (LN). The prevalence (frequency of reads compared to all reads in percent) is compared between inflammatory lung tissue (irAE, y-axis) and the tumor from patient 1 (left panel), patient 2 (middle panel) and all analyzed patients (right panel). Analysis by one-way ANOVA, # p < 0.05.
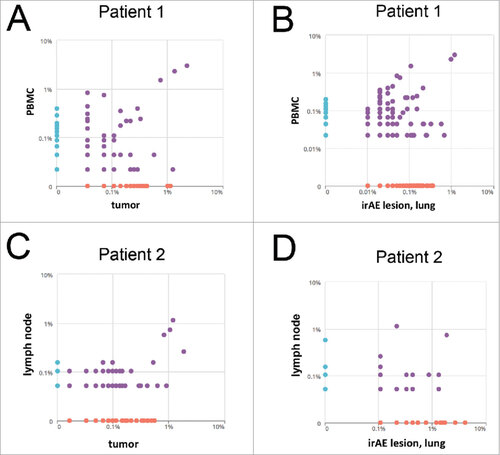
Prevalent TIL and irAE lesion-infiltrating clones are circulating in the periphery. Recent analysis has shown that tumor-reactive clones can be found in the periphery.Citation16 We therefore wanted to test if frequent T cell clones in irAE lesions could also be found in the periphery. Analysis and comparison of the frequency of T cell clones of tumor lesions and irAE lesions was performed to study if common and potentially relevant clones can be found in the peripheral blood or also secondary lymphoid organs (lymph node). In patient 1, we detected the most prevalent clones in the metastatic lesion, the pulmonary irAE lesion but also in the peripheral blood ( and ). Similarly, several prevalent clones found in the tumor were also detected in secondary lymphoid organs of patient 2 including the lymph node ( and ).
Figure 4. Frequency of tumor and irAE-infiltrating T cells in the peripheral blood. (A, B) Prevalence of T cell clones in peripheral blood compared to the metastasis (A) or pulmonary irAE lesion (B) in patient 1. (C, D) Frequency in percent of T cell clones in the lymph node compared to the frequency in the tumor (C) or the pulmonary lung lesion (D).
Discussion
Our report is the first analysis of the TCR repertoire in pulmonary inflammatory lesions from patients treated with PD1 inhibitors. We demonstrate a significant overlap of the T cell repertoire in irAE lesions and TILs. This underlines that the most frequent T cell clones in the tumor and irAEs are shared. In support of this a recent report of two patients with fatal myocarditis identified a strong expression of muscle-associated proteins including troponin and desmin in the tumor.Citation17 Our data support a similar possibility, i.e. that T cells in our patients react to shared antigens that can be found in the tumor and also in the lung. Bioinformatical approaches have recently been published that could allow antigen determination by sequencing the TCR by two independent groups.Citation18,19 Using such methodology, the antigens of prevalent clones in tumors and also in irAE lesions could be potentially defined. Recent studies have also shown an expansion of autoreactive T cell clones in the periphery upon treatment with CTLA-4 blocking antibodies in patients with prostate cancer.Citation20 Not only an expansion of clones was seen but also a diversification of T cells was detected in prostate cancer patients treated with CTLA-4 blocking antibodies and experiencing irAEsCitation21, which supports the hypothesis that newly developed reactivity to shared auto-antigens may play role in the generation of inflammatory complications in patients undergoing checkpoint blockade. An important question with regard to shared antigens is if identified frequent clones that are most likely tumor-reactive are causative for irAE lesions or inflammation in target organs of irAEs driven by other T cell clones attracts tumor specific T cells into these organs as a non-specific secondary event. These activated tumor-specific T cells may then further augment the initial local inflammation.
In general, the exact mechanism of how irAEs in patients treated with checkpoint blockade develop remains largely unknown.Citation22 Other potential mechanisms of irAE development include an exacerbation of previous subclinical autoimmune syndromes, which would be revealed by lowering the threshold for immune activation by the treatment with checkpoint inhibitors.Citation23,24 In one case, a patient who developed severe myositis upon treatment with nivolumab and ipilimumab had pre-existing anti-striated muscle antibodies.Citation23 In another case, a patient with metastatic NSCLC developed a cerebral vasculitis with pre-existing anti-nuclear antibodies (ANAs) and anti-endothelial antibodies.Citation24 However, larger cohorts have not yet been analyzed and it remains unknown if screening for autoantibodies could be used as a biomarker to identify patients at risk for irAEs. An early phase trial with anti-CTLA-4 blocking antibodies showed also a dependence of irAE development on polymorphisms of the CTLA-4 gene.Citation25 Treatment with checkpoint inhibitors including CTLA-4 blocking antibodies can induce epitope spreading.Citation21,22,26 Epitope spreading could enhance anti-tumor responses but also the generation of inflammatory complications of checkpoint blockade. Epigenetic reprogramming of T cells is also possibly involved in a break of peripheral tolerance towards autoantigens and could support the development of irAEs. Regulatory T cells can be reprogrammed to IL-17 producing cells (TH17 cells) that might induce or support autoimmune diseases including psoriasis or autoimmune hepatitis.Citation27
We performed histological analysis for T cell markers in patient 1 and 2. We found in both patients that more T cells were activated and the CD4 to CD8 ratio was decreased. The analysis has clear limitations and while the irAE lesions are both from lungs, the tumor tissue was obtained from lung (patient 1) or from the liver (patient 2) with a significant different microenvironment. Moreover, the tumor tissue from patient 1 was nearly completely necrotic, which precludes spatial analysis between tumor cells and infiltrating immune cells. To understand the immune infiltrates better, histological analysis should be performed on a larger and less heterogeneous sample collection.
Our second important finding is that tumor-specific T cells recirculate via the bloodstream to distant organs (in this case the lung) which are target organs of irAEs. Recent studies found that anti-tumor T cells can be identified in the periphery.Citation16,28 We also found the most prevalent clones were circulating in the peripheral blood of patients. This fact could also be taken into account when trying to further investigate the specificity of these clonotypes. Understanding the mechanisms how similar T cell clones are recirculating to the tumor and organs affected by immune-related side effects could help to identify a biomarker to early detect toxicity in patients undergoing checkpoint blockade.
Taken together, our study shows that there is a significant overlap of T cell clones infiltrating tumor lesions and pulmonary irAE lesions. In addition, prevalent clones can also be found in the peripheral blood. While these findings are hypothesis generating and can be the basis for future work on the loss of immune tolerance in patients treated with checkpoint inhibitors, further studies are needed to elucidate exact mechanisms involved in irAE formation and potential targets to prevent these important side effects of checkpoint inhibition without affecting anti-tumor efficacy.
Material and methods
Patients and sample preparation
Ethics approval was obtained from the local ethical committee to analyze the tissue and blood samples (Ethikkomission Nordwest- und Zentralschweiz, EK321/10 and UBE 15–106). Written informed consent was obtained from all patients prior to the sample collection. PBMCs were isolated by Ficoll density gradient and genomic DNA was prepared. DNA was isolated from paraffin embedded formalin fixed tissue and sent for TCR analysis. The DNA content was analyzed by a Nanodrop2000.
TCR sequencing and sample analysis
TCR analysis of tissue and PBMC samples was performed from isolated DNA. The CDR3 region was amplified and sequenced by the immunoSEQ Assay (Adaptive Biotechnologies, Seattle, WA) as previously described.Citation29 Data analysis was performed using the immunoSEQ Analyzer (Adaptive Biotechnologies, Seattle, WA).
Histology and immunohistochemistry
Staining of serial tissue sections was performed with antibodies against CD3 (clone 2GV6, Ventana), CD4 (clone SP36, Venatana), CD8 (clone SP57, Ventana), FoxP3 (clone SP97, Abcam), PD-1 (clone NAT105, Ventana), PD-L1 (clone SP263, Ventana), granzyme B (polyclonal, Ventana), perforin (clone MRQ-23, Ventana) and TIA (clone TIA-1, Biocare Medical) according to standard protocols. Slides were scanned using a Philips Ultra Fast Scanner 300 (Philips Digital Pathology Solutions, Hamburg Germany). Digital image analysis was carried out by a board-certified pathologist (VHK) using HALO™ v.2.0.1145.19 (Indica Labs, Corrales, NM 87048, USA). In brief, digital annotations were placed on strict serial sections of tumor tissue and the corresponding irAE lesions. Areas of necrosis and unspecific staining were excluded. Color deconvolution was used to optimize software recognition of cell nuclei and the 3,3′-diaminobenzidine (DAB) reaction product. Detection of marker-positive cells was optimized and visually controlled for each marker. Machine learning assisted tissue classification was carried out to eliminate pigment and unspecific staining where appropriate. Whole slide analysis was performed. Total cell counts and marker-positive cells were quantified using the HALO™ cytonuclear algorithm, normalized to tissue area and recorded (Figure S1).
Statistical analysis
Statistical analysis was performed with GraphPad Prism version 6.05. Statistical tests are noted in the figure legends and p-values <0.05 were considered as statistically significant.
2017ONCOIMM0670R-f05-z-4c.pdf
Download PDF (256.9 KB)Acknowledgments
This work was supported by funding from the Lichtenstein Foundation (to H.L.), Schoenmakers Foundation (to H.L.), the Goldschmidt-Jacobson Foundation (to H.L), and the Swiss National Science Foundation (320030_162575 to AZ). Most importantly, we also thank all the patients that allowed the use of their material and made this work possible.
References
- Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27(4):450–61. doi:10.1016/j.ccell.2015.03.001. PMID:25858804
- Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541(7637):321–30. doi:10.1038/nature21349. PMID:28102259
- Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348(6230):56–61. doi:10.1126/science.aaa8172. PMID:25838373
- Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348(6230):69–74. doi:10.1126/science.aaa4971. PMID:25838375
- Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Cowey CL, Lao CD, Schadendorf D, Dummer R, Smylie M, Rutkowski P, et al. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N Engl J Med. 2015;373(1):23–34. doi:10.1056/NEJMoa1504030. PMID:26027431
- Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, Chow LQ, Vokes EE, Felip E, Holgado E, et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N Engl J Med. 2015;373(17):1627–39. doi:10.1056/NEJMoa1507643. PMID:26412456
- Brahmer J, Reckamp KL, Baas P, Crino L, Eberhardt WE, Poddubskaya E, Antonia S, Pluzanski A, Vokes EE, Holgado E, et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. N Engl J Med. 2015;373(2):123–35. doi:10.1056/NEJMoa1504627. PMID:26028407
- Ferris RL, Blumenschein G, Jr., Fayette J, Guigay J, Colevas AD, Licitra L, Harrington K, Kasper S, Vokes EE, Even C, et al. Nivolumab for Recurrent Squamous-Cell Carcinoma of the Head and Neck. N Engl J Med. 2016;375(19):1856–67. doi:10.1056/NEJMoa1602252. PMID:27718784
- Bellmunt J, de Wit R, Vaughn DJ, Fradet Y, Lee JL, Fong L, Vogelzang NJ, Climent MA, Petrylak DP, Choueiri TK, et al. Pembrolizumab as Second-Line Therapy for Advanced Urothelial Carcinoma. N Engl J Med. 2017;376(11):1015–26. doi:10.1056/NEJMoa1613683. PMID:28212060
- Gotwals P, Cameron S, Cipolletta D, Cremasco V, Crystal A, Hewes B, Mueller B, Quaratino S, Sabatos-Peyton C, Petruzzelli L, et al. Prospects for combining targeted and conventional cancer therapy with immunotherapy. Nat Rev Cancer. 2017;17(5):286–301. doi:10.1038/nrc.2017.17. PMID:28338065
- Hofmann L, Forschner A, Loquai C, Goldinger SM, Zimmer L, Ugurel S, Schmidgen MI, Gutzmer R, Utikal JS, Göppner D, et al. Cutaneous, gastrointestinal, hepatic, endocrine, and renal side-effects of anti-PD-1 therapy. Eur J Cancer. 2016;60:190–209. doi:10.1016/j.ejca.2016.02.025. PMID:27085692
- Zimmer L, Goldinger SM, Hofmann L, Loquai C, Ugurel S, Thomas I, Schmidgen MI, Gutzmer R, Utikal JS, Göppner D, et al. Neurological, respiratory, musculoskeletal, cardiac and ocular side-effects of anti-PD-1 therapy. Eur J Cancer. 2016;60:210–25. doi:10.1016/j.ejca.2016.02.024. PMID:27084345
- Michot JM, Bigenwald C, Champiat S, Collins M, Carbonnel F, Postel-Vinay S, Berdelou A, Varga A, Bahleda R, Hollebecque A, et al. Immune-related adverse events with immune checkpoint blockade: a comprehensive review. Eur J Cancer. 2016;54:139–48. doi:10.1016/j.ejca.2015.11.016. PMID:26765102
- Champiat S, Lambotte O, Barreau E, Belkhir R, Berdelou A, Carbonnel F, Cauquil C, Chanson P, Collins M, Durrbach A, et al. Management of immune checkpoint blockade dysimmune toxicities: a collaborative position paper. Ann Oncol. 2016;27(4):559–74. doi:10.1093/annonc/mdv623. PMID:26715621
- Koelzer VH, Rothschild SI, Zihler D, Wicki A, Willi B, Willi N, Voegeli M, Cathomas G, Zippelius A, Mertz KD. Systemic inflammation in a melanoma patient treated with immune checkpoint inhibitors-an autopsy study. J Immunother Cancer. 2016;4:13. doi:10.1186/s40425-016-0117-1. PMID:26981243
- Huang AC, Postow MA, Orlowski RJ, Mick R, Bengsch B, Manne S, Xu W, Harmon S, Giles JR, Wenz B, et al. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature. 2017;545(7652):60–5. doi:10.1038/nature22079. PMID:28397821
- Johnson DB, Balko JM, Compton ML, Chalkias S, Gorham J, Xu Y, Hicks M, Puzanov I, Alexander MR, Bloomer TL, et al. Fulminant Myocarditis with Combination Immune Checkpoint Blockade. N Engl J Med. 2016;375(18):1749–55. doi:10.1056/NEJMoa1609214. PMID:27806233
- Dash P, Fiore-Gartland AJ, Hertz T, Wang GC, Sharma S, Souquette A, Crawford JC, Clemens EB, Nguyen THO, Kedzierska K, et al. Quantifiable predictive features define epitope-specific T cell receptor repertoires. Nature. 2017;547(7661):89–93. doi:10.1038/nature22383. PMID:28636592
- Glanville J, Huang H, Nau A, Hatton O, Wagar LE, Rubelt F, Ji X, Han A, Krams SM, Pettus C, et al. Identifying specificity groups in the T cell receptor repertoire. Nature. 2017;547(7661):94–8. doi:10.1038/nature22976. PMID:28636589
- Subudhi SK, Aparicio A, Gao J, Zurita AJ, Araujo JC, Logothetis CJ, Tahir SA, Korivi BR, Slack RS, Vence L, et al. Clonal expansion of CD8 T cells in the systemic circulation precedes development of ipilimumab-induced toxicities. Proc Natl Acad Sci U S A. 2016;113(42):11919–24. doi:10.1073/pnas.1611421113. PMID:27698113
- Oh DY, Cham J, Zhang L, Fong G, Kwek SS, Klinger M, Faham M, Fong L. Immune Toxicities Elicted by CTLA-4 Blockade in Cancer Patients Are Associated with Early Diversification of the T-cell Repertoire. Cancer Res. 2017;77(6):1322–30. doi:10.1158/0008-5472.CAN-16-2324. PMID:28031229
- June CH, Warshauer JT, Bluestone JA. Is autoimmunity the Achilles' heel of cancer immunotherapy? Nat Med. 2017;23(5):540–7. doi:10.1038/nm.4321. PMID:28475571
- Bilen MA, Subudhi SK, Gao J, Tannir NM, Tu SM, Sharma P. Acute rhabdomyolysis with severe polymyositis following ipilimumab-nivolumab treatment in a cancer patient with elevated anti-striated muscle antibody. J Immunother Cancer. 2016;4:36. doi:10.1186/s40425-016-0139-8. PMID:27330809
- Läubli H, Hench J, Stanczak MA, Heijnen I, Papachristofilou A, Frank S, Zippelius A, Stenner-Liewen F. Cerebral vasculitis mimicking intracranial metastatic progression of lung cancer during PD-1 blockade. J Immunother Cancer. 2017; 5:46. doi:10.1186/s40425-017-0249-y.
- Sanderson K, Scotland R, Lee P, Liu D, Groshen S, Snively J, Sian S, Nichol G, Davis T, Keler T, et al. Autoimmunity in a phase I trial of a fully human anti-cytotoxic T-lymphocyte antigen-4 monoclonal antibody with multiple melanoma peptides and Montanide ISA 51 for patients with resected stages III and IV melanoma. J Clin Oncol. 2005;23(4):741–50. doi:10.1200/JCO.2005.01.128. PMID:15613700
- Kwek SS, Dao V, Roy R, Hou Y, Alajajian D, Simko JP, Small EJ, Fong L. Diversity of antigen-specific responses induced in vivo with CTLA-4 blockade in prostate cancer patients. J Immunol. 2012;189(7):3759–66. doi:10.4049/jimmunol.1201529. PMID:22956585
- Miossec P, Kolls JK. Targeting IL-17 and TH17 cells in chronic inflammation. Nat Rev Drug Discov. 2012;11(10):763–76. doi:10.1038/nrd3794. PMID:23023676
- Gros A, Parkhurst MR, Tran E, Pasetto A, Robbins PF, Ilyas S, Prickett TD, Gartner JJ, Crystal JS, Roberts IM, et al. Prospective identification of neoantigen-specific lymphocytes in the peripheral blood of melanoma patients. Nat Med. 2016;22(4):433–8. doi:10.1038/nm.4051. PMID:26901407
- Carlson CS, Emerson RO, Sherwood AM, Desmarais C, Chung MW, Parsons JM, Steen MS, LaMadrid-Herrmannsfeldt MA, Williamson DW, Livingston RJ, et al. Using synthetic templates to design an unbiased multiplex PCR assay. Nat Commun. 2013;4:2680. doi:10.1038/ncomms3680. PMID:24157944
