ABSTRACT
Purpose: To investigate the immunoprevalence of SALL4-derived peptides in healthy volunteers and cancer patients. Experimental Design: A multistep approach including prediction algorithms was used to design in silico SALL4-derived peptides theoretically able to bind on common HLA-DR and HLA-A/B molecules. The presence of T-cell responses after a long term T-cell assay (28 days) against SALL4 was monitored in 14 healthy donors and the presence of T-cell responses after a short term T-cell assay (10 days) was monitored in 67 cancer patients using IFN-γ ELISPOT assay. A T-cell clone specific for the immunoprevalent A18 K-derived peptide was isolated, characterized and used as a tool to characterize the natural processing of A18 K. Results: A SALL4 specific T-cell repertoire was present in healthy donors (8/14) and cancer patients (29/67) after short term T-cell assay. We further identified two immunoprevalant SALL4-derived peptides, R18 A and A18 K, which bind MHC-class II. In parallel, an A18 K specific Th1 clone recognized monocyte derived Dendritic Cell (moDC) loaded with SALL4 containing cell lysate. The level of IFN-γ secreted by specific T-cell clone was greater in presence of moDC loaded with SALL4 containing cell lysate (49.23 ± 14.02%) than with moDC alone (18.03 ± 3.072%) (p = 0.0477) Conclusion: These results show for the first time immunogenicity of SALL4 oncogenic protein-derived peptides, especially A18 K and R18 A peptides and make them potential targets for personalized medicine. Thus, SALL4 possess major characteristics of a tumor antigen.
Introduction
Spalt (sal) genes were first identified in Drosophila melanogaster in which they have important roles in homeotic specifications.Citation1,Citation2 In human, homologue spalt genes, so called Sal Like (SALL), are split up in four paralogue groups, SALL1, SALL2, SALL3 and SALL4.Citation3 SALL4 encodes a C2 H2 zinc finger transcription factor of unusual but characteristic structure.Citation4 In adult, a major role of SALL4 protein is to regulate hematopoiesis by conferring to hematopoietic stem cells their capacity of self-renewal and pluripotency.Citation5,Citation6 Furthermore, SALL4 protein can be detected in spermatogonial, adipose and embryonic stem cells during early development.Citation7
In 2006, SALL4 protein has been described for the first time as an oncogene.Citation8 Subsequently, aberrant SALL4 expression and deregulated functions in solid tumors like hepatocellular carcinoma, gastric cancers, colorectal cancers, lung cancers, germ cell tumors and hematological malignancies have been reported.Citation9-16 SALL4 has been involved in the leukemogenic mechanism at the origin of Burkitt's lymphoma and acute myeloid leukemia as its inhibition induced significant growth suppression and massive apoptosis of leukemic cells.Citation6,Citation16 Functional roles of SALL4 in metastasis and drug resistance have been described in a panel of solid tumors.Citation17-20 Moreover, in gastric cancers, overexpression of SALL4 enhanced the growth and metastasis of cancer cells and conferred them stem cells' properties.Citation12 Indeed, cells stably transfected with SALL4 gene express stem cell markers and become multipotent.Citation12 Altogether this rational sustains SALL4's role in the generation, maintenance and proliferation of cancer stem cells, as suggested by Tatetsu H and colleagues.Citation21
Moreover, SALL4 can be re-expressed in a subgroup hepatocellular carcinoma and was recognized as a marker for a progenitor subclass with an aggressive phenotype.Citation11 More generally, aberrant amplification of SALL4 among patients with solid cancer is also an unfavorable predictor of survival expectancy.Citation22
Based on SALL4's previously described characteristics, it appeared to be a potential appealing tumor antigen. Indeed, according to Cheever MA, SALL4 protein has some dominant criterions of an ideal tumor antigen, which are its expression in stem cells, its oncogenic properties and its overexpression in cancer cells.Citation23 Nevertheless, SALL4's immunogenicity has not been documented yet. Then, we performed a comprehensive analysis of SALL4 T-cells specific responses in healthy donors and patients with gastro-intestinal cancers, germ cell tumors or chronic myeloid leukemia. By combining long and short-term T-cell assays we demonstrated that it exists a specific SALL4-repertoire in both healthy donors and cancer patients. Two immunoprevalent peptides were identified, A18 K and R18 A. Finally, we isolated one CD4 T cell clone, which secreted IFN-γ in presence of moDC loaded with SALL4 expressing tumor cells, showing the capacity of A18 K peptide to be naturally processed and presented by moDC.
Results
Identification of HLA-A/B and HLA-DR restricted SALL4 derived epitopes
The potential implication of SALL4 protein as an antigen for the adaptive immune system led us to determine if SALL4-derived peptides display immunogenic properties. We first confirmed the absence of SALL4 in healthy adult tissues using GTEx resource database by looking at SALL4 gene expression in human healthy tissues (A) and the presence of SALL4 protein in several gastro-intestinal tumor cell lines (B).Citation24 SALL4 expression in germ cell tumor has been demonstrated in 2009 and is actually a very good marker widely and routinely used in immunohistochemistry for the diagnostic of this tumor because of its specificity, its sensibility.Citation13 In chronic myeloid leukemia, using immunohistochemistry staining, SALL4 protein expression was mostly detected in blast crisis (75%) and not in chronic phase.Citation25 SALL4 expression contributes to the survival of leukemic cells and was previously established in primary leukemic cells.Citation26 Then, the amino acid sequence of SALL4 was screened to identify the existence of specific T-cell epitopes able to bind multiple HLA-DR or HLA-A/B molecules using results obtained from 3 algorithms (Syfpeithi, NetMHCIIpan3.1 and NetMHCpan3) in order to optimize peptide selection. Some of the most commonly expressed alleles encoded by HLA-DRB1 genes were included (DRB1#0101, DRB1#0301, DRB1#0401, DRB1#0701, DRB1#1101 and DRB1#1501). Actually, 85% of people in Europe and in the United State possess at least one of these molecules. As well, commonly expressed alleles encoded by HLA-A/B genes were included (HLA-A#01:01, HLA-A#02:01, HLA-A#03:01, HLA-A#24:02, HLA-A#26:01, HLA-B#07:02, HLA-B#08:01).Citation27 First, we selected seven 9-mer and two 10-mer peptides' sequences that scored high for at least one HLA-A or B allele commonly found in humans in peptide selection's software. (). Then, we decided to select one 15-mer, two 18-mer, one 20-mer and two 25-mer promiscuous peptides' sequences that scored high for multiple HLA-DR alleles commonly found in humans in peptide selection's software (). As a result, we retained fifteen peptides on the basis of their theoretical ability determined in silico to bind to HLA molecules: six promiscuous peptides were designed to bind to HLA-DR molecules and nine peptides were designed to bind to HLA-A/B molecules.
Figure 1. SALL4 expression in human healthy adult tissues and gastro-intestinal tumor cell lines. (A) The absence of SALL4 gene expression in human healthy adult tissues was determined using GTEx resource database. (B) SALL4 protein expression by western blotting in PBMC and HeLa cell line (control), in colorectal tumor cell lines (HT29, Colo 205 and Colo320), in gastric tumor cell line (MKN45) and in pancreatic tumor cell lines (BXPC3, Bespac7, C7, 5 and 12).
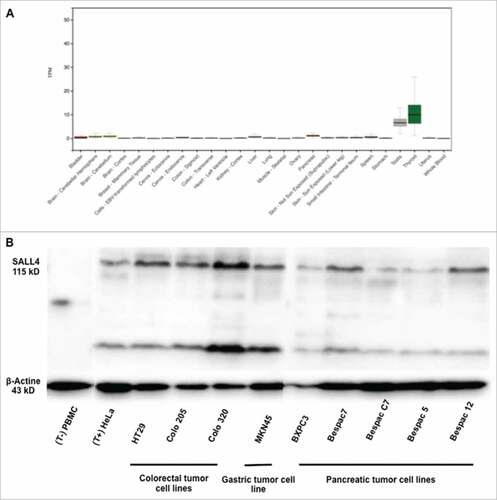
Table 1. Characteristics of HLA-A/B SALL4-specific-restricted peptides predicted by immune informatics database. SALL4 amino acid sequences were submitted to two prediction algorithms (NetMHCpan3 and SYPEITHI) and peptides binding prediction to HLA-A/B were classified as strong (+++), moderate (++), weak (+) or null (−).
Table 2. Characteristics of HLA-DR SALL4-specific-restricted peptides predicted by immune informatics database. SALL4 amino acid sequences were submitted to two prediction algorithms (NetMHCII2.2 and SYPEITHI) and peptides binding prediction to HLA-DR were classified as strong (+++), moderate (++), weak (+) or null (−).
Presence of SALL4-specific T-cells in healthy donors after long term T-cell assay
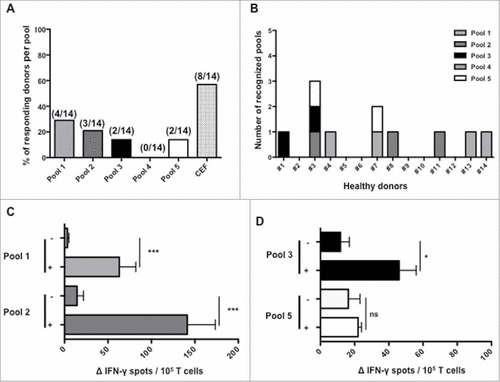
To determine the presence of a SALL4-specific T-cell repertoire among 14 healthy donors, SALL4-derived peptides were used to stimulate T-cells in vitro using long-term T-cell assays. SALL4-specific-T-cells were revealed using IFN-γ-ELISPOT assay. As shown in A, among the 14 healthy donors tested, 28%, 21%, 14% and 14% had a T-cell response directed against peptides from pools 1, 2, 3 and 5 respectively. Altogether, a SALL4-specific repertoire was present in 8 out of 14 healthy donors (B). Two of them, HD3 and HD7, presented multiple responses for respectively 3 and 2 peptides' pools. Mean intensities of positive responses for HLA-DR restricted peptides from pool 1 and 2 were respectively 63 +/− 19 and 141 +/− 32 IFN-γ spots per 105 seeded cells (C). Mean intensities of positive responses for HLA-A and/or B restricted peptides from pool 3 and 5 were respectively 46 +/− 10 and 22 +/− 2 IFN-γ spots (D). Peptides from pool 4 failed to induce an immune response. These results implied that it exists precursor T-cells specific for SALL4-protein epitopes in human peripheral T-cell repertoire. We observed that several SALL4-derived peptides were immunogenic especially peptides from pool 1 and 2 that induce more frequent and intense responses.
Figure 2. Immune responses induced by SALL4 derived peptide's pools in T lymphocytes from healthy donors. PBMC were cultured during 28 days with the 5 pools of SALL4 derived peptides (15 µg/mL). T cell reactivity against peptide's pools was detected by IFN-γ ELISPOT assay as described in materials and methods. (A) Frequency of responding healthy donors (8/14) according to SALL4 derived peptide's pool. (B) Number and detailed of each recognized pools within the 14 healthy donors. Magnitude of T cell responses against SALL4 derived peptides for (C) pool 1 (p < 0.001), pool 2 (p < 0.001), (D) pool 3 (p < 0.05) and pool 5 (not significant). The CEF viral peptide pool was a positive control in ELISPOT IFN-γ assay.
SALL4-specific T-cell responses in healthy donors and cancer patients after short term T-cell assay
Our first results showed that SALL4-derived peptides induced immunological responses in human healthy donors. We next decided to investigate the existence of SALL4 specific spontaneous memory immunity among cancer patients and healthy donors. Constitutive SALL4 gene expression has been associated with gastro-intestinal cancers, germ-cell tumors and hematological malignancies.Citation8,Citation11,Citation28 On the basis of the broad expression of SALL4 oncoprotein in cancers, we carried out a comprehensive analysis of SALL4 specific T-cell responses in sixty-seven cancer patients who had a gastro-intestinal cancer (n = 28), a germ-cell tumor (n = 13) or a chronic myeloid leukaemia (n = 26). A short term in vitro stimulation was performed to amplified T-cell before IFN-γ ELISPOT assay. Among healthy donors, 4 out of 14 (28.6%) and 8 out of 14 (57.1%) had a T-cell response observed respectively in pool 1 and 2 (A). Since immunological responses achieved using peptides from pool 3, 4 and 5 were rare and with a limited intensity (C), we decided to further analyze SALL4 immunogenicity in cancer patients using peptides from pool 1 and 2. As shown in B, 16 out of 67 (23.9%) and 17 out of 67 (25.4%) cancer patients had a T-cell response observed respectively in experiments using peptides from pool 1 and 2. Response rates both in healthy donors and cancer patients according to peptide pools are presented in . Median intensities of positive responses for healthy donors were respectively 149 [14 – 374] and 196 [21 – 280] IFN-γ spots for pools 1 and 2 (A). Median intensities of positive responses for cancer patients were respectively 64 [15 – 352] and 126 [17 – 316] IFN-γ spots for pools 1 and 2 (B). The frequency and quality of antiviral T cell responses in both healthy donors and cancer patients were acceptable. We concluded from these experiments that it exists both in healthy donors and cancer patients, T-cells specific for SALL4-derived peptides. It appeared therefore appropriate to investigate which peptides were immunogenic.
Figure 3. Magnitude and frequency of positive responses induced by SALL4 derived peptide's pools in healthy donors and cancer patients. PBMC were cultured during 8 days with either the five pools of SALL4 derived peptides (15 µg/mL) (healthy donors) or pool 1 and 2 (cancer patients). T cell reactivity against the 5 pool of peptides was detected by IFN-γ ELISPOT assay as described in materials and methods. Results represented specific and positive IFN-γ spots after subtraction of background. Responses were positive when IFN-γ spots were more than 10 and more than 2-fold the background. Magnitude and frequency of spontaneous T cell responses against SALL4 derived peptides in (A) healthy donors (n = 14, controls) and (B) cancer patients (n = 28 GI cancer, n = 26 CML and n = 13 germinal cancer). The CEF viral peptide pool was a positive control in ELISPOT IFN-γ assay. GI, Gastro Intestinal; CML, Chronic Myeloid Leukemia.
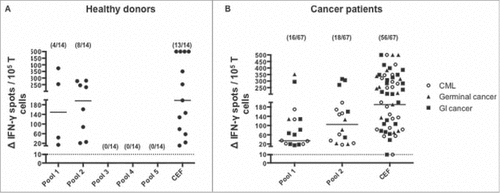
Table 3. Mean, median and frequencies of T-cells responses in healthy donors and cancer patients according to peptides pools.
R18 A and A18 K peptides induce T-cell responses in healthy donors and gastro-intestinal cancer patients
Since intense immune responses were observed following stimulation of human Peripheral Blood Mononuclear Cells (PBMC) with peptides derived from pools 1 and 2, the immunogenicity of individual peptides from these pools was analyzed.
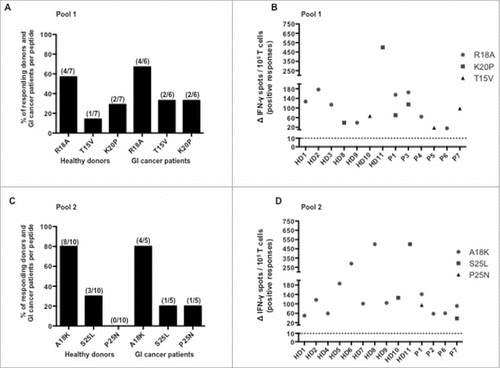
For this purpose, previous experiments were reproduced using separately peptides from pool 1 (R18 A, T15 V and K20P), 2 (A18 K, S25 L and P25 N) and PBMC from 11 healthy donors (never tested before) and 7 cancer patients with a gastro-intestinal cancer who previously had positive responses. HLA-DR genotyping of several healthy donors and cancer patients has been done and detailed in . As shown in A, the frequency of positive responses to R18 A peptide was higher than the one of T15 V and K20P peptides both in healthy donors and cancer patients. Considering peptides from pool 1 among healthy donors, R18 A, T15 V and K20P recognition frequencies were respectively 57%, 14% and 29% (A). Among cancer patients, R18 A, T15 V and K20P recognition frequencies were respectively 67%, 33% and 33% (A). As shown in C, the frequency of positive responses to A18 K peptide was higher than the one of S25 L and P25 N peptides in both healthy donors and cancer patients. Considering peptides from pool 2, A18 K, S25 L and P25 N recognition frequencies were respectively 80%, 30% and 0% among healthy donors and 80%, 20% and 20% among cancer patients (C). Healthy donors and cancer patients' responses to each peptide(s) are presented in B and in D. Altogether, we have shown that SALL4 includes peptides recognized by T lymphocytes from both healthy donors and cancer patients. We have further evidenced the presence of SALL4 specific responses. Moreover, our results identified two promiscuous SALL4-derived peptides, R18 A (SALL4321–339) and A18 K (SALL4718–735), which appeared to be more immunogenic and might be included to assess the role of CD4 SALL4 immunity in most patients.
Table 4. HLA-DR genotyping of several healthy donors and cancer patients.
Figure 4. Frequency of responding healthy donors and GI cancer patients against SALL4 derived peptides and magnitude of responses induced by these peptides. PBMC were cultured during 8 days with distinct SALL4 derived peptides (5 µg/mL). T cell reactivity against SALL4 derived peptides pools was detected by IFN-γ ELISPOT assay as described in materials and methods. The results represented specific IFN-γ spots after subtraction of background. Responses were positive when IFN-γ spots were more than 10 and more than 2-fold the background. Each result presented in the dot plot related to the ELISPOT assays are the mean of duplicates. (A) Frequency of positive responses against R18 A, T15 V and K20P peptides among healthy donors (n = 7, controls) and GI cancer patients (n = 6) who had at least one positive response. (B) Corresponding magnitude of spontaneous T cell responses against R18 A, T15 V and K20P peptides. (C) Frequency of positive responses against A18 K, S25 L and P25 N peptides among healthy donors (n = 10, controls) and GI cancer patients (n = 5) who had at least one positive response. (D) Corresponding magnitude of spontaneous T cell responses against A18 K, S25 L and P25 N peptides. GI, Gastro Intestinal.
Generation of A18 K-specific CD4 T-cell clone
SALL4 peptides were initially selected to bind to HLA-DR molecules (Human Leucocyte Antigen) in a promiscuous manner thus presented to CD4 T-cells in most patients. To assess the potential interest of SALL4-derived peptides for cell therapy and therapeutic anticancer vaccine, we decided to investigate if A18 K and R18 A peptides might induce the production of specific T-cell clones. For this purpose, PBMC from healthy donors and cancer patients in whom a positive response to pool 1 and 2 was previously observed, were stimulated using either A18 K or R18 A peptides for 9 days and an IFN-γ ELISPOT was performed. A18 K or R18 A specific T-cells were isolated using an IFN-γ capture method. Following an amplification step, T-cells were cloned by limiting dilution. One CD4 T-cell clone specific for A18 K, HD5.1, was isolated from HD5's PBMC (data not shown). HD5.1 T-cell clone restriction was evaluated using HLA-DR, HLA-DP and HLA-DQ blocking antibodies. HD5.1's reactivity was strongly inhibited only by HLA-DR blocking antibodies indicating its HLA-DR restriction (A). To further substantiate the ability of HD5.1 T-cell clone to be activated by A18 K in a HLA-DR dependent manner, an EBV-induced B-cell line (Epstein Barr Virus) was generated from PBMC of the same donor used to propagate this clone (so far referred as B-EBV HD5).Citation29 To determinate the HLA-DR allele restriction of the T-cell clone HD5.1 a moderate to high-resolution HLA typing (data not shown), was performed on the B-EBV HD5-cell autologous cell line. Three HLA-DR alleles were identified: HLA-DRB1#04, HLA-DRB1#16 and HLA-DRB4#01:03. A tumor cell line harboring similar restriction was used (COLO 205) to confirm the HLA-DRB1#04 restriction of the A18 K specific CD4 T-cell clone (B).
Figure 5. Functional and phenotypic characteristics of A18 K specific HD5.1 clone. PBMC from a healthy donor in whom a positive response to pool 2 was previously observed were cultured with distinct peptides from pool 2 during 8 days (5 µg/mL). Using the single cell level dilution method, one CD4 T cell clone specific for A18 K peptide, HD5.1, was isolated. (A) HLA-II restriction of HD5.1 T cell clone was assessed by performing an IFN-γ intracellular staining in presence of anti-HLA-DP/DQ/DR antibodies. Percentage of IFN-γ producing T-cell clones (left panel) and mean frequencies of IFN-γ secretion (right panel) was shown. (B) Assessment of HD5.1 clone activation using cells lines harboring similar HLA-DR restriction, Colo205 (stimulated 48 hours using IFN-γ, 1000 UI/mL) and B-EBV HD5, previously loaded with A18 K peptide, by performing an IFN-γ intracellular staining. HD5.1 T cell clone secreted INF-γ in presence of B-EBV HD5 (84.33 +/− 6.70%) and Colo 205 (47.75 +/− 13.45%) loaded with A18 K peptide, whereas not without the peptide loaded on the two cell lines (p < 0.001). (C) Functional avidity of HD5.1 T cell clone was evaluated after stimulation with a range of the indicated increasing A18 K peptide concentrations (XX as irrelevant peptide). (D) HD5.1 T cell clone chemokine receptor pattern was determined by flow cytometry using CXCR3, CCR4, CCR6, CRTh2 and CXCR5 antibodies. (E) Polarization of HD5.1 T-cell clone responses. Detection of cytokine production by DIAplex assay in supernatant after 15 h of culture in presence of A18 K peptide. All data are representative of at least 3 independent experiments.
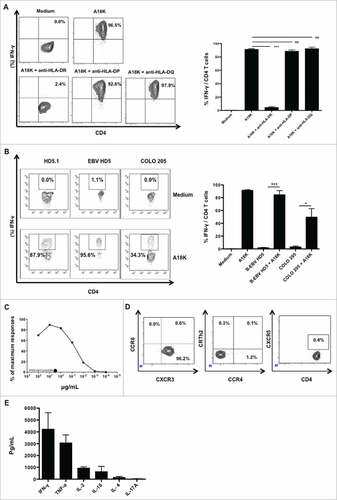
HD5.1 avidity was next determined by incubating HD5.1 clone overnight with a range of A18 K peptide concentration. As shown in C, half-maximal IFN-γ secretion was observed at 0.1 μg/mL To characterize CD4 T-cell polarization, chemokine receptor pattern was first determined using flow cytometry. HD5.1 clone mainly expressed the chemokine receptor CXCR3 and did not express CCR4, CCR6, CRTh2 or CXCR5 (D). To confirm the Th1 polarization, HD5.1 clone was stimulated with A18 K peptide and the cytokine secretion was evaluated. The clone produced IFN-γ and TNF-α, few IL-2 and IL-10 and negligible amount of IL-4 and IL-17 A (E). Thus HLA-DRB1#04-restricted and SALL4-specific Th1 cells could be isolated from PBMC.
SALL4 derived A18 K peptide is naturally processed by antigen presenting cells
To address the natural processing of A18 K, we selected the HLA-DRB1#04 negative cell line HeLa which express high levels of SALL4 (A, B).
Figure 6. A18 K peptide was naturally processed and presented by moDC. moDC were generated from Ficoll-isolated PBMCs from healthy donors selected according to their HLA-DR typing (similar to the clone), as described in material and methods. At day 5, immature moDC were incubated with SALL4 lysate for 8 hours. Mature moDC were generated using LPS. Pulsed cells were washed twice and added at HD5.1 T cell clone overnight. HD5.1 clone's activation was assessed by performing an IFN-γ intracellular staining. (A) SALL4 expression by qRT-PCR in PBMC (control) and HeLa cell line. (B) SALL4 protein expression by western blotting in PBMC (control) and HeLa cell line. (C) HD5.1 T cell clone secreted INF-γ in presence of moDC alone (18.03 +/− 3.07%), moDC loaded with PBMC lysate protein (24.73 +/− 3.42%) and moDC loaded with HeLa lysate protein (49.23 +/− 14.02%) (p < 0.05). Percentage of IFN-γ producing T-cell clones (left panel) and mean frequencies of IFN-γ secretion (right panel) was shown. All data are representative of at least 3 independent experiments.
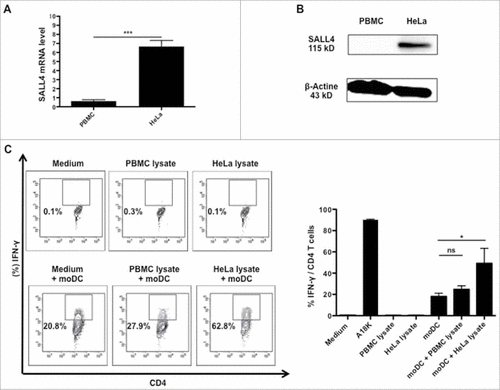
Natural presentation of A18 K peptide by moDC was investigated using HeLa tumor cell lysate containing high level of SALL4 protein (A, B). moDC were generated during 5 days from HLA-DRB1#04 positive PBMC of healthy donors. HeLa lysate protein was applied for 24 hours on immature moDC and LipoPolySaccharide (LPS) (1μg/mL) was added the last 6 hours of culture in the medium. Then, HD5.1 clone was added and co-cultured overnight with moDC. HD5.1 T-cell's activation was assessed by performing an intracellular IFN-γ staining as shown in C. HD5.1 clone secreted IFN-γ in presence of moDC alone (18.03 +/− 3.07%), moDC loaded with PBMC lysate protein (24.73 +/− 3.42%) and moDC loaded with HeLa lysate protein. Clone HD5.1 cultured in presence of moDC loaded with HeLa lysate protein secreted significantly more IFN-γ (49.23 +/− 14.02%) than clone HD5.1 cultivated with moDC alone (p = 0.0477). We observed from these experiments that immature moDC loaded with HeLa lysate containing SALL4 protein appropriately processed the peptide A18 K. Obviously, A18 K appeared to be an immunogenic peptide that can be naturally processed and presented by moDC.
Discussion
SALL4, which functions as a driver of stem cell homeostasis, is widely expressed in various solid and hematological malignancies. During natural development, its expression gradually decreases with tissue differentiation status, to finally become marginal in somatic tissue and restricted to germinal, adipose and hematological stem cells. At the time of early development, SALL4 deficiency results in embryonic lethality. Actually, SALL4 appeared to be a major transcription factor that maintains self-renewal and pluripotency of stem cells but also cancer stem cells.Citation30, Citation31 SALL4 possesses some characteristics of an ideal tumor antigen, supporting its potential use as a target for cancer therapy or as a cancer biomarker. We therefore investigated its immunogenicity using the reverse antigen identification strategy. A large pre-existing T-cell repertoire in healthy donors and cancer patients was found. Our results led to the identification of two immunoprevalent SALL4-derived peptides (A18 K and R18 A). The promiscuous binding of these peptides among different HLA-DR proteins will promote the development of specific immunomonitoring protocols to better decipher the role of SALL4 immunity. Finally, using an A18 K specific T-cell clone we demonstrated that A18 K peptide could be naturally processed and presented by moDC.
Multiple sequence alignment ruled out the possibility that HLA-DR specific SALL4 peptides share homology with another human or microbial protein. A18 K specific T-cells were found in both healthy donors and cancer patients. The SALL4 sequence 718–735, which stands for A18 K peptide, is quite similar to orthologs found in farm animals (cow (72%), horse (100%), pork (89%)) and pets (cat (84%), dog (83%)), which could be a source of cross-reactivity by contact or ingestion, as previously suggested by Chevaleyre C et al. concerning the antigen CCNB1.Citation32 SALL4 protein or cross-reactive peptides do not exist within plants, fungi or bacteria that are therefore not plausible source of sensitization. Otherwise, SALL4-specific memory T-cells could have been primed by endogenous SALL4 after alteration of its expression in the periphery. Indeed, an important T-cell or tissue regeneration process could lead to the abnormal overexpression of SALL4 protein leading to T-cell activation. Another hypothesis of the mechanism leading to spontaneous memory SALL4 repertoire is the role of SALL4 during intracellular infections. Memory SALL4 specific T-cell could have been activated by cross reactivity with viral antigens as suggested by the detection of memory T-cells specific to HIV in healthy donors without any previous exposure.Citation33,Citation34 However, no sequence analogy with virus or bacteria was identified here.
Another explanation for the existence of a SALL4-memory repertoire in healthy donors could be the activation of endogenous SALL4 in human cells driven by viral proteins. Consequently, we have investigated the possible interaction between EBV infection and SALL4 expression. Epstein – Barr virus (EBV) is an intracellular virus with latent potency, affecting more than 90% of the human population worldwide. Two viral components are required for the replication of EBV genome: OriP and EBNA 1.Citation35 Upon infection, EBV establishes different types of latency with different spectra of latent gene expression, notably EBNA1 protein.Citation36 The latent persistence of the EBV proteins promotes antigen-mediated memory T-cell homeostasis and enables a high frequency of peripheral memory T-cells as observed with SALL4 specific lymphocytes. Moreover, β-catenin pathway, which is critical for cell polarity and differentiation, is up regulated in EBV infected B cells and epithelial cells.Citation37 A TCF/LEF consensus sequence was reported in the SALL4 promoter region.Citation38 Thus, SALL4 expression could be activated by LEF1 or TCF4E, upon EBV infection and during EBV latency. However, we failed to identify a direct link between EBV infection and SALL4 expression since transformation of 6 B-cell lines derived from different donors using EBV did not modulate SALL4 protein or RNA levels (data not shown).
The involvement of SALL4 during intracellular infection spreading was recently unraveled in chronic Hepatitis Virus B (HVB)-related hepatocarcinoma.Citation39 Indeed, HVB replication was correlated with SALL4 gene demethylation and an increased transcription level, suggesting that viral infection might transiently induce SALL4 expression. A direct link between viral infections and SALL4 immunity infections remains to be validated.
Ideal targets for cancer immunotherapy are protein silenced in healthy tissues, overexpressed in tumor cells and directly involved in tumor survival and dissemination. According to Cheever, immunogenicity is also an essential characteristic for a tumor antigen to become a target as part of an immunotherapeutic strategy.Citation23 We demonstrated the immunogenicity of SALL4 protein, particularly two CD4 T-cell epitopes, making it a very interesting tumor antigen and thus a potential target in anticancer vaccine or transgenic T-cell receptor immunotherapeutic strategies. Thus, as an example, we can mention hTERT protein, which has emerged as a clinically relevant tumor antigen in immunotherapeutic strategies particularly cancer vaccine.Citation40 Indeed, Godet Y. et al. identified and characterized immunogenic CD4 T-cell epitopes defined as universal cancer peptides (UCP).Citation41 UCP peptides are currently tested in anticancer clinical trials (NCT02818426, NCT01660529). Another exemple of immunogenic tumor antigen is the apoptosis inhibitor protein survivin that induce specific class-II HLA-restricted T-cell responses.Citation42 Indeed, survivin could be considered as a valuable tumor antigen for immune-based clinical approaches and has already been tested in clinical-trials.Citation43, Citation44
In conclusion, our work demonstrates the immunogenicity of SALL4 and the presence of immunity towards this protein both in healthy donors and cancers patients. We characterized the transcription factor SALL4 as a new tumor antigen. Indeed, it possesses important characteristics of a tumor antigen that are its expression in stem cells, its oncogenicity, its overexpression in cancer cells and now its immunogenicity. As a new tumor antigen, SALL4 could be used as a new target for cancer immunotherapy. Furthermore, above the natural presentation of SALL4 to CD4 T-cells, our study also identified promiscuous peptides suitable for the development of a specific immunomonitoring bioassay or to propagate Th1 CD4 T-cells in most cancer patients. Further investigations are expected to determine the prognostic value of SALL4-related Th1 immunity in cancers patients.
Patients and methods
Patients and healthy donors
Chronic myeloid leukemia patients (n = 26) were recruited from the department of Hematology at the University Hospital of Besançon from May 2015 to June 2015. Gastro-intestinal (n = 28) and germ cell tumor (n = 13) patients were recruited from the department of oncology at the University Hospital of Besançon from March 2014 to June 2015. Patients' characteristics are detailed in . All patients were enrolled after the signature of informed consent, in accordance with the French regulation and after approval by the local and national ethics committee. Blood was collected after cancer specific treatment. Blood cells from anonymous healthy donors were collected at the Etablissement Français du Sang (EFS, Besançon, France) as apheresis kit preparations after the signature of informed consent and following EFS guidelines. HLA-DR genotyping was done by using the Olerup SSP DRB1 typing kit (Olerup, Sweden). PBMC from healthy donors and cancer patients were isolated by density centrifugation on Ficoll-Hyperpaque gradients (Sigma-Aldrich), washed twice, respectively in PBS and RPMI-1640 medium and frozen in 2 mL cryogenic vials from 1.106 to 1.107 PBMC per vial using a fetal calf serum 10% DMSO cryomedia. Vials were placed at −80 °C.
Table 5. Baseline clinical characteristics of patients.
Cell culture
Authenticated COLO 205, HT29 and Colo 320, colon adenocarcinoma cells, HeLa cervix adenocarcinoma cells, MKN45 gastric tumor cell and BXPC3 pancreas adenocarcinoma cells were obtained from the ATCC immediately amplified to constitute liquid nitrogen stocks and (upon thawing) never passed for more than 3 weeks before use in experimental determinations. Bespac cell lines were generated in our laboratory from ascites of patients with a pancreas adenocarcinoma. All cell lines were negative for known human's pathogens, including mycoplasma. All cells were periodically authenticated by morphologic inspection. Tumor cells were cultured in DMEM (Fisher) or in RPMI (Fisher) supplemented with 10% FBS, 1% penicillin and 1% streptomycin. In order to express HLA-DR, COLO 205 cells were stimulated 48 hours using IFN-γ 1000 UI/mL (Peprotech) before realizing co-culture with HD5.1 T-cell clone.
Epitope selection and synthetic peptides
The fifteen peptides derived from SALL4 were predicted to bind multiples HLA-A/B and HLA-DR molecules using SYFPETHI, NetMHCpan 3.0 and NetMHCIIpan 3.1 software (access via www.syfpeithi.de/, www.cbs.dtu.dk/services/NetMHCpan/ and www.cbs.dtu.dk/services/NetMHCIIpan/)Citation45. Nine peptides were retained based on their highest predictive score to bind to HLA-A/B molecules (). Six peptides were retained based on their highest predictive score to bind to HLA-DR molecules (). All synthetic peptides (>75% purity) were purchased from ProImmune LTD (Oxford, England).
ELISPOT
Assessment of antigen-specific T-cell responses in healthy donors (long term T-cell assay)
Responses were assessed by IFN-γ ELISPOT after a long-term in vitro stimulation of fresh or thawed PBMC (viability > 50%) with SALL4-derived peptides during 28 days. Ficoll-isolated PBMCs from healthy donors were cultured with 5 µg/mL of SALL4-derived peptides in pool of 3 peptides in a 24 well plate (4.106 cells per well) ( and ). Recombinant interleukin 7 (IL-7) (5 ng/mL; Peprotech, 200–07) was added at day 1. Recombinant IL-2 (20 UI/mL, Novartis) was added at day 3, 6, 10, 16, 20, and 24. At day 14, pools of SALL4-derived-peptides were added again to their assigned wells as previously described to stimulate T-cells a second time. For the recall response against viruses, cells were similarly cultured with a mix of 32 peptides from influenza virus, Epstein-Barr virus and cytomegalovirus to a concentration equal to 2 µg/mL (CEF, Cellular Technology Limited, PA-CEF-001). At day 27, pools of SALL4-derived-peptides were added to their assigned wells. At Day 28, the specificity of T-cell was investigated by IFN- γ ELISPOT (Diaclone, 856 051 020P) as detailed below.
Assessment of antigen-specific T-cell responses in healthy donors and cancer patients (short term T-cell assay)
Responses were assessed by IFN-γ ELISPOT after a short-term in vitro stimulation of fresh or thaw PBMC (viability > 50%) with SALL4-derived peptide during 8 days. Recombinant IL-7 (5 ng/mL) was added at day 1. Recombinant IL-2 (20 UI/mL) was added at day 3 and 6. At day 8, pools of SALL4-derived-peptides were added to their assigned wells. At Day 9, the specificity of T-cell lines was investigated by IFN- γ ELISPOT, as detailed below.
ELISPOT assay
IFN-γ ELISPOT was conducted as previously described.Citation41 Briefly, T-cells (105 per well) were cultured in anti-human IFN-γ monoclonal antibody pre-coated ELISPOT plate with the five pools of SALL4-derived peptides (5 µg/mL) in X-vivo 15 medium (Lonza) for 18 hours at 37 °C. Cells cultured with medium alone or PHA (5 µg/mL; Sigma-Aldrich) were used as negative and positive controls, respectively. The IFN-γ spots were revealed following the manufacturer's instructions (Diaclone, 856 051 020P). The number of specific T-cells expressed as spot forming cells/105 cells was calculated after subtracting negative control values (background). Spot-forming cells were counted using the C.T.L. Immunospot system (Cellular Technology Limited). Responses were positive when IFN-γ spots were more than 10 and more than two times the background. All the experiments were conducted in duplicates. Each result presented in the dot plot related to the ELISPOT assays is the mean of the duplicates
CD4 T-cell clone isolation, amplification and characterization
SALL4-reactive T-cells were isolated by IFN-γ T-cell sorting (Miltenyi Biotec, 130-054-201), according to manufacturer's instructions. SALL4-specific HD5.1 T-cell clone was generated by limiting dilution and amplified after stimulation by PHA in presence of irradiated allogenic PBMCs, B-EBV cell line, and 150 UI/mL of IL-2 according to previously described procedure.Citation46 Functional analyses of HD5.1 T-cell clone were done by using intracytoplasmic IFN-γ staining and DIAplex Human Th1/Th2 complet (Diaclone, 880.140.007) and DIAplex IL-17 A cytokine assay (Diaclone, 880.940.001).
Western blotting
Cells were lysed in sample buffer (2% sodium dodecyl sulfate (SDS) in 125 mM Tris HCl, pH 6.8). An equivalent protein amount, extracted from 1.106 to 5.106 cells, was separated by electrophoresis on 7.5% SDS–polyacrylamide gels and transferred to Polyvinylidene difuoride membranes (GE Healthcare). Blots were then blocked for 1 hour in 6% milk before incubation with specific antibodies as follows: mouse anti-human SALL4 (1/10 000) (R&D systems, MAB6374) and rabbit anti-actin (1/10 000) (Cell Signaling, #8457 L). Blotted proteins were detected and quantified on a bioluminescence imager and BIO-1D advanced software (Vilber-Lourmat) after blots were incubated with a horseradish peroxidase–conjugated appropriate secondary antibody (Beckman Coulter).
RNA isolation, reverse transcription, and real‐time quantification
Total RNA was extracted using the RNeasy Total RNA Isolation kit (Qiagen, 74106), following manufacturer instructions. One microgram of total RNA was used as template for cDNA synthesis performed using a high-capacity RNA to cDNA kit (Applied Biosystem, 4387406). Qualitative RT-PCR (BIO-RAD) was performed using the MyTaq DNA polymerase ready-to-use master mix (Bioline, BIO-25042). Specific primers used were 5′ -TGC AGC AGT TGG TGG AGA AC-3′ (forward) and 5′-TCG GTG GCA AAT GAG ACA TTC- 3′ (reverse) (Eurofins Genomics). Data were normalized to ABL protein levels (Eurofins Genomics). Samples were realized in duplicate. Relative expressions were calculated with the following equation: ΔCt (Sample) = Ct (SALL4) – Ct (ABL). Relative quantity = 2−ΔΔCt x100. PCR products were analyzed by agarose gel electrophoresis followed by ultraviolet detection.
Generation of monocyte derived dendritic cells (moDC)
Ficoll-isolated PBMCs from healthy donors selected according their HLA-DR typing were seeded into cell culture flasks (10.106 cells/mL) in RPMI-1640 10% human serum 1% penicillin streptomycin. After 1.5 hours of incubation at 37 °C, non-adherent T-cells were removed and adherent remaining monocytes were cultured with IL-4 at 1000 UI/mL (Peprotech, 200–04) and GM-CSF at 1000 UI/mL (Peprotech, 300–03). For differentiation and survival of moDC medium renewal was performed after 3 days of culture. At day 5, immature moDC were collected and incubated with SALL4 positive (HeLa) or negative (allogeneic PBMC) cell lysate for 8 hours. Mature moDC were generated after addition of 1 µg/mL LPS (Sigma-Aldrich, L8274), during 6 hours. Pulsed cells were washed twice and added at 5.104 per micro-tube to 5.105 HD5.1 clone T-cell in 1000 µL of X-vivo 15 and Golgi-Plug over-night at 37 °C (ratio moDC:HD5.1 T-cell clone, (2:1) or (1:1)). An intracytoplasmic IFN-γ staining was performed the day after.
Flow cytometry
Surface staining was performed using Pacific Blue-CD3 (BD Pharmingen, 558117) and FITC-CD4 (Diaclone, 954.031.010). Chemokine receptors expression was investigated performing surface staining with BV510-CCR6 (BD Horizon, 563241), PE-CCR4 (BD, 551120), BV510-CXCR5 (BD Horizon, 551120), APC-CXCR3 (BD, 550967) and V450-CrTH2 (BD, 561661). Monoclonal antibodies with matched isotypes (BV510 mouse IgG1, PE mouse IgG1, APC mouse IgG1 and V450 mouse IgG1 antibodies) were used to exclude nonspecific fluorescence. For intracytoplasmic cytokine staining, the cells were fixed and permeabilized using Cytofix/CytoPerm Plus KIT (BD Bioscience, 555028) before staining with APC-IFN-γ (BD Pharmingen, 554702). The samples were directly acquired after finishing the staining on FACS Canto II (BD Bioscience) and data analyzed with DIVA software. The PMT voltages were adjusted using unstained cell for all parameters. The experiments were performed on a median PBMC cell count of 1.106.
Statistics
Statistical analyses were carried out with Prism 5 software. The level of significance was set at P < 0.05 for all tests (#P<0.05, ##P<0.01, ###P<0.001). Variables were expressed as a mean ± SD or median [min – max] and tested with Student t test.
Abbreviations
| EBV | = | Epstein Barr Virus |
| HLA | = | Human Leucocyte Antigen |
| HBV | = | Hepatitis Virus B |
| LPS | = | LipoPolySaccharide |
| moDC | = | MOnocyte derived Dendritic Cell |
| PBMC | = | Peripheral Blood Mononuclear Cells |
| SALL4 | = | SaL Like 4 |
Disclosure of potential conflicts of interest
The authors report no conflict of interest.
Authors' contributions
Conception and design: Borg Christophe, Adotévi Olivier and Godet Yann,
Development of methodology: Borg Christophe, Godet Yann and Mercier-Letondal Patricia
Acquisition of data: Kroemer Marie, Spehner Laurie, Mercier-Letondal Patricia, Boullerot Laura
Analysis and interpretation of data: Kroemer Marie, Spehner Laurie, Mercier-Letondal Patricia, Boullerot Laura
Writing, review and/or revision of the manuscript: Kroemer Marie, Spehner Laurie, Borg Christophe
Administrative, technical, or material support: Borg Christophe, Adotévi Olivier, Kim Stefano, Jary Marine, Ferrand Christophe, Nguyen Thierry and Larosa Fabrice
Study supervision: Borg Christophe and Godet Yann
References
- Frei E, Schuh R, Baumgartner S, Burri M, Noll M, Jürgens G, Seifert E, Nauber U, Jäckle H. Molecular characterization of spalt, a homeotic gene required for head and tail development in the Drosophila embryo. EMBO J. 1988;7(1):197–204.
- Jürgens G. Head and tail development of the Drosophila embryo involves spalt, a novel homeotic gene. EMBO J. 1988;7(1):189–96.
- Sweetman D, Münsterberg A. The vertebrate spalt genes in development and disease. Dev Biol. 2006; 293(2):285–93. doi:10.1016/j.ydbio.2006.02.009.
- Kohlhase J, Schuh R, Dowe G, Kühnlein RP, Jäckle H, Schroeder B, Schulz-Schaeffer W, Kretzschmar HA, Köhler A, Müller U, et al. Isolation, Characterization, and Organ-Specific Expression of Two Novel Human Zinc Finger Genes Related to theDrosophilaGenespalt. Genomics. 1996;38(3):291–8. doi:10.1006/geno.1996.0631.
- Gao C, Kong NR, Li A, Tatetu H, Ueno S, Yang Y, He J, Yang J, Ma Y, Kao GS, et al. SALL4 is a key transcription regulator in normal human hematopoiesis. Transfusion. 2013;53(5):1037–49. doi:10.1111/j.1537-2995.2012.03888.x.
- Yang J, Chai L, Gao C, Fowles TC, Alipio Z, Dang H, Xu D, Fink LM, Ward DC, Ma Y. SALL4 is a key regulator of survival and apoptosis in human leukemic cells. Blood.2008; 112(3):805–13. doi:10.1182/blood-2007-11-126326.
- Xiong J. SALL4: engine of cell stemness. Curr Gene Ther. 2014;14(5):400–11. doi:10.2174/1566523214666140825125138.
- Ma Y, Cui W, Yang J, Qu J, Di C, Amin HM, Lai R, Ritz J, Krause DS, Chai L. SALL4, a novel oncogene, is constitutively expressed in human acute myeloid leukemia (AML) and induces AML in transgenic mice. Blood. 2006;108(8):2726–35. doi:10.1182/blood-2006-02-001594.
- Camparo P, Comperat EM. SALL4 is a useful marker in the diagnostic work-up of germ cell tumors in extra-testicular locations. Virchows Arch Int J Pathol. 2013;462(3):337–41. doi:10.1007/s00428-012-1353-5.
- Kobayashi D, Kuribayashi K, Tanaka M, Watanabe N. Overexpression of SALL4 in lung cancer and its importance in cell proliferation. Oncol Rep. 2011;26(4):965–70.
- Yong KJ, Gao C, Lim JSJ, Yan B, Yang H, Dimitrov T, Kawasaki A, Ong CW, Wong K-F, Lee S, et al. Oncofetal gene SALL4 in aggressive hepatocellular carcinoma. N Engl J Med. 2013;368(24):2266–76. doi:10.1056/NEJMoa1300297.
- Zhang L, Xu Z, Xu X, Zhang B, Wu H, Wang M, Zhang X, Yang T, Cai J, Yan Y, et al. SALL4, a novel marker for human gastric carcinogenesis and metastasis. Oncogene. 2014; 33(48):5491–500. doi:10.1038/onc.2013.495.
- Cao D, Li J, Guo CC, Allan RW, Humphrey PA. SALL4 is a novel diagnostic marker for testicular germ cell tumors. Am J Surg Pathol. 2009;33(7):1065–77. doi:10.1097/PAS.0b013e3181a13eef.
- Ardalan Khales S, Abbaszadegan MR, Abdollahi A, Raeisossadati R, Tousi MF, Forghanifard MM. SALL4 as a new biomarker for early colorectal cancers. J Cancer Res Clin Oncol. 2015;141(2):229–35. doi:10.1007/s00432-014-1808-y.
- Liu J, Wang L, Yang A, Jiang P, Wang M. Up-regulation of SALL4 associated with poor prognosis in gastric cancer. Hepatogastroenterology. 2014;61(133):1459–64.
- Cui W, Kong NR, Ma Y, Amin HM, Lai R, Chai L. Differential expression of the novel oncogene, SALL4, in lymphoma, plasma cell myeloma, and acute lymphoblastic leukemia. Mod Pathol Off J U S Can Acad Pathol Inc. 2006;19(12):1585–92.
- Li A, Jiao Y, Yong KJ, Wang F, Gao C, Yan B, Srivastava S, Lim GSD, Tang P, Yang H, et al. SALL4 is a new target in endometrial cancer. Oncogene. 2015;34(1):63–72. doi:10.1038/onc.2013.529.
- Liu L, Zhang J, Yang X, Fang C, Xu H, Xi X. SALL4 as an Epithelial-Mesenchymal Transition and Drug Resistance Inducer through the Regulation of c-Myc in Endometrial Cancer. PloS One. 2015;10(9):e0138515. doi:10.1371/journal.pone.0138515.
- Yanagihara N, Kobayashi D, Kuribayashi K, Tanaka M, Hasegawa T, Watanabe N. Significance of SALL4 as a drug-resistant factor in lung cancer. Int J Oncol. 2015;46(4):1527–34. doi:10.3892/ijo.2015.2866.
- Chen Y-Y, Li Z-Z, Ye Y-Y, Xu F, Niu R-J, Zhang H-C, Zhang Y-J, Liu Y-B, Han B-S. Knockdown of SALL4 inhibits the proliferation and reverses the resistance of MCF-7/ADR cells to doxorubicin hydrochloride. BMC Mol Biol. 2016;17:6. doi:10.1186/s12867-016-0055-y.
- Tatetsu H, Kong NR, Chong G, Amabile G, Tenen DG, Chai L. SALL4, the missing link between stem cells, development and cancer. Gene. 2016;584(2):111–19. doi:10.1016/j.gene.2016.02.019.
- Cheng J, Gao J, Shuai X, Tao K. Oncogenic protein SALL4 and ZNF217 as prognostic indicators in solid cancers: a meta-analysis of individual studies. Oncotarget. 2016;7(17):24314–25. doi:10.18632/oncotarget.8237.
- Cheever MA, Allison JP, Ferris AS, Finn OJ, Hastings BM, Hecht TT, Mellman I, Prindiville SA, Viner JL, Weiner LM, et al. The prioritization of cancer antigens: a national cancer institute pilot project for the acceleration of translational research. Clin Cancer Res. 2009;15(17):5323–37. doi:10.1158/1078-0432.CCR-09-0737.
- Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, Hasz R, Walters G, Garcia F, Young N, et al. The Genotype-Tissue Expression (GTEx) project. Nat Genet 2013;45(6): 580–5. doi:10.1038/ng.2653.
- Lu J, Ma Y, Kong N, Alipio Z, Gao C, Krause DS, Silberstein LE, Chai L. Dissecting the role of SALL4, a newly identified stem cell factor, in chronic myelogenous leukemia. Leukemia 2011;25(7):1211–3. doi:10.1038/leu.2011.65.
- Gao C, Dimitrov T, Yong KJ, Tatetsu H, Jeong H, Luo HR, Bradner JE, Tenen DG, Chai L. Targeting transcription factor SALL4 in acute myeloid leukemia by interrupting its interaction with an epigenetic complex. Blood 2013;121(8):1413–21. doi:10.1182/blood-2012-04-424275.
- Kollman C, Maiers M, Gragert L, Müller C, Setterholm M, Oudshoorn M, Hurley CK. Estimation of HLA-A, -B, -DRB1 Haplotype Frequencies Using Mixed Resolution Data from a National Registry with Selective Retyping of Volunteers. Hum Immunol. 2007;68(12):950–8. doi:10.1016/j.humimm.2007.10.009.
- Cao D, Humphrey PA, Allan RW. SALL4 is a novel sensitive and specific marker for metastatic germ cell tumors, with particular utility in detection of metastatic yolk sac tumors. Cancer 2009;115(12):2640–51. doi:10.1002/cncr.24308.
- Miller G. Immortalization of human lymphocytes by Epstein-Barr virus. Yale J Biol Med. 1982;55(3):305–10.
- Jeong H-W, Cui W, Yang Y, Lu J, He J, Li A, Song D, Guo Y, Liu BH, Chai L. SALL4, a stem cell factor, affects the side population by regulation of the ATP-binding cassette drug transport genes. PloS One 2011;6(4):e18372. doi:10.1371/journal.pone.0018372.
- Oikawa T, Kamiya A, Zeniya M, Chikada H, Hyuck AD, Yamazaki Y, Wauthier E, Tajiri H, Miller LD, Wang XW, et al. Sal-like protein 4 (SALL4), a stem cell biomarker in liver cancers. Hepatol. 2013;57(3):1469–83. doi:10.1002/hep.26159.
- Chevaleyre C, Benhamouda N, Favry E, Fabre E, Mhoumadi A, Nozach H, Marcon E, Cosler G, Vinatier E, Oudard S, et al. The Tumor Antigen Cyclin B1 Hosts Multiple CD4 T Cell Epitopes Differently Recognized by Pre-Existing Naive and Memory Cells in Both Healthy and Cancer Donors. J Immunol. 2015;195(4):1891–901. doi:10.4049/jimmunol.1402548.
- Su LF, Kidd BA, Han A, Kotzin JJ, Davis MM. Virus-specific CD4(+) memory-phenotype T cells are abundant in unexposed adults. Immunity. 2013;38(2):373–83. doi:10.1016/j.immuni.2012.10.021.
- Campion SL, Brodie TM, Fischer W, Korber BT, Rossetti A, Goonetilleke N, McMichael AJ, Sallusto F. Proteome-wide analysis of HIV-specific naive and memory CD4(+) T cells in unexposed blood donors. J Exp Med. 2014;211(7):1273–80. doi:10.1084/jem.20130555.
- Leight ER, Sugden B. EBNA-1: a protein pivotal to latent infection by Epstein-Barr virus. Rev Med Virol. 2000;10(2):83–100. doi:10.1002/(SICI)1099-1654(200003/04)10:2%3c83::AID-RMV262%3e3.0.CO;2-T.
- Van Zuylen WJ, Rawlinson WD, Ford CE. The Wnt pathway: a key network in cell signalling dysregulated by viruses. Rev Med Virol. 2016;26(5):340–55. doi:10.1002/rmv.1892.
- Everly DN, Kusano S, Raab-Traub N. Accumulation of cytoplasmic beta-catenin and nuclear glycogen synthase kinase 3beta in Epstein-Barr virus-infected cells. J Virol. 2004; 78(21):11648–55. doi:10.1128/JVI.78.21.11648-11655.2004.
- Böhm J, Sustmann C, Wilhelm C, Kohlhase J. SALL4 is directly activated by TCF/LEF in the canonical Wnt signaling pathway. Biochem Biophys Res Commun. 2006; 348(3):898–907. doi:10.1016/j.bbrc.2006.07.124.
- Fan H, Cui Z, Zhang H, Mani SK, Diab A, Lefrancois L, Fares N, Merle P, Andrisani O. DNA demethylation induces SALL4 gene re-expression in subgroups of hepatocellular carcinoma associated with Hepatitis B or C virus infection. Oncogene. 2017;36(17):2435–45 doi:10.1038/onc.2016.399.
- Kobayashi H, Celis E. Peptide epitope identification for tumor-reactive CD4 T cells. Curr Opin Immunol. 2008;20(2):221–7. doi:10.1016/j.coi.2008.04.011.
- Godet Y, Fabre E, Dosset M, Lamuraglia M, Levionnois E, Ravel P, Benhamouda N, Cazes A, Pimpec-Barthes FL, Gaugler B, et al. Analysis of Spontaneous Tumor-Specific CD4 T-cell Immunity in Lung Cancer Using Promiscuous HLA-DR Telomerase-Derived Epitopes: Potential Synergistic Effect with Chemotherapy Response. Clin Cancer Res. 2012;18(10):2943–53. doi:10.1158/1078-0432.CCR-11-3185.
- Casati C, Dalerba P, Rivoltini L, Gallino G, Deho P, Rini F, Belli F, Mezzanzanica D, Costa A, Andreola S, et al. The apoptosis inhibitor protein survivin induces tumor-specific CD8+ and CD4+ T cells in colorectal cancer patients. Cancer Res. 2003;63(15):4507–15.
- Mehrotra S, Britten CD, Chin S, Garrett-Mayer E, Cloud CA, Li M, Scurti G, Salem ML, Nelson MH, Thomas MB, et al. Vaccination with poly(IC:LC) and peptide-pulsed autologous dendritic cells in patients with pancreatic cancer. J Hematol Oncol 2017;10(1):82–95. doi:10.1186/s13045-017-0459-2.
- Fenstermaker RA, Ciesielski MJ, Qiu J, Yang N, Frank CL, Lee KP, Mechtler LR, Belal A, Ahluwalia MS, Hutson AD. Clinical study of a survivin long peptide vaccine (SurVaxM) in patients with recurrent malignant glioma. Cancer Immunol Immunother. 2016; 65(11):1339–52. doi:10.1007/s00262-016-1890-x.
- Kobayashi H, Celis E. Peptide epitope identification for tumor-reactive CD4 T cells. Curr Opin Immunol. 2008;20(2):221–7. doi:10.1016/j.coi.2008.04.011.
- Godet Y, Desfrançois J, Vignard V, Schadendorf D, Khammari A, Dreno B, Jotereau F, Labarrière N. Frequent occurrence of high affinity T cells against MELOE-1 makes this antigen an attractive target for melanoma immunotherapy. Eur J Immunol. 2010;40(6):1786–94. doi:10.1002/eji.200940132.
