ABSTRACT
Shedding of ADAM10 substrates, like TNFα, MICA or CD30, is reported to affect both anti-tumor immune response and antibody-drug-conjugate (ADC)-based immunotherapy. Soluble forms of these molecules and ADAM10 can be carried and spread in the microenvironment by exosomes released by tumor cells. We reported new ADAM10 inhibitors able to prevent MICA shedding in Hodgkin lymphoma (HL), leading to recognition of HL cells by cytotoxic lymphocytes.
In this paper, we show that the mature bioactive form of ADAM10 is released in exosome-like vesicles (ExoV) by HL cells and lymph node mesenchymal stromal cells (MSC). We demonstrate that ADAM10 inhibitors are released in ExoV by MSC or HL cells, endocytosed by bystander cells and localized in the endolysosomal compartment in HL MSC. ExoV released by HL cells can enhance MICA shedding by MSC, while ExoV from MSC induce TNFα or CD30 shedding by HL cells. Of note, ADAM10 sheddase activity carried by ExoV is prevented with the ADAM10 inhibitors LT4 and CAM29, pretreating either the ExoV-producing or the ExoV-receiving cells. In particular, both inhibitors reduce CD30 shedding maintaining the anti-tumor effects of the ADC Brentuximab-Vedotin or the anti-CD30 Iratumumab on HL cells.
Thus, spreading of ADAM10 activity due to ExoV can result in the release of cytokines, like TNFα, a lymphoma growth factor, or soluble molecules, like sMICA or sCD30, that potentially interfere with host immune surveillance or immunotherapy. ADAM10 blockers can interfere with this process, allowing the development of anti-lymphoma immune response and/or efficient ADC-based or human antibody-based immunotherapy.
KEYWORDS:
Introduction
ADAMs (A Disintegrin And Metalloproteinase) family includes transmembrane proteins with protease activity exerted through a catalytic metalloproteinase domain on a wide panel of substrates.Citation1,Citation2 When this process targets transmembrane molecules, including growth factors, cytokines, receptors and their ligands or cell adhesion molecules, it is called ectodomain shedding and leads to the release of soluble bioactive molecules.Citation1,Citation2 Some of them, such as tumor necrosis factor (TNF)α, a substrate for ADAM17 and ADAM10, are involved in the pathogenesis and development of several cancers.Citation3-6 Moreover, overexpression of ADAM10 or ADAM17 relates with parameters of tumor progression (size, grade, metastasis and lymph node involvement).Citation6,Citation7 Thus, ADAMs have been proposed as both biomarkers and therapeutic targets for cancer,Citation7,Citation8 while ADAM10 or ADAM17 inhibitors with anti-tumor effects have been described.Citation9-11
The sheddase activity of ADAMs can also target the so-called “stress molecules”, like the MHC-class-I related MICA and MICB, and the UL16 binding proteins (ULBPs), expressed on cancer cell surface and responsible for inducing an immune response against tumor cells.Citation12,Citation13 This mechanism has been evidenced in chronic lymphocytic leukemia, acute myeloid leukemia, non-Hodgkin and Hodgkin's lymphomas (HL).Citation14-17 In particular, we described overexpression of ADAM10 in the lymph node microenvironment in HL, together with impaired stimulation of T lymphocytes with anti-tumor activity.Citation18 Likewise, CD30 shedding due to ADAM10 activity, has been reported to decrease the efficiency of targeted lymphoma cell killing obtained with monoclonal antibodies in vitro and this effect can be prevented by the use of the inhibitor GI254023X.Citation19 We also developed inhibitors with high specificity for ADAM10 to enhance efficiency and selectivity of action; exposure of HL cells to these inhibitors significantly increases their sensitivity to lymphocyte-mediated killing.Citation20,Citation21
Bioactive ADAM10 can be released in extracellular microvesicles (EV, 150–800 nm of size) and exosomes (<150 nm) as well, derived either by tumor or by neighboring stromal cells, which are potentially able to spread the ADAM10 enzymatic activity in the microenvironment.Citation22,Citation23 Indeed, EV contain multiple cytoplasmic or membrane molecules derived from the budding process, including growth factors, cytokines and metalloproteases, that can interact with EV recipient cells and deliver signals promoting tumor invasiveness and metastasis.Citation24,Citation25 Since the process of EV release is particularly active in proliferating cells, such as cancer cells, it has further been proposed that EV and exosomes can play a role in promoting and maintaining tumor cell survival. Of note, exosomes have been described to down-regulate NKG2D expression on natural killer and T lymphocytes, representing a powerful mechanism of immune escape.Citation26,Citation27 Thus, it is worth inhibiting ADAM10 enzymatic activity conveyed by the EV exchange between stromal and cancer cells.
In this paper, we show that: i) ADAM10 mature form is released in exosome-like vesicles (ExoV) by HL cells and lymph node mesenchymal stromal cells (MSC); ii) fluorescent ADAM10 inhibitors localize in the endolysosomal compartment in HL MSC, are released in ExoV and are in turn endocytosed by bystander cells; iii) ADAM10 sheddase activity carried by ExoV is prevented by the specific inhibitors LT4 and CAM29; iv) further, LT4 and CAM29 reduce CD30 shedding maintaining Brentuximab-Vedotin (Btx Ved) and Iratumumab effects in Hodgkin lymphoma cells.
Results
ADAM10 mature form is released in ExoV by HL cells and lymph node MSC. ADAM10 evaluated in post nuclear supernatants (cells) or in the exosome-like enriched fraction (ExoV) of HL cells, is mainly present as the mature form of about 68kDa after prodomain removal. ExoV were also positive for the specific exosomal marker CD81 and the ADAM10 substrate MICA/B, the latter detectable in L428 and, to a lesser extent, in L540 cells (). The low MW band (35k Da) visible in ExoV was already reported as an ADAM10 cleavage product.Citation28 Transmission electron microscopy (TEM) analysis of L428 or L540 ExoV (, left panels) revealed the prevalence of small vesicles ranging from 50 to 150 nm in diameter, while flow cytometry showed ADAM10 reactivity in L428 ExoV conjugated with latex beads (, right panel). ADAM10 was also detected in MSC and HL cells isolated from the same HL lymph node (LN) (MSC773 and RS773 cells), as well as in their ExoV and conditioned medium (CM) (). ADAM10 expression on both MSC and Reed-Sternberg cells has been detected at the tumor site in HL (Suppl. ).Citation18 Likewise, ADAM10 was detected by western blot (WB) in ExoV purified from the sera of 6 HL patients (Suppl. ).
Figure 1. ADAM10 mature form in ExoV is released by MSC and HL cells. Panel A: Post nuclear supernatants (cells) or exosome-like vesicles (ExoV), prepared from the conditioned medium (CM) of L428, L540 or KMH2 HL cells (107) by differential centrifugation, were lysed, subjected to western blot, probed with the anti-ADAM10 antiserum or the anti-CD81 or the anti-MICA/B mAb, followed by HRP-labeled antibodies and developed with ECL. MW markers (kDa) are indicated on the left. Panel B: Transmission electron microscopy (TEM) analysis of L428 or L540 cell ExoV (left panels). Flow cytometry analysis (right panel) of L428 ExoV conjugated overnight with latex beads (4 μm) and stained with the anti-ADAM10 antibody followed by an APC-conjugated antiserum (grey histogram) or with the APC-antiserum alone (white histogram). Panel C: ExoV and CM from MSC773 or RS773 HL cells were subjected to western blot for ADAM10 as in panel A. Panel D: TEM images of stromal ExoV from MSC773. Panel E: MSC773 cells were silenced with specific (A10) or non-targeting (NT) siRNA pool. Cell lysates, or lysates from ExoV purified from NT or A10 MSC supernatants, were subjected to western blot as in panel A and probed with anti-ADAM10 mAb or mAbs against the exosomal markers CD81 and CD63, as in panel A, or the anti-β-actin mAb. MW markers (kDa) are indicated on the left.
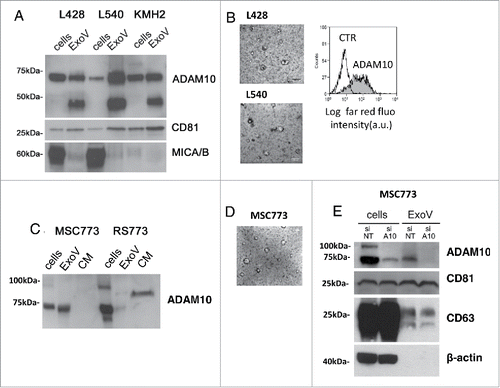
An ADAM10 soluble form (sADAM10) is apparently present in the CM of RS773 cells as a band of MW intermediate between mature ADAM10 and the precursor form (). The evidence that ADAM10, besides initiating intramembrane proteolysis of several substrates, is itself subject to a proteolytic cascade is growing. MSC ExoV were also analyzed upon TEM visualization, as shown in (average dimension: 60% 50–100 nm, 40% 100–200 nm). ADAM10 silencing in MSC773 cells determined a decrease both in cytoplasmic and in ExoV content, confirming the role of stromal ExoV as ADAM10 extracellular carriers. On the other hand, the exosomal markers CD81 and CD63 are unaffected by ADAM10 silencing (). Moreover, ADAM10 silencing in MSC773 resulted in reduced ADAM10 surface expression (Suppl. ) and sMICA shedding (Suppl. ).
Figure 2. ADAM10 inhibitors localize in the endolysosomal compartment in HL MSC. Panel A: Subcellular fractions (L, E, M, SN, as indicated in the text) were obtained from 107 MSC16412 as described in Materials and Methods, either untreated (left blot) or upon 48 h exposure to LT4 (right blot) and Western Blot was performed with the indicated antibodies: anti-ADAM10 ectodomain, anti-LAMP-1, anti-cathepsin D, anti-Rab7 (late endosomes, lysosomes) or anti-EEA1 (endosomes) mAbs, as indicated, followed by secondary anti-mouse HRP-labeled antibodies and ECL development. Panel B: MSC16412 were incubated with CAM29 (from10 µM to 1 µM, light grey histograms) 20 min at RT, washed and stained with the anti-ADAM10 mAb followed by APC-GAM (dark grey histograms) or APC GAM alone (white histograms), washed and run on a FACS. Results are shown as Log far red fluorescence intensity (a.u.) vs number of cells. One representative experiment out of four is shown. Panel C: MSC16412 and MSC 773 were treated with 10 µM LT4 or CAM29 o.n. at 37°C, then RNA was extracted and real-time RT-PCR for ADAM10 performed as described in Materials and Methods. Results are expressed as relative mRNA expression values. Panel D: MSC16412 seeded on 0.2 mm thin round glass slides were incubated for 24 h with Rab5GFP (2 µL) or for 1 h with LysoTracker Red (50 nM); after washes, slides were stained with CAM36 or CAM50 (5 µM for 60 min at RT). Panel E: MSC16412 seeded on glass slides were treated with to 10 µM nocodazole (NZ) for 1 h at 37°C, prior to exposure to CAM36. Samples were washed and analyzed by FV500 FluoView Confocal Laser Scanning Microscope System (Olympus), with PlanApo 40x NA1.00 (E) or 60x NA1.40 (D) oil objectives; the data were analyzed with FluoView 4.3b computer program. Images were taken in sequence mode to avoid cross-talk among fluorochromes and shown in pseudocolor (green or red) or bright field, as indicated.
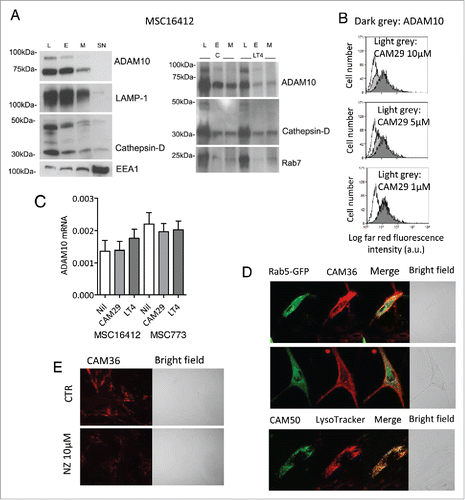
ADAM10 inhibitors localize in the endolysosomal compartment in HL MSC. As shown in (left blot), the analysis of subcellular fractions isolated from MSC16412 showed that the precursor and the mature form of ADAM10 is mainly localized in the endolysosomal compartment (lysosome, L and endosome, E enriched fractions), identified by LAMP-1 and cathepsin-D, or by EEA1, respectively with a detectable amount in the membrane (M) fraction, but not in the cytosol (SN). The presence of EEA1 in the SN due to dissociation from early endosomes (EE) has been described.Citation29 To determine the subcellular distribution of the ADAM10 inhibitors that we reported to be active on HL cells,Citation20,21 we performed a series of experiments using the previously described LT4, the newly synthesized CAM29, both showing a IC50 selective for ADAM10 (Supplemental Data and Suppl.Table 1), or CAM29 conjugated with cyanine 5.5 (Cy5.5, CAM36) or with fluorescein isothiocyanate (FITC, CAM50).Citation30 First, upon exposure to LT4 (), and CAM29 (not shown), ADAM10 time-dependent compartmentalization in endolysosomes increased in MSC16412, possibly interfering with ADAM10 stability due to retention in the degradative pathway while decreasing membrane localization (, right blot). Second, exposure of MSC16412 () or MSC773 (not shown) to CAM29 led to a reduction of surface reactivity of the anti-ADAM10 mAb and this interference was dose-dependent (), thus supporting that the inhibitor induces a conformational change and/or a subcellular redistribution of ADAM10 that becomes unavailable for antibody binding. Conversely, treatment with ADAM10 inhibitors did not alter significantly ADAM10 mRNA levels in either MSC () or L540 and L428 (not shown).
Then, MSC16412 () or MSC773 (not shown) were pre-incubated with either Rab5-GFP, specific for early endosomes (EE), or LysoTracker to identify lysosomes (L) and late endosomes (LE), followed by staining with CAM36, and analyzed by confocal microscopy. First, both membrane and intracellular vesicular distribution of CAM36 can be observed, with limited co-localization with Rab5-GFP, mainly in sub-membrane areas or, at a lesser extent, in perinuclear microvesicles (, upper and middle panels, yellow points in the merged images). On the other hand, MSC16412 () or MSC773 (not shown) pre-incubated with LysoTracker Red and then stained with CAM50, displayed an intense co-localization of the inhibitor in intracellular microvesicles (, lower panels, yellow points in the merge images) that can be defined as the endolysosomal compartment. When MSC16412 were pretreated with nocodazole (NZ), that interferes with microtubule polymerization and endocytosis, a reduction in CAM36 uptake was observed (). These data strongly suggest that the ADAM10 inhibitors bind to the enzyme and mostly share its localization, both at the cell membrane and in the subcellular trafficking compartments.
ADAM10 inhibitors are released in ExoV and endocytosed by bystander cells. To further evaluate the localization of ADAM10 inhibitors, we performed transmission electron microscopy (TEM) on MSC16412 exposed for 24 h to the gold-conjugated CAM29 (CAM49) compound.Citation30 As shown in , CAM49 was detectable in microvesicles, one appearing as a blebbing structure (, quadrant in a enlarged in b), indicative of membrane internalization by endocytosis.Citation31 Thus, we set up a series of experiments using ExoV purified from conditioned medium of MSC or HL cell lines.
Figure 3. ADAM10 inhibitors are released in ExoV and endocytosed by bystander cells. Panel A: a) TEM image of MSC16412 showing the nucleus (n), the endoplasmic reticulum (er), vacuoles (v) and lysosomes (l) and, close to the cytoplasmic membrane, microvesicles (mv) with gold aggregates. b) Magnification of the quadrant in a). Bar = 500 nm. Panel B: L428 cells, incubated with Rab5-GFP 24 h before addition of ExoV obtained from CAM36-treated MSC16412, analyzed by confocal microscopy after 4 h; arrows indicate areas of co-localization (yellow dots) of ExoV-CAM36 (red) with Rab5-GFP (green). Panel C and D: MSC16412 seeded on 0.2 mm thin round glass slides were incubated for 24 h with Rab5-GFP (2 µL, panel C) or for 1 h with LysoTracker DND99 (50 nM, panel D) together with Syto16, 1 µM; then ExoV obtained from CAM36-treated L428 were added for 4 h and samples analyzed by confocal microscopy (as indicated). Samples were observed with PlanApo 40x NA1.00 (B,C, D upper) or 60x NA1.40 (D lower, enlargement of D upper) oil objectives and data analyzed with FluoView 4.3b computer program (Olympus). Results are shown as bright field or pseudocolor images. Lower row in panel D: enlargement of the square in the upper row. Arrows: colocalization areas.
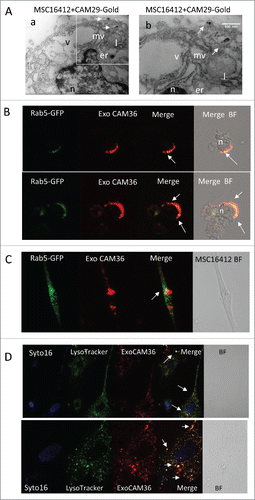
ExoV obtained from MSC16412 ( and ) or MSC773 (not shown), either untreated or exposed to CAM36 for 24 h, were first conjugated with latex beads and analyzed by flow cytometry (). ExoV derived from MSC16412 exposed to CAM36 (Cy5.5) were detected as far-red fluorescent events, compared to ExoV from untreated MSC16412 (, upper quadrant, dark grey vs light grey histograms); moreover, ExoV from untreated MSC16412 expressed ADAM10, as evidenced by the specific mAb (, lower quadrant, dark grey histogram), thus indicating that ADAM10 inhibitors localize in ExoV where ADAM10 is detectable as well. In other experiments, MSC16412-derived ExoV were added (15 µg/105cells) for 24 h to L428 cells: of note, ExoV from CAM36-treated MSC16412 were endocytosed by L428 cells, as detectable by flow cytometry (, upper quadrant, black vs light grey histograms) and by confocal microscopy (); in particular, areas of co-localization (yellow dots) of ExoV-CAM36 (red) with Rab5-GFP (green) are present in L428 cells. Likewise, ExoV from CAM36-treated L428 cells were endocytosed by MSC16412 (15 µg/105cells) as evidenced by flow cytometry (, lower quadrant, black vs light grey histograms) and by confocal microscopy ( and ). shows that ExoV-CAM36 (red) are partially co-localized (yellow dots) with Rab5-GFP (green) that identifies early endosomes; moreover, in , colocalization (yellow) with the lysosomal compartment marker LysoTracker (green) is depicted. The dark grey histogram in , upper quadrant, indicates L428 cells incubated with ExoV from MSC16412 exposed to the unrelated Cy5.5 compound ST178.
Figure 4. ADAM10 inhibitors are released in ExoV and endocytosed by bystander cells: effects on sMICA and TNFα shedding. Panel A: Upper quadrant: ExoV, from MSC16412 exposed to CAM36 (Cy5.5) for 24 h were conjugated with latex beads (dark grey histograms) compared with beads-conjugated ExoV from untreated MSC16412 (light grey histogram as in upper quadrant; white: beads alone). Lower quadrant: ExoV, obtained from untreated MSC16412 were conjugated with latex beads (light grey histograms) and stained with anti-ADAM10 mAb followed by APC-GAM (dark grey histogram; white: latex beads plus APC-GAM). Panel B: Upper quadrant: L428 cells added of ExoV obtained from untreated (light grey histogram) or CAM36-treated MSC16412 (black histogram; white histogram: L428 without ExoV) for 24 h. Dark grey histograms: L428 cells incubated with ExoV (15 µg/106cells) from MSC16412 exposed to the unrelated Cy5.5 compound ST178. Lower quadrant: ExoV (15 µg/106cells) from untreated (light grey histogram) or CAM36-treated (black histogram) L428 cells were added for 24 h to MSC16412 (white histogram: MSC16412 without ExoV). Panels A and B: samples were run on a CyAn ADP Analyzer and results, representative of four experiments, are expressed as Log far red fluorescence intensity (a.u.) vs number of cells. Panel C: sMICA (pg/ml/105 cells) in the SN of MSC773 cultured for 48 h without or with ExoV (15, 30 or 60 µg for 48 h) from L428 (black circles) or L540 (white circles) HL cell lines. Panel D: sMICA (pg/ml/105 cells) in the SN of MSC773 before or after the addition of ExoV (10 or 30 µg for 48 h) from L428 untreated or pretreated with 10 µM LT4, as indicated. ExoV30 µg RPMI: sMICA content in the SN of ExoV cultured alone. Panel E: TNFα content (pg/ml/105 cells) in the SN of L428 cells cultured for 48 h without or with ExoV (30 µg) released by MSC773, untreated or pretreated with 10 µM LT4 (left histograms) or 10 µM CAM29 (right histograms). Some samples were prepared using L428 cells exposed for 24 h to either 10 µM LT4 (left histograms) or 10 µM CAM29 (right histograms), before addition of MSC-derived ExoV. •p< 0.05, ••p<0.01, •••p<0.001 vs Nil; ###p<0.001 vs ExoV C; ***p<0.001 vs Nil in L428 cells unexposed to CAM29.
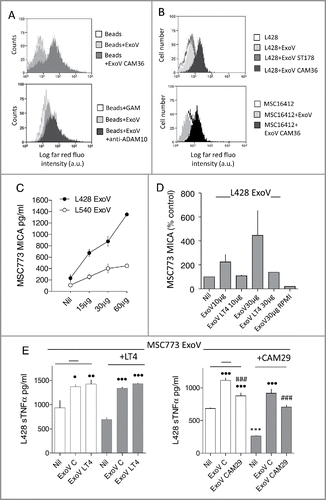
ADAM10 in ExoV from MSC and HL cells is active and inhibited by LT4 or CAM29. To evaluate the ADAM10 activity carried by ExoV and verify the blocking by specific inhibitors, we used ExoV purified from L428 or L540 cells or MSC773, untreated or cultured in the presence of LT4 or CAM29 (10 µM) for 48 h. MICA was chosen as substrate for ADAM10 in MSC because these cells do not express CD30 (,) and do not produce TNFα.Citation32 CD30 (see below) and TNFα were chosen for HL cell lines as they are expressed by these cells (,) and are substrates for ADAM10 and/or ADAM17.Citation33,Citation34
Figure 5. ExoV contribute to CD30 shedding and are inhibited by LT4 or CAM29. Panels A and B: L428 cells or MSC16412 (A) or latex beads-conjugated ExoV from L428 or MSC16412 (B), were stained with the anti-CD30 mAb followed by PE-GAM (grey histograms) or PE-GAM alone (white histograms), washed and run on a FACS. Results are shown as Log red fluorescence intensity (a.u.) vs number of cells. Panel C: sCD30 (pg/ml/105 cells) in the SN of L428 cells cultured for 48 h without (Nil) or with ExoV (30 µg) from MSC16412 untreated (ExoV C) or treated with 10 µM LT4 (ExoV LT4) or 10 µM CAM29 (ExoV CAM29), as indicated. Some samples were prepared using L428 cells exposed for 24 h to 10 µM LT4 (as indicated in the left histogram), before addition of MSC-derived ExoV. *p<0.05 ExoV C vs Nil; #p<0.05 ExoV LT4 vs ExoV C. Panel D: sCD30 (pg/ml/105 cells) in the SN of MSC16412 before (Nil) or after the addition of ExoV (50 or 100 µg for 48 h) from L428 untreated or pretreated with 10 µM LT4, as indicated. In some samples MSC16412 were exposed for 24 h to 10 µM LT4, as indicated, before addition of MSC-derived ExoV. •••p<0.01 vs Nil; ***p<0.001 vs ExoV C matched samples; ##p<0.01, ###p<0.001 vs matched samples without LT4. Panel E: sCD30 content (pg/ml/105 cells) in the SN of L540 cells cultured for 48 h without (Nil) or with 10 µM LT4 or 10 µM CAM29, pretreated or not with nocodazole (Nz), as indicated. Left: *p<0.05, **p<0.01 vs Nil. Right: *p<0.05 Nz vs Nil; **p<0.01 LT4/CAM29 vs Nil: #p<0.05 Nz+LT4 vs LT4;°p<0.05 Nz+LT4 vs Nz;°°p<0.01 Nz+CAM29 vs Nz. Panel F: L540 cells were incubated with LT4 or CAM29 (10 µM, black histograms), as indicated, washed and stained with the anti-CD30 mAb followed by PE-GAM (grey histograms) or PE-GAM alone (white histograms), washed and run on a FACS. Results are shown as Log red fluorescence intensity (a.u.) vs number of cells.
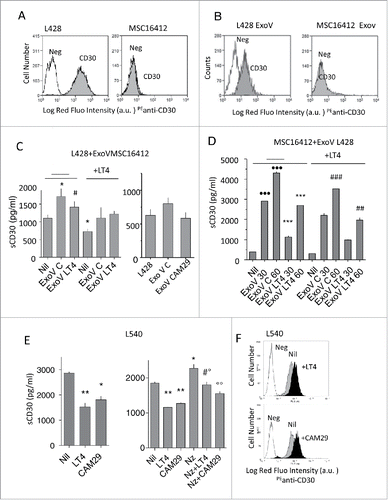
As shown in , ExoV from L428 and, to a lesser extent, those released by L540 cells, were able to increase the shedding of sMICA by MSC773 cells and this effect was dose-dependent. Of note, ExoV released by L428 cells pretreated with the ADAM10 inhibitor LT4 did not enhance sMICA shedding by MSC773 (). In turn, ExoV from untreated MSC773 could increase the shedding of TNFα by L428 cells (, left and right graphs, central white columns). In this case, ExoV from LT4 treated MSC were also effective (, left graph, right white column). On the other hand, ExoV from MSC773 pretreated with CAM29 were less effective in increasing TNFα release ( right graph, right white and grey columns); likewise, the effect of MSC773-derived ExoV on TNFα shedding was lower when L428 cells were pretreated with CAM29 (, right panel, central and right grey columns).
ADAM10 inhibitors reduce the shedding and maintain the expression of CD30 on HL cells increasing the biological effect of the ADC Btx Ved and the humanized anti-CD30 Iratumumab. As CD30 is a reported substrate for ADAM10 and is also a target for immunotherapy,Citation19,Citation32,Citation34,Citation35 we first analyzed the effect of ExoV on CD30 shedding from L540 and L428 cells and the possible counteracting action of ADAM10 inhibitors. In and expression of CD30 on L428 cells and their ExoV is shown, compared to MSC16412 and related ExoV that are CD30 negative. shows that ExoV from MSC16412 were able to increase the shedding of sCD30 by L428 cells. In turn, ExoV released by MSC16412 pretreated with the ADAM10 inhibitor LT4 or CAM29 (both at 10 µM concentration; left and right graph, respectively) were less effective in enhancing sCD30 shedding by L428. When L428 cells were treated with LT4 before exposure to MSC-derived ExoV, the increase in sCD30 shedding was less evident (, left graph). Noteworthy, ExoV obtained from L428 cells added to MSC16412 resulted in a detectable, dose-dependent sCD30 shedding (), conceivably of vesicular origin, as a result of the interaction between L428-derived ExoV, carrying CD30, and cell membrane ADAM10 on CD30-negative MSC (). It is of interest that CD30 release from ExoV produced by LT4-treated L428 cells was less efficient; also, the exposure of LT4-pretreated MSC to L428-derived ExoV resulted in a lower sCD30 shedding (). Exposure of L540 cells to either LT4 or CAM29 (10 µM for 48 h) could not only reduce sCD30 release (, left), but also enhance CD30 surface expression (). Of note, when endocytosis was inhibited by nocodazole, the ADAM10 inhibitors effectively reduced the expected increase in CD30 shedding due to prolonged permanence of ADAM10 and its substrate at the level of the cell membrane (, right). A similar effect was detectable using ExoV from MSC silenced for ADAM10 (Suppl. , for L540 and L428, and D for L540).
We next verified whether the two ADAM10 inhibitors could help to maintain the anti-tumor effect of the anti-CD30 antibody-drug conjugate (ADC) Btx Ved. First, L428, L540 and KMH2 cell lines were exposed to serial dilution of BtxVed (10 to 0.01 µg/ml) and the variation in the intracellular ATP content, as a parameter of cell viability, was evaluated at 72 h and 96 h. Suppl. shows that L540 was the most sensitive cell type to BtxVed, with a reduction of ATP content detectable with a lower drug concentration (>50% at 1 µg/ml after 72 h; in Suppl. , data are reported as number of cells/ml at the same time points, thus indicating that the decrease in ATP intracellular content parallels a reduction in the number of cells). To determine whether ADAM10 inhibitors can enhance BtxVed effect, L428, L540 and KMH2 cells were exposed for 12 h to 10 µM LT4 or CAM29 and cultured in the presence (or absence) of BtxVed (10 to 0.01 µg/ml). As shown in , both LT4 and CAM29 reduced the ATP intracellular content by 15% in either HL cell line after 72 h of culture. Of note, the two ADAM10 inhibitors produced an increase of about 25% in the efficiency of BtxVed to reduce ATP content in L428 and L540 cells (, central and right panels); this effect was more evident on KMH2 cell line, where the effect of BtxVed was almost doubled even at the lowest ADC concentration (, left panel). Apoptosis was then evaluated in parallel samples of HL cell lines, exposed to serial dilution of BtxVed alone, as above, or after exposure to 10 µM LT4 or CAM29, by staining with annexin V (AV) and propidium iodide (PI), followed by FACS analysis. Apoptotic cells were identified as AV+PI+ and AV+PI−, while necrotic cells as AV− PI+. LT4 was able to increase significantly the pro-apoptotic effect of BtxVed at low doses (0.1 or 1 µg/ml) on KMH2 cells (); on the other hand, the effect on L428 and L540 cell lines was observed as an increase in necrotic cells percentage upon treatment with 0.1 or 1 µg/ml BtxVed ().
Figure 6. Effect of ADAM10 inhibitors on HL cell proliferation and BtxVed biological activity. Panel A: L428, KMH2 and L540 cells, either untreated or pretreated for 12 h with 10 µM LT4 or 10 µM CAM29, as indicated, were cultured for 72 h in the presence of BtxVed (10 to 0.1 µg/ml). Then intracellular ATP content was measured with the CellTiter-Glo(R) Luminescent Cell Viability Assay. Results are expressed as arbitrary units of luminescence x103 and are mean ± SD from 3 independent experiments. *p<0.001 vs Nil (BtxVed alone). Panel B and C: KMH2, L428 and L540 cells cultured as in panel A were stained with annexin V (AV) and propidium iodide (PI), followed by FACS analysis. Results are expressed as percentage of apoptotic cells (panel B) identified as AV+PI+ and AV+PI− or percentage of necrotic cells (panel C) as AV− PI+ and are mean with SD from 3 experiments. *p<0.001 vs Nil (BtxVed alone). Panel D: cytolytic activity exerted by PBMC, isolated from 6 different donors, against KMH2, L428 and L540 cells, labeled with BATDA, at the E:T ratio of 40:1 in the presence or absence of the anti-CD30 human antibody Iratumumab (Ira, 0.1 µg/ml). After 2 h the fluorescence emitted by lysed cells was analyzed by the time-resolved fluorometer VICTOR2 (PerkinElmer). Results are expressed as percentage of specific release.
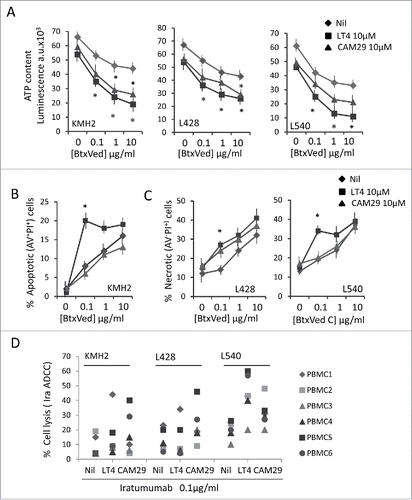
In another series of experiments, we assessed the potential ability of ADAM10 inhibitors to enhance the antibody-dependent cellular cytotoxicity (ADCC) of HL cells. To this aim, we used the human therapeutic anti-CD30 mAb Iratumumab (Ira), also known as MDX-060,Citation35 at the lowest dose (0.1 µg/ml) displaying a minimum activity, based on our preliminary results and reported data,Citation35,Citation36 in an ADCC assay against KMH2, L428 and L540 HL cell lines with peripheral blood mononuclear cells (PBMC) as effectors. As shown in , pre-treatment of HL target cells with either LT4 or CAM29 led to an increase in the Ira-induced ADCC (Nil in the figure, cell lysis in the absence of Ira <5%, not shown). This was evident for all PBMC donors against L540, with higher efficacy of LT4 (: mean with Ira only 20%, with LT4 44%, with CAM29 31%); in the case of KMH2 and L428 donor 1 was more responsive to LT4-treated cells (PBMC1) and donor 5 and 6 (PBMC5, PBMC6) to CAM29-treated HL cells ().
Thus, ADAM10 specific inhibitors are able to decrease the release of sCD30, increase its surface expression and maintain, or increase, the biological effect of the anti-CD30 ADC BtxVed or the human anti-CD30 Iratumumab on HL cells.
Discussion
In this paper, we show that: i) ADAM10 mature form is released in exosome-like vesicles (ExoV) by HL cells and lymph node mesenchymal stromal cells (MSC); ii) fluorescent ADAM10 inhibitors localize in the endolysosomal compartment in HL MSC, are released in ExoV and are in turn endocytosed by bystander cells; iii) ADAM10 sheddase activity carried by ExoV is prevented by the specific inhibitors LT4 and CAM29; iv) in particular LT4 and CAM29 reduce CD30 shedding enhancing the anti-tumor BtxVed effects and Ira-mediated ADCC on Hodgkin lymphoma cells.
First, we report that the bioactive mature form of ADAM10 can be released by HL cells and lymph-node (LN) MSC in extracellular microvesicles enriched in exosome-like particles (ExoV, 50 to 200 nm of size); according to the literature, these microvesicles are potentially able to spread the ADAM10 enzymatic activity in the microenvironment.Citation22,Citation23 Of note, we demonstrate that specific ADAM10 inhibitors, that we reported to block cellular ADAM10 and increase the sensitivity of HL cells to lymphocyte-mediated killing,Citation20,Citation21 can also bind to exosomal ADAM10 and interfere with its enzymatic function. In particular, we show that the fluorescent ADAM10 inhibitors CAM50 and CAM36, localize in endolysosomes in HL LN MSC and bind to the enzyme, both at the cell membrane and in the subcellular compartments. The unlabeled compound CAM29, showing the same structure of CAM36 and CAM50 without fluorochromes, can both prevent the recognition of ADAM10 by a specific antibody at the surface of MSC and cross-inhibit the binding of CAM36 and CAM50. Of note, these inhibitors can also cause the retention of ADAM10 in the endolysosomal compartment, leading to enhanced degradation, thus reducing its membrane localization.
Using fluorescent CAM36, we also demonstrated that ADAM10 inhibitors are released in ExoV, either by HL cells or MSC, and are in turn endocytosed by bystander MSC or HL cells; this would imply that it is possible to interfere with ADAM10 enzymatic activity conveyed by the EV exchange between stromal and cancer cells. Indeed, in our experiments, co-culture of MSC with ExoV released by HL cells resulted in an increase of MICA shedding; however, ExoV collected from HL cells that were pretreated with LT4, were not effective. When ExoV obtained from MSC were added to HL cells, the release of TNFα raised, unless either the ExoV-producing MSC or the ExoV-recipient HL cells were pretreated with ADAM10 inhibitors. In this case CAM29 was much more efficient than LT4; this is possibly due to the conserved ability of CAM29 to inhibit also ADAM17 (Suppl. Table 1), that is reported as the major TNFα converting enzyme.Citation8,Citation21 The different sensitivity of the HL cell lines to LT4 or CAM29 (L428 being more sensitive to CAM29) might also be explained on the basis of a differential expression and intracellular trafficking of tetraspannins as transporters of ADAM10 in the different HL cell lines (this hypothesis is currently under investigation).Citation37 Another hypothesis is that the kinetics of LT4-ExoV fusion with the plasma membrane is more rapid than CAM29-ExoV, due the steric hindrance of CAM29 and a possible different conformational change of vesicular ADAM10 that favors its permanence on the plasma membrane, thus allowing more visible inhibition.
As CD30 is a substrate for ADAM10 and also a target for immunotherapy,Citation19,Citation33-36 we analyzed the effect of ExoV on CD30 shedding from HL cells and investigated the ability of ADAM10 inhibitors to counteract this process. ExoV released by MSC could indeed enhance sCD30 shedding by HL cells and this process was prevented by pretreating with ADAM10 inhibitors either HL cells before co-incubation with ExoV, or MSC during the release of ExoV. It is of interest that a dose-dependent sCD30 increase in culture media (CM) was detected also when ExoV obtained from HL cells were added to MSC. As ExoV bind to the cell membrane and are internalized by MSC, sCD30 shedding is conceivably a result of the interaction between HL-derived ExoV, carrying CD30, and ADAM10 on MSC cell-membrane, in keeping with other reports.Citation33 This is further supported by the finding that HL-derived ExoV were much less effective on MSC pretreated with the ADAM10 inhibitor LT4. Of note, we also show that the two ADAM10 inhibitors, besides showing themselves a partial inhibitory effect on HL cell proliferation, can help to maintain and enhance the anti-tumor effect of the anti-CD30 ADC BtxVed and the Iratumumab-mediated ADCC of HL target cells ().
Figure 7. Proposed model for the role of ADAM10 inhibitors in maintaining BtxVed and Iratumumab biological effect. Panel A: HL malignant cells, expressing CD30, in the lymphoid tissue are in contact with MSC. Shedding of sCD30 as a consequence of ADAM10 enzymatic activity, either present at the surface of HL or MSC, or carried by ExoV, leads to decreased membrane-associated CD30 expression and impairment of BtxVed or Iratumumab (Ira) binding to HL cells. Panel B: LT4 and CAM29, either inside HL cells or MSC, or carried by ExoV, would inhibit cellular or exosomal ADAM10, reducing sCD30 release, increasing CD30 surface expression and allowing BtxVed or Ira recognition of HL cells and anti-tumor biological effect.
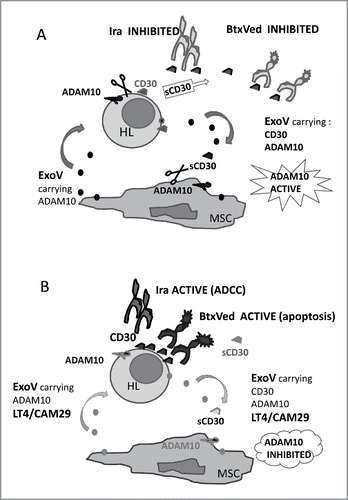
Thus, the cross-talk occurring between stromal and cancer cells in the HL microenvironment, due to exosome-like microvesicles and ADAM10 activity, can result in the release of cytokines, as TNFα, described as lymphoma growth factor,Citation34 or soluble molecules, such as sMICA or sCD30, that potentially interfere with host immune response, or with ADC-based immunotherapy.Citation14-18,Citation33 ADAM10 blockers can interfere with such process, allowing the development of an anti-lymphoma immune response and/or an efficient ADC-based or humanized Ab-immunotherapy ().Citation19-21,Citation33-36,Citation38
Methods
Cell cultures. The HL cell line L428 or KMH2, from pleural effusion, and L540, from bone marrow of HL patients, (DSMZ GmbH, Braunschweig, Germany), and the RS773 cell line obtained from a HL lymph node (LN) were maintained in RPMI 1640 complete medium supplemented with 10% FCS and glutamine (2 mM).Citation18 The LN MSC773 and MSC16412 were cultured in MEM-α (GIBCO) supplemented with 1% Chang medium, glutamine (2 mM), 1% penicillin/streptomycin. Stabilization of primary HL and MSC cultures was performed as described.Citation18,Citation20
ADAM10 inhibitors. LT4 was synthesized as described,Citation21 and CAM29, showing a IC50 selective for ADAM10 (Supplemental Data and Suppl.Table 1), or CAM29 conjugated with cyanine 5.5 (Cy5.5, CAM36) or with fluorescein-isothiocyanate (FITC, CAM50), were newly synthesized.Citation30 As reported in our previous publication,Citation21 LT4 is a sulfonamido-based hydroxamate that inhibits ADAM10 by interacting with its catalytic domain by chelating the zinc ion, critical for both substrate binding and cleavage. CAM29 is its PEGylated analogue used to prepare the fluorescent probes and the CAM49 gold nanoprobe prepared by conjugating multivalent gold nanoparticles with CAM29.Citation30 Compound ST178, a Cy5.5-N-isopropyl amide unable to bind ADAM10, was used as a negative control for the fluorescent probes.
The ADAM10 inhibitors used on the different HL cell lines, or on isolated MSC, at 2.5 up to 10 μM concentration for 48, 72 and 96 h (and the solvent DMSO at the same dilutions and time points) did not exert any toxic effect, as assessed by evaluating the mitochondrial potential upon staining with the dual emission fluorescent probe JC-1 (Molecular Probes, Life Technologies Italia, Monza) and did not induce directly necrosis or apoptosis, evaluated by annexin V/ PI staining (not shown). MMP and ADAMs inhibition assays were performed as described (Suppl. Data).Citation39,Citation40
Small interfering RNA (siRNA) transfection and real-time RT-PCR. ON-TARGET plus SMART pool for human ADAM10 (100 nM, Dharmacon CarloErba, Milan, Italy) was used to knockdown the expression of ADAM10; siCONTROL non-targeting (NT) siRNA pool (Dharmacon) was used as negative control. MSC773 (2.5 × 105) were transfected in serum-free Opti-MEM medium (Gibco, Life Technologies) with Lipofectamine 2000 (Invitrogen, Life Technologies) following the manufacturer's instructions. ADAM10 expression by western blot was analyzed 72 h after transfection in MSC773 and in the ExoV released by the same cells, purified as described below.
MSC773 cells, untreated or silenced, were then stained with the anti-ADAM10 mAb, followed by isotype-specific APC-GAM and run on a CyAn ADP Analyzer. In some experiments, ExoV from MSC773, untreated or silenced with ADAM10 siRNA or with NT siRNA pool were added to L540 or L428 cells (10 µg/105 cells) for 24 h, then SN were collected for sCD30 ELISA and cells were stained with the anti-CD30 mAb, followed by APC-GAM, and run on a CyAn ADP Analyzer.
Total RNA was isolated from controls and treated cells using the RNeasy Mini kit (Qiagen Italia, Milan) according to the manufacturer's instructions. mRNA expression was analyzed by quantitative real-time reverse transcription-PCR, by using the following specific primers: ADAM10 sense 5′-ATG TTT TCA TGC GGT GCA GAT and antisense 5′-GTA ATA CTG CCC ACC AAT GAG C; 18 S sense 5′-ACACGGACAGGATTGACAGATT and antisense 5′-AGACAAATCGCTCCACCAACTA. cDNA amplification and relative expression values were obtained as described.Citation41
ELISA for sMICA, TNFα and sCD30. Conditioned culture media (CM) were collected from cell cultures (either MSC or HL cell lines), untreated or after 48 h exposure to the ADAM10 inhibitors (LT4, CAM29, 10 μM). In some experiments, the release of ADAM10 substrates by MSC773 cells, untreated (WT), or silenced with ADAM10 siRNA, or with NT siRNA pool for 48 h was measured in the CM by ELISA.
The anti-MICA mAbs AMO1 and BAMO3 were from Immatics Biotechnologies (Tubingen, Germany) and the ELISA detection kit for sCD30 (Human CD30 Picokine ELISA kit) from Boster Bio (TebuBio, Milan, Italy). Anti-mouse IgG2 a HRP was from Southern Biotechnology (Birmingham, AL). Plates were developed with 3,3′,5,5′-tetramethylbenzidine (Sigma-Aldrich, Milan, Italy) and read at a OD450nm. Results are expressed as pg/ml and referred to a standard curve obtained with the MICA/Fc chimera (R&D System) or the standard CD30 contained in the specific kit. TNFα was measured after treatment for 1 h of SN with 1 N HCl followed by 1 N NaOH with a TNFα specific kit (PeproTech, London, UK). Results normalized to a standard curve are expressed as pg/ml.
Cell proliferation and evaluation of apoptosis/necrosis. Cell proliferation was assessed at 72 or 96 h, upon treatment of KMH2, L428 or L540 cell lines with LT4 or CAM29 (10 μM) or with BtxVed (10 to 0.01 µg/ml), provided by the Pharmacy Service of our Institute, by measuring the intracellular ATP content with the CellTiter-Glo(R) Luminescent Cell Viability Assay (Promega Italia, Milan, Italy) according to the manufacturer's instruction. In some experiments, HL cell lines were exposed for 12 h to 10 µM LT4 or 10 µM CAM29, prior to BtxVed treatment. Results are expressed as arbitrary units (a.u.) of luminescence. Parallel samples were used for cell count evaluation by FACS. Apoptosis was evaluated in HL cell lines, treated with serial dilution of BtxVed alone, as above, or after 12 h exposure to 10 µM LT4 or CAM29, by staining intact cells with annexin-V (AV) and propidium iodide (PI), (Sigma-Aldrich), followed by FACS analysis. Necrotic cells were identified as AV− PI+, apoptotic cells as AV+PI+ and AV+PI−, living cells as AV−PI−.
ADCC assay. The DELFIA EuTDA cytotoxicity reagents have been used (PerkinElmer, Wellesley, MA, USA). KMH2, L428 or L540 cell lines, untreated or exposed for 12 h to 10 µM LT4 or CAM29, were labelled with bis-acetoxymethyl 2,2 ′,6 ′,2“-terpyridine-6,6”-dicarboxylate (BATDA) for 20 min at 37°C, washed and co-incubated in 96-well round bottom plates with PBMC, isolated from 6 different buffy coats, at the E:T ratio of 40:1 in the presence or absence of the anti-CD30 human antibody Iratumumab (Ira, Creative Biolabs, Shirley, NY, USA), at the lowest dose (0.1 µg/ml) displaying a minimum activity, based on our preliminary results and reported data.Citation35,Citation36 After 2 h, 20 µl of supernatant from each well were transferred to flat-bottomed plates and 200 µl of Europium solution was added. The fluorescence emitted by the binding of Europium with the ligand TDA was analyzed by the time-resolved fluorometer VICTOR2 (PerkinElmer). The percentage of specific release was calculated applying the following formula: experimental release (counts)-spontaneous release (counts)/maximum release (counts)-spontaneous release (counts). Maximum and spontaneous release were calculated according to the manufacture's instruction.
Subcellular fractionation. Lysosomal, endosomal and membrane fractions were isolated by differential centrifugation following published proceduresCitation42,Citation43 with slight modifications (see Supplemental Data). Briefly, cell pellets obtained from 107 MSC16142 or MSC773 were washed in PBS, suspended in 1 ml homogenization buffer (250 mM sucrose, 0.5 mM EGTA), 20 mM Hepes-KOH (pH7) and passed sequentially through 21G1/2 and 25G needles. The homogenate was centrifuged 10 min at 1000 xg (nuclear fraction). Post-nuclear supernatants (cells) were centrifuged in an Eppendorf 5417 R refrigerated minicentrifuge at 8000 xg for 20 min (L: lysosome-enriched fraction); supernatants were further ultracentrifuged in TL-100 ultracentrifuge 10 min at 50000 xg (E: endosome-enriched fraction), 90 min at 100000 xg (M: membrane-enriched fraction) and the last recovered as supernatant (SN). All fractions were suspended in RIPA buffer with protease inhibitors (Sigma-Aldrich), stored at −20°C, or processed for western blot analysis. Protein content was assessed by the Lowry DC protein Assay (BioRad, Wadfirt, UK).
Western Blot. Total cell lysates were prepared in RIPA buffer supplemented with 1 mM orthovanadate. Protein quantification was performed with the DC Protein Assay (Bio-Rad). Samples (10–20 µg of proteins) of either cell lysates or subcellular fractions, were resolved on 4–20% gradient SDS-polyacrylamide Tris-Glycine gels and transferred on PVDF membranes (GE-Healthcare, Milan, Italy). Western Blot analysis was performed with the following antibodies: mouse monoclonal (mAb) anti-ADAM10 ectodomain (non-reducing conditions) clone 163003, R&D Systems (Milano, Italy); rabbit polyclonal anti ADAM10 ab1997, Abcam (Cambridge, UK); anti-MICA/B mAb (BAMO1, BAMOMAB GmbH, Gräfelfing, Germany); anti-CD81 mAb clone 5A6, or CD63 clone H5C6 (BioLegend, San Diego, CA, USA). For the identification of subcellular fractions by specific markers the following antibodies were used: anti-cathepsin-D (Calbiochem, Merck Millipore, Vimodrone, Milan, Italy); anti-EEA1 (mAb BD Biosciences, Milano, Italy); anti-lysosome-associated membrane glycoprotein-1 (LAMP-1) 1D4B (Developmental Studies Hybridoma Bank, Iowa City, Iowa) anti-Rab7 (B-3, Santa Cruz Biotechnology, Heilderberg, Germany); secondary anti-mouse and anti-rabbit horseradish peroxidase (HRP)-labeled antibodies were from Cell Signaling Technology (Danvers, MA). Immobilon™ Western Chemiluminescent HRP Substrate (ECL) was from Merck Millipore.
Immunohistochemistry. Paraffin-embedded samples from 10 HL patients, obtained from the Unit of Pathology, IRCCS Policlinico San Martino, Genoa, under conventional diagnostic procedures, provided informed consent and approval by the institutional ethical committee (IRB approval 0026910/07, renewal 03/2009, and 14/09/15). were analyzed (4 are shown) for the expression in situ of ADAM10, CD30 and transglutaminase (TG)II. Immunohistochemistry was performed on 6-μm-thin sections, treated with Peroxo-Blok (Novex, Life Technologies) to quench endogenous peroxidase, followed by Ultra Blok reagent (Ultravision Detection System, BioOptica, Milan Italy). The following antibodies were added: polyclonal rabbit anti-ADAM10 antiserum (1:100, Abcam), anti-CD30 mAb (2 μg/mL), polyclonal rabbit anti-TGII antiserum (1:100, Thermo Scientific) and an isotypic unrelated antibody was used as negative control (Dako Cytomation). Biotinylated goat anti-rabbit or goat anti-mouse antiserum (BioOptica) was then added, followed by HRP-conjugated avidin (Thermo Scientific) and the reaction developed using 3,3′-diaminobenzidine (DAB) as chromogen. Slides were counterstained with hematoxylin, cover-slipped with Eukitt (Bio Optica), and analyzed under a Leica DM MB2 microscope equipped with a charged coupled device (CCD) camera (Olympus DP70 with a 20x or 40x objective).
ExoV isolation from MSC or HL cell lines and HL patients' sera. L428 or L540 HL cell lines or MSC16412 and MSC773 were cultured in serum-free medium for 48 h; then ExoV were isolated following published procedures,Citation44 with slight modifications described in detail in the Supplemental Data section. Briefly, the conditioned medium (CM) from T175 flasks containing 90% confluent MSC (2.8 × 107) or from HL cells (4 × 107), was centrifuged at 300 xg for 15 min and 2000 xg for 20 min to remove live and dead cells, and cell debries, respectively. The supernatant was concentrated 10-fold and dialyzed against PBS with Amicon Ultra-4 or Ultra-15 devices (NMWL 100 kDa) (Merck Millipore) at 3000 xg, centrifuged at 20000 xg for 20 min and at 100000 xg for 120 min in a TL-100 Ultracentrifuge equipped with TLA-100.3 rotor (Beckman Coulter, Milan, Italy). The pellet containing purified ExoV was washed with PBS by centrifugation at 100000 xg, to remove contaminating protein aggregates, and suspended in RIPA buffer with protease inhibitors for western blot, or in PBS for cell cultures or transmission electron microscopy. ExoV, purified from 6 HL patients' sera (IRB as above), following the same procedure, were subjected to WB for ADAM10 and CD81 as described above. Protein concentration was measured by the DC Protein assay. ExoV size and concentration was verified by as described in Supplemental Methods.
Latex beads conjugation of ExoV and FACS analysis. Aldehyde sulfate latex beads (Life Technologies) were washed with 1 µM MES buffer pH 6.0, collected by centrifugation (3000 xg, 20 min), mixed (3 × 105 bead/sample) with 15, 30 or 60 µg of ExoV, purified from MSC 16412 of L428 cells and incubated overnight at RT in a vertical tube rotator. The reaction was stopped with PBS-Glycine (final concentration 100 mM) and beads were washed with PBS, 3% FCS, 0.01% NaN3, incubated with anti-ADAM10 antibody for 30 min at 4°C, washed and stained with APC-conjugated secondary antibody. 100000 beads were analyzed on a CyAn ADP Analyzer (Beckman Coulter, Milano, Italy) set according to Thery et al. (Suppl. Data and Suppl. ).Citation42 In other experiments, 2.5 × 104 MSC16412 or MSC773 were pre-incubated with CAM29 (10 µM or 1 µM) 20 min at RT, washed, stained with the anti-ADAM10 ectodomain mAb followed by APC-conjugated goat anti-mouse (GAM) anti-serum and run on a CyAn ADP Analyzer (Beckman Coulter). Results are reported as Log red fluorescence intensity (arbitrary units, a.u.) vs cell number. In some experiments ExoV, obtained from MSC16412 or MSC773 exposed to CAM36 for 24 h, were coupled to latex beads and analyzed by a CyAn ADP Analyzer (see Supplemental Data). Other ExoV samples, and the relative producing L428, L540 or MSC16412 cells, were stained with the anti-CD30 mAb followed by PE-GAM (grey histograms) or PE-GAM alone (white histograms), and analyzed by FACS.
Co-culture of MSC or HL cells with ExoV. For functional experiments, 10, 15, 30 or 60 µg of ExoV obtained from HL cell lines L428 or L540, either untreated or exposed to 10 µM LT4 or CAM29 for 24 h, were added to sub-confluent MSC773 or MSC16412 seeded in flat bottom 96 w plates and placed at 37°C in a 5%CO2 incubator; in some samples, MSC773 or MSC16412 were exposed to 10 µM LT4 or CAM29 for 24 h before adding L428 or L540-derived ExoV. In other experiments, 30 µg of ExoV produced by MSC773 or MSC16412, either untreated or pretreated for 24 h with 10 µM LT4 or CAM29, were added to L428 or L540 cells (106/ml) in 96 w or 24 w plates and incubated at 37°C; in some samples, L428 or L540 cells were exposed to 10 µM LT4 or CAM29 for 24 h before addition of MSC-derived ExoV. After 48 h of culture, CM were collected, centrifuged at 100000 xg for 90 min to remove ExoV and analyzed for sMICA, sCD30 or TNFα by ELISA. Another series of experiments were performed with ExoV, obtained from MSC16412 exposed to CAM36, or the unrelated Cy5.5 compound ST178, for 24 h; these ExoV were either conjugated with latex beads (see below) or added (15 µg/105cells) for 2 h to L428 cells previously seeded on 0.2 mm thin round glass slides and cultured for 24 h. In turn, ExoV from CAM36-treated (24 h) L428 were added to MSC16412 previously seeded on glass slides as above; then all samples were analyzed by either FACS analysis, after detachment from the slides, or confocal microscopy (see below).
Confocal Microscopy. 2.5 × 104 MSC (either MSC16412 or MSC773) seeded on 0.2 mm thin round glass slides were incubated for 24 h with Rab5GFP (CellLight Reagents BacMam 2.0, Thermo Fisher, 2 µL) or 1 h with LysoTracker DND99 (Thermo Fisher, 50 nM). After extensive washes, slides were stained with CAM36 or CAM50 at 5 µM for 60 min at RT. Some samples were treated with 10 µM nocodazole (Sigma-Aldrich) for 1 h at 37°C prior to exposure to CAM36. In other experiments, L428 or MSC16412, labelled with either Rab5-GFP or LysoTracker DND99, were exposed to CAM36-ExoV (15μg/105 cells) as described above and incubated 4 h (to optimize images) at 37°C before performing confocal microscopy. Samples were then analyzed by FV500 (FluoView confocal Laser Scanning Microscope System, Olympus Europe GMBH, Hamburg, Germany) equipped with an Argon laser to excite carboxyfluoscein and a He-Neon red laser at 633 nm to excite cyanine 5 dye associated to a IX81 motorized microscope (Olympus). Samples were observed with PlanApo 40x NA1.00 or 60x NA1.40 oil objectives and data analyzed with FluoView 4.3b computer program (Olympus). Each image has been taken in sequence mode to avoid cross-contribution of each fluorochrome. Results are shown as bright field or pseudocolor images.
Transmission electron microscopy. ExoV-containing pellet (50–100μg of protein) was resuspended in 50 to 100 μl of 2% PFA and 5 μl were deposited on Formvar-carbon coated EM grids for 20 min, then transferred to a 50-μl drop of 1% glutaraldehyde for 5 min. Grids were washed 8 times with 100μl drop of distilled water for 2 min and transferred to a 50μl drop of uranyl-oxalate solution, pH 7, for 5 min. Samples were then contrasted in a solution of uranyl oxalate, pH 7, embedded in a mixture of 4% uranyl acetate and 2% methyl cellulose in a ratio of 100μl/900μl, transferred to a 50-μl drop of methyl cellulose-UA for 10 min on ice and air dried. ExoV were observed under a transmission electronic microscope Zeiss Leo EM 900 at 80 kV. For TEM analysis of MSC16412 exposed to gold-conjugated CAM29 (CAM49) overnight at 2.5 µM concentration, cells were processed as describedCitation30 (see Supplemental Methods).
Statistical analysis. Data are presented as mean ± SE. The values have been tested for Gaussian distribution by the D'Agostino-Pearson test. Statistical analysis was performed, using two-tailed unpaired Student's t test or One way ANOVA with Tuckey's multicomparison test by the GraphPad Prism 5 Version 5.03 computer software. The cut-off value of significance is indicated in each figure legend.
Disclosure of potential conflict of interest
No potential conflict of interest were disclosed.
supp_data.zip
Download Zip (2.8 MB)Additional information
Funding
References
- Edwards DR, Handsle MM, Pennington CJ. The ADAM metalloproteinases. Mol Aspects Med. 2008;29(5):258–89. doi: 10.1016/j.mam.2008.08.001. PMID:18762209.
- Reiss K., and Saftig P. The “A Disintegrin And Metalloprotease” (ADAM) family of sheddases: Physiological and cellular functions. Semin Cell Dev Biol. 2009;20(2):126–37. doi: 10.1016/j.semcdb.2008.11.002. PMID:19049889.
- Blobel CP. ADAMs: key components in EGFR signalling and development. Nature Rev Cancer. 2005;6(1):32–43. doi: 10.1038/nrm1548. PMID:15688065.
- Rocks N, Paulissen G, El Hour M, Quesada F, Crahay C, Gueders M, Foidart JM, Noel A, Cataldo D. Emerging roles of ADAM and ADAMTS metalloproteinases in cancer. Biochimie. 2008;90(2):369–79. doi: 10.1016/j.biochi.2007.08.008. PMID:17920749.
- Duffy MJ, McKiernan E, O'Donovan N, McGowan P. Role of ADAMs in cancer formation and progression. Clin Cancer Res. 2009;15(4):1140–44. doi: 10.1158/1078-0432.CCR-08-1585. PMID:19228719.
- Murphy G. The ADAMs: Signalling scissors in the tumor microenvironment. Nature Rev Cancer. 2008;8:929–41. doi: 10.1038/nrc2459. PMID:19005493.
- Duffy MJ, Mullooly M, O'Donovan N, Sukor S, Crown J, Pierce A, McGowan PM. The ADAMs family of proteases: new biomarkers and therapeutic targets for cancer? Clin Proteomics. 2011;8:9–13. doi: 10.1186/1559-0275-8-9. PMID:21906355.
- Saftig P, Reiss K. The “A Disintegrin And Metalloproteases” ADAM10 and ADAM17: novel drug targets with therapeutic potential? Eur J Cell Biol. 2011;90:527–35. doi: 10.1016/j.ejcb.2010.11.005. PMID:21194787.
- Zhou BB, Peyton M, He B, Liu C, Girard L, Caudler E, Lo Y, Baribaud F, Mikami I, Reguart N, et al. Targeting ADAM-mediated ligand cleavage to inhibit HER3 and EGFR pathways in non-small cell lung cancer. Cancer Cell. 2006;10(1):39–50. doi: 10.1016/j.ccr.2006.05.024. PMID:16843264.
- Witters L, Scherle P, Friedman S, Fridman J, Caulder E, Newton R, Lipton A Synergistic inhibition with a dual epidermal growth factor receptor/HER-2/neu tyrosine kinase inhibitor and a disintegrin and metallo proteinase inhibitor. Cancer Res. 2008;68:7082–89. doi: 10.1158/0008-5472.CAN-08-0739. PMID:18757423.
- Moss ML, Stoeck A, Yan W, Dempsey PJ. ADAM10 as a target for anti-cancer therapy. Curr Pharm Biotechnol. 2008;9:2–8. doi:10.2174/138920108783497613. PMID:18289051.
- Waldhauer I, Steinle A. Proteolytic release of soluble UL16-binding protein 2 from tumor cells. Cancer Res. 2006; 66(5):2520–26. doi:10.1158/0008-5472.CAN-05-2520. PMID:16510567.
- Waldhauer I, Goehlsdorf D, Gieseke F, Weinschenk T, Wittenbrink M, Ludwig A, Stevanovic S, Rammensee HG, Steinle A. Tumor-associated MICA is shed by ADAM proteases. Cancer Res. 2008;68(15):6368–6376. doi: 10.1158/0008-5472.CAN-07-6768. PMID:18676862.
- Nausch N, Cerwenka A. NKG2D ligands in tumor immunity. Oncogene. 2008;27:5944–58. doi: 10.1038/onc.2008.272. PMID:18836475.
- Salih HR, Antropius H, Gieseke F, Lutz SZ, Kanz L, Rammensee HG, Steinle A. Functional expression and release of ligands for the activating immunoreceptor NKG2D in leukemia. Blood. 2003;102:1389–96. doi: 10.1182/blood-2003-01-0019. PMID:12714493.
- Poggi A, Venturino C, Catellani S, Clavio M, Miglino M, Gobbi M, Steinle A, Ghia P, Stella S, Caligaris-Cappio F et al. Vδ1 T lymphocytes from B-CLL patients recognize ULBP3 expressed on leukemic B cells and up-regulated by trans-retinoic acid. Cancer Res. 2004;64:9172–9. doi: 10.1158/0008-5472.CAN-04-2417. PMID:15604289.
- Catellani S, Poggi A., Bruzzone A., Dadati P., Ravetti JL., Gobbi M., Zocchi MR. Expansion of Vdelta1 T lymphocytes producing IL-4 in low-grade non-Hodgkin lymphomas ex pressing UL-16-binding proteins. Blood. 2007;109:2078–85. doi: 10.1182/blood-2006-06-028985. PMID:16973957.
- Zocchi MR, Catellani S, Canevali P, Tavella S, Garuti A, Villaggio B, Zunino A, Gobbi M, Fraternali-Orcioni G, Kunkl A, et al. High ERp5/ADAM10 expression in lymph node microenvironment and impaired NKG2D-ligands recognition in Hodgkin lymphomas. Blood. 2012; 119:1479–89. doi: 10.1182/blood-2011-07-370841. PMID:22167753.
- Eichenauer DA, Simhadri VL, von Strandmann EP, Ludwig A, Matthews V, Reiners KS, von Tresckow B, Saftig P, Rose-John S, Engert A, Hansen HP. ADAM10 inhibition of human CD30 shedding increases specificity of targeted immunotherapy in vitro. Cancer Res. 2007;67(1):332–8. doi: 10.1158/0008-5472.CAN-06-2470. PMID:17210715.
- Zocchi M.R, Camodeca C, Nuti E, Rossello A, Venè R, Tosetti F, Dapino I, Costa D, Musso A, Poggi A. (2016) ADAM10 new selective inhibitors reduce NKG2D ligand release sensitizing Hodgkin lymphoma cells to NKG2D-mediated killing. Oncoimmunology. 2015;5(5):e1123367. doi: 10.1080/2162402X.2015.1123367. PMID:27467923.
- Camodeca C, Nuti E, Tepshi L, Boero S, Tuccinardi T, Stura E A, Poggi A, Zocchi MR, Rossello A. Discovery of a new selective inhibitor of A Disintegrin And Metalloprotease 10 (ADAM10) able to reduce the shedding of NKG2D ligands in Hodgkin's lymphoma cell models. Eur J Med Chem. 2016;111:193–201. doi: 10.1016/j.ejmech.2016.01.053. PMID:26871660.
- Stoeck A, Keller S, Riedle S, Sanderson MP, Runz S, Le Naour F Gutwein P, Ludwig A, Rubinstein E, Altevogt P. A role for exosomes in the constitutive and stimulus-induced ectodomain cleavage of L1 and CD44. Biochem J. 2006;393(Pt3):609–18. doi:10.1042/bj20051013. PMID:16229685.
- Chitadze G, Lettau M, Bhat J, Wesch D, Steinle A, Fürst D, Mytilineos J, Kalthoff H, Janssen O, Oberg HH, et al. Shedding of endogenous MHC class I-related chain molecules A and B from different human tumor entities: heterogeneous involvement of the “a disintegrin and metalloproteases” 10 and 17. Int J Cancer. 2013;133(7):1557–66. doi: 10.1002/ijc.28174. PMID:23526433.
- Théry C, Boussac M, Véron P, Ricciardi-Castagnoli P, Raposo G, Garin J, Amigorena S. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J Immunol. 2001; 166(12):7309–18. doi:10.4049/jimmunol.166.12.7309. PMID:11390481.
- Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–9. doi: 10.1038/ncb1596. PMID:17486113.
- Lundholm M, Schröder M, Nagaeva O, Baranov V, Widmark A, Mincheva-Nilsson L, Wikström P. Prostate tumor-derived exosomes down-regulate NKG2D expression on natural killer cells and CD8+ T cells: mechanism of immune evasion. PLoS One. 2014;9(9):e108925. doi: 10.1371/journal.pone.0108925. PMID:25268476.
- Filipazzi P, Bürdek M, Villa A, Rivoltini L, Huber V. Recent advances on the role of tumor exosomes in immunosuppression and disease progression. Semin Cancer Biol. 2012;22(4):342–9. doi: 10.1016/j.semcancer.2012.02.005. PMID:22369922.
- Escrevente C, Morais VA, Keller S, Soares CM, Altevogt P, Costa J. Functional role of N-glycosylation from ADAM10 in processing, localization and activity of the enzyme. Biochim Biophys Acta. 2008;1780(6):905–13. doi: 10.1016/j.bbagen.2008.03.004. PMID:18381078.
- Waugh MG. Raft-like membranes from the trans-Golgi network and endosomal compartments. Nat Protoc. 2013;8(12):2429–39. doi: 10.1038/nprot.2013.148. PMID:24202556.
- Camodeca C, Nuti E, Tosetti F, Poggi A, D'Arrigo C, Zocchi MR, Rossello A. Synthesis and in vitro evaluation of ADAM-10 and ADAM-17 selective bioimaging probes. Bioconjugate Chem 2018, Submitted.
- Norman LL, Brugués J Sengupta K, Sens P, Aranda-Espinoza H. Cell blebbing and membrane area homeostasis in spreading and retracting cells. Biophys J. 2010 Sep 22;99(6):1726–33. doi: 10.1016/j.bpj.2010.07.031. PMID:20858416.
- Van Den Berk LC Jansen BJ, Siebers-Vermeulen KG, Roelofs H, Figdor CG, Adema GJ, Torensma R. Mesenchymal stem cells respond to TNF but do not produce TNF. J. Leukoc Biol. 2010;7(2):283–9. doi: 10.1189/jlb.0709467. PMID:19897767.
- Hansen HP, Trad A, Dams M, Zigrino P, Moss M, Tator M, Schön G, Grenzi PC, Bachurski D, Aquino B, et al. CD30 on extracellular vesicles from malignant Hodgkin cells supports damaging of CD30 ligand-expressing bystander cells with Brentuximab-Vedotin, in vitro. Oncotarget. 2016;7(21):30523–35. doi: 10.18632/oncotarget.8864. PMID:27105521.
- Nakayama S, Yokote T, Tsuji M, Akioka T, Miyoshi T, Hirata Y, Hiraoka N, Iwaki K, Takayama A, Nishiwaki U, et al. Expression of tumour necrosis factor-α and its receptors in Hodgkin lymphoma. Br J Haematol. 2014;167(4): 574–77. doi: 10.1111/bjh.13015. PMID:25039986.
- Borchmann P, Treml JF, Hansen H, Gottstein C, Schnell R, Staak O, Zhang HF, Davis T, Keler T, Diehl V, et al. The human anti-CD30 antibody 5F11 shows in vitro and in vivo activity against malignant lymphoma. Blood. 2003;102(10):3737–42. doi: 10.1182/blood-2003-02-0515. PMID:12881320.
- Cardarelli P, Moldovan-Loomis MC, Preston B, Black A, Passmore D, Chen T-H, Chen S, Liu J, Kuhne MR, Srinivasan M, et al. In vitro and in vivo characterization of MDX-1401 for therapy of Malignant lymphoma. Clin Cancer Res. 2009;15(10):3376–3383. doi: 10.1158/1078-0432.CCR-08-3222. PMID:19401346.
- Noy PJ, Yang J, Reyat JS, Matthews AL, Charlton AE, Furmston J, Rogers DA, Rainger GE, Tomlinson MG. TspanC8 Tetraspanins and A Disintegrin and Metalloprotease 10 (ADAM10) Interact via Their Extracellular Regions:evidence for distinct binding mechanisms for different TspanC8 proteins. J Biol Chem. 2016;291(7):3145–57. doi: 10.1074/jbc.M115.703058. PMID:26668317.
- Borchmann S, von Tresckow B, Engert A. Current developments in the treatment of early-stage classical Hodgkin lymphoma. Curr Opin Oncol. 2016; 28(5):377–83. doi: 10.1097/CCO.0000000000000314. PMID:27455136.
- Nuti E, Casalini F, Avramova SI, Santamaria S, Fabbi M, Ferrini S, Marinelli L, La Pietra V, Limongelli V, Novellino E, et al. Potent Arylsulfonamide Inhibitors of Tumor Necrosis Factor-α Converting Enzyme Able to Reduce Activated Leukocyte Cell Adhesion Molecule Shedding in Cancer Cell Models. J Med Chem. 2010; 53(6):2622–35. doi: 10.1021/jm901868z. PMID:20180536.
- Neumann U, Kubota H, Frei K, Ganu V, Leppert D. Characterization of Mca-Lys-Pro-Leu-Gly-Leu-Dpa-Ala-Arg-NH2, a fluorogenic substrate with increased specificity constants for collagenases and tumor necrosis factor converting enzyme. Anal Biochem. 2004;328(2):166–173. doi: 10.1016/j.ab.2003.12.035. PMID:15113693.
- Benelli R, Monteghirfo S, Balbi C, Barboro P, Ferrari N. Novel antivascular efficacy of metronomic docetaxel therapy in prostate cancer: hnRNP K as a player. Int J Cancer 2009;124:2989–96. doi:10.1002/ijc.24305. PMID:19319982.
- Colombo MI, Lenhard JM, Mayorga LS, Stahl PD. Reconstitution of endosome fusion: identification of factors necessary for fusion competency. Methods Enzimol. 1992;219:32–44. doi:10.1016/0076-6879(92)19007-S. PMID: 1488005.
- Waugh MG. Raft-like membranes from the trans-Golgi network and endosomal compartments. Nat Protoc. 2013;8(12):2429–39. doi: 10.1038/nprot.2013.148. PMID:24202556
- Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;Chapter 3:Unit 3. 22. doi: 10.1002/0471143030.cb0322s30. PMID:18228490.
