ABSTRACT
Nivolumab is an anti-PD1 antibody, given in second-line or later treatment in advanced non-small cell lung cancer (NSCLC). The objective of this study was to describe the predictive value of circulating tumor DNA (ctDNA) on the efficacy of nivolumab in advanced NSCLC. We prospectively included all consecutive patients with advanced NSCLC treated with nivolumab in our Department between June 2015 and October 2016. Plasma samples were obtained before the first injection of nivolumab and at the first tumor evaluation with nivolumab. ctDNA was analyzed by Next-Generation Sequencing (NGS), and the predominant somatic mutation was followed for each patient and correlated with tumor response, clinical benefit (administration of nivolumab for more than 6 months), and progression-free survival (PFS). Of 23 patients, 15 had evaluable NGS results at both times of analysis. ctDNA concentration at the first tumor evaluation and ctDNA change correlated with tumor response, clinical benefit and PFS. ROC curve analyses showed good diagnostic performances for tumor response and clinical benefit, both for ctDNA concentration at the first tumor evaluation (tumor response: positive predictive value (PPV) at 100.0% and negative predictive value (NPV) at 71.0%; clinical benefit: PPV at 83.3% and NPV 77.8%) and the ctDNA change (tumor response: PPV 100.0% and NPV 62.5%; clinical benefit: PPV 100.0% and NPV 80.0%). Patients without ctDNA concentration increase >9% at 2 months had a long-term benefit of nivolumab. In conclusion, NGS analysis of ctDNA allows the early detection of tumor response and long-term clinical benefit with nivolumab in NSCLC.
Introduction
Lung cancer is the leading cause of cancer-related death in the world.Citation1 Recently, immune checkpoint inhibitors (ICI) have shown efficacy in the treatment of non-small cell lung cancer (NSCLC). Nivolumab is an anti-PD1 antibody, which was tested in a second-line setting in advanced NSCLC. In the pivotal randomized phase III trials, overall response rate (ORR) was 20% and disease control rate (DCR) was 50%.Citation2,Citation3 Of interest, some patients experienced a prolonged (i.e., more than 6 months) clinical benefit with nivolumab. However, no biomarker has been currently identified to predict the efficacy of nivolumab in second-line treatment or more. Programmed death-ligand 1 (PDL1) immunohistochemistry (IHC) has been often used. A recent study has shown the benefit of another ICI (pembrolizumab) in first-line treatment in high-PD-L1 expression (≥50%) advanced NSCLC, compared to platinum-based chemotherapy (hazard ratio (HR) for overall survival at 0.60; 95%CI 0.41–0.89).Citation4 PD-L1 IHC was associated with ORR in Chekmate-057, but not in Chekmate-017.Citation2,Citation3 Moreover, patients with negative PDL1 expression in IHC may however experience tumor response with nivolumab. There is an urgent need to identify new biomarkers associated with nivolumab efficacy.
The monitoring of circulating tumor DNA (ctDNA) may be an interesting tool in this setting. For cytotoxic chemotherapy, several studies have failed to prove a predictive role of ctDNA concentration for response.Citation5,Citation6 However, in case of oncogenic addiction, the monitoring of the mutated allelic fraction can be used to follow the efficacy of targeted therapies and look for the appearance of resistance mutation, especially in EGFR-mutated NSCLC.Citation7-12 Next-generation sequencing (NGS) is also feasible in ctDNA.Citation13 With regard to ICIs, recent reports suggested that the decrease of the ctDNA concentration during treatment was associated with tumor response.Citation14-16 However, most of these studies used digital-droplet Polymerase chain reaction (ddPCR) to monitor only one specific mutation.
In this study, we tested the hypothesis that the change in ctDNA concentration as measured using large-cancer gene panel screening by NGS between baseline and evaluation at 2 months correlated with tumor response and clinical benefit of nivolumab in advanced NSCLC.
Results
Patient characteristics
Between June 2015 and October 2016, 34 patients received nivolumab in second-line or later treatment for an advanced NSCLC. From them, 20 patients had plasma samples for both C1 nivolumab and M2. Of these 20 patients, 15 (75%) had interpretable data in NGS at the two time-points and were used for analyses. There were 9 men (60.0%), 14 current or former smokers (93.0%) with 35 packet-years (median), 11 stage IV NSCLC (73.0%) and 10 non-squamous carcinoma (66.7%). On 15 patients, 14 had evaluable tumor sample for PD-L1 expression assessment (immunohistochemistry, IHC). Seven patients (50.0%) and 2 patients (14.3%) had PD-L1 ≥1% and ≥50% expression on tumor cells, respectively. The patients received nivolumab mainly in second-line treatment (n = 13, 86.7%) and less often in fourth-line treatment (n = 2, 13.3%). The median number of nivolumab injections was 13 (4–20). The overall response rate was 33.3%. After a median follow-up of 17.3 months, 10 patients (66.7%) exhibited tumor progression, of whom 7 (46.7%) died. Median PFS for the 15 patients with nivolumab was 4.1 months and median OS 10.5 months. The tumor evaluation was performed after 4 injections of nivolumab, i.e., at 2 months of treatment, for all patients, except for 3 patients with clinical progression after only 1 injection of nivolumab. These 3 patients had plasma samples taken after 1 injection of nivolumab, and were included in the analyses. describes the population characteristics, and the comparison between patients with objective response (OR) at the first tumor evaluation and patients without OR, and between patients with clinical benefit (i.e., nivolumab given > 6 months) and patients without clinical benefit. No difference was seen in terms of clinical and pathological features between the groups.
Table 1. Patients' characteristics and comparison between patients with tumor response and without tumor response, and between patients with clinical benefit of nivolumab and patients without clinical benefit of nivolumab.
displays the PD-L1 status and the mutations found in NGS for each patient.
Table 2. PDL-L1 expression in immunohistochemistry and mutations found in NGS for the 15 patients.
ctDNA concentration at baseline
The ctDNA concentration at baseline (median) was 0.006 ng/µl (0.003–0.013). The ctDNA concentration at baseline was not different between patients with OR and patients without OR: 0.005 ng/µl (0.001–0.007) versus 0.009 ng/µl (0.004–0.029), respectively (p = 0.501) (). No difference was observed between ctDNA at baseline according to subsequent clinical benefit, with a ctDNA at baseline for patients with clinical benefit at 0.005 ng/µl (0.001–0.008) versus 0.012 ng/µl (0.006–0.046) for patients without clinical benefit of nivolumab (p = 0.148) (). Baseline ctDNA concentrations were no different according to PD-L1 expression in IHC: median 0.007 ng/µl (0.006–0.012) in PD-L1 ≥1% patients versus 0.005 ng/µl (0.002–0.036) in PD-L1 <1% patients (p = 0.701); median 0.007 ng/µl (0.004–0.010) in PD-L1 ≥50% patients versus 0.007 ng/µl (0.005–0.018) in PD-L1 <50% patients (p = 0.927). Baseline ctDNA concentrations were not correlated with baseline tumor burden (p = 0.371) (Supplementary ).
ctDNA concentration at the first evaluation
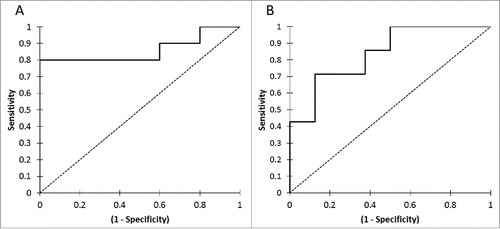
At the first tumor evaluation, the ctDNA concentration was statistically different between patients with OR versus patients without OR: 0.001 ng/µl (0.0006–0.001) versus 0.007 ng/µl (0.003–0.078), respectively (p = 0.032) (). ROC curve analysis determined a positivity threshold at 0.002 ng/µl, allowing the detection of an absence of OR in case of ctDNA > 0.002 ng/µl with a sensibility of 80%, a specificity of 100%, a positive predictive value at 100% and a negative predictive value at 71% (AUC = 0.86) (). Overall response rate (ORR) was 71.4% if ctDNA < 0.002 ng/µl, versus 0.0% if ctDNA > 0.002 ng/µl at the first tumor evaluation (p = 0.003).
Similar results were observed for clinical benefit, with lower ctDNA concentration at the first tumor evaluation in patients with clinical benefit compared with patients without clinical benefit: 0.001 ng/µl (0.0004–0.003) versus 0.008 ng/µl (0.004–0.131), respectively (p = 0.032) (). ROC curve analysis determined a positivity threshold at 0.006 ng/µl, allowing the detection of an absence of clinical benefit in case of ctDNA > 0.006 ng/µl with a sensibility of 71.4%, a specificity of 87.5%, a positive predictive value at 83.3% and a negative predictive value at 77.8% (AUC = 0.839) ().
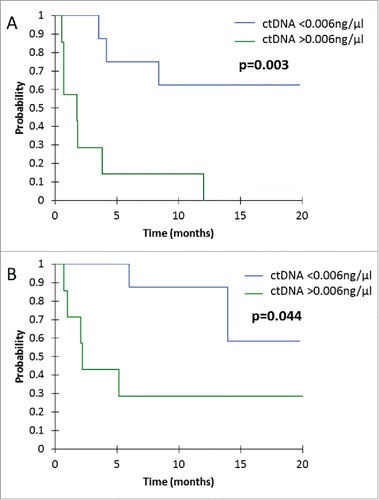
Median PFS for patients with ctDNA > 0.006 ng/µl at first tumor evaluation was 1.8 months, versus not-reached for patients with ctDNA < 0.006 ng/µl (p = 0.003) (). Median OS for patients with ctDNA > 0.006 ng/µl at first tumor evaluation was 2.2 months, versus not-reached for patients with ctDNA < 0.006 ng/µl (p = 0.044) ().
Figure 2. Kaplan-Mayer survival curves for PFS (A) and OS (B), according to ctDNA concentration at the first tumor evaluation.
ctDNA concentrations at the first evaluation were no different according to PD-L1 expression in IHC: median 0.003 ng/µl (0.000–0.012) in PD-L1 ≥1% patients versus 0.006 ng/µl (0.001–0.085) in PD-L1 <1% patients (p = 0.695); median 0.007 ng/µl (0.004–0.011) in PD-L1 ≥50% patients versus 0.005 ng/µl (0.000–0.031) in PD-L1 <50% patients (p = 0.926). ctDNA concentrations at the first tumor evaluation were not correlated with tumor burden at the first tumor evaluation (p = 0.543) (Supplementary ).
Evolution of the ctDNA concentration
The change in the ctDNA concentration was strongly associated with tumor response. All the patients with tumor response (n = 5/5) had a decrease in ctDNA concentration, whereas 60% (n = 6/10) of patients without tumor response had an increase in the ctDNA concentration (p = 0.025) (). In patients with tumor response, the median of the relative change in the ctDNA was −87.8% (−91.4% – −45.9%), whereas in patients without tumor response at first tumor evaluation, it was +34.1% (−39.8% – +188.5%) (p = 0.032). The corresponding ROC curve analysis defined a threshold of 30% of ctDNA decrease at first tumor evaluation, allowing the detection of an absence of OR if ctDNA relative evolution >−30%, with a sensitivity at 70%, a specificity at 100%, a positive predictive value at 100% and a negative predictive value at 62.5% (AUC = 0.860) ().
Figure 3. ROC curves for PFS (A) and OS (B) according to relative evolution of ctDNA concentration between baseline and the first tumor evaluation.
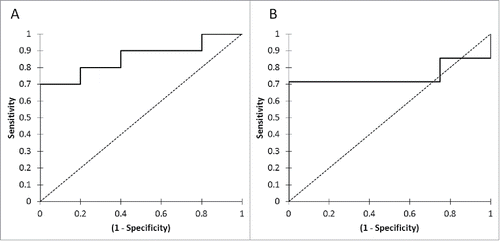
For the prediction of clinical benefit with nivolumab, 87.5% (n = 7/8) patients with clinical benefit had a decrease of their ctDNA concentration at the first tumor evaluation, whereas 71.4% (n = 5/7) of patients without clinical benefit with nivolumab had an increase of their ctDNA concentration (p = 0.020) (). In patients with clinical benefit, the median of the relative change in ctDNA concentration was −45.8% (−63.3% – −28.3%), whereas in patients without tumor response at first tumor evaluation, it was +166.6% (−14.0% – +229.2%) (p = 0.118). The corresponding ROC curve analysis defined a threshold of 9% of ctDNA increase at first tumor evaluation, allowing the detection of an absence of clinical benefit with nivolumab if ctDNA concentration relative evolution >9%, with a sensitivity at 71.4%, a specificity at 100%, a positive predictive value at 100% and a negative predictive value at 80% (AUC = 0.750) ().
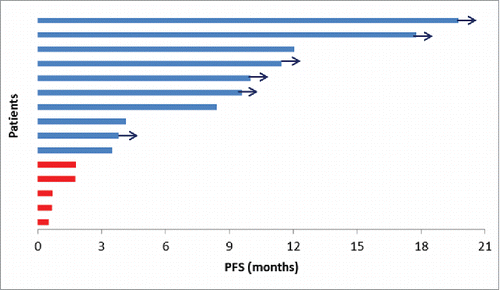
Median PFS for patients with ctDNA increase >9% at first tumor evaluation was 0.7 months, versus 12.0 months for patients with ctDNA decrease (p < 0.001) (). Median OS for patients with ctDNA increase >9% at first tumor evaluation was 2.1 months, versus not-reached for patients with ctDNA decrease (p < 0.001) (). The majority of patients without increase of ctDNA >9% at the first tumor evaluation had a prolonged use and benefit of nivolumab, as shown in .
Figure 4. Kaplan-Mayer survival curves for PFS (A) and OS (B), according to relative evolution of ctDNA concentration between baseline and the first tumor evaluation. Threshold at +9% to define ctDNA augmentation.
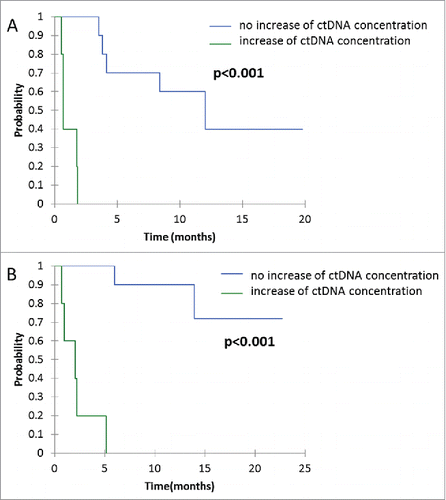
Figure 5. PFS according to the relative change in ctDNA concentration between baseline and the first tumor evaluation for each patients. Red bars indicate an increase >9% of ctDNA concentration; blue bars indicate an absence of increase >9% of ctDNA concentration. Blue arrows indicate ongoing treatment at the time of the analysis.
Concerning the other mutations found in NGS (), their concentrations had the same evolution as the predominant mutation selected for follow-up, except for one patient with a minor mutated clone (SMAD4 mutation), which concentration increased whereas others (PTEN mutation and CTNNB1 mutation) decreased between baseline and the first evaluation.
Discussion
In this study, we showed that the ctDNA concentration at the time of first tumor evaluation and the relative change in this concentration was associated with tumor response, PFS and OS. Moreover, we were able to predict at the time of the first evaluation which patients would have a sustained clinical benefit of nivolumab.
The use of biomarkers to predict the response to ICI remains challenging, especially in second-line or later treatment for advanced NSCLC. PDL1 IHC, often used in clinical trials, is poorly associated with response to nivolumab in this setting.Citation2,Citation3 Moreover, PDL1 IHC raises several issues: different antibodies used in different trials depending on the ICI, different types of analyses (tumor cells with or without immune cells), different positivity thresholds, and tests often performed on small biopsies, whereas PDL1 staining is heterogeneous in the tumor.Citation17 Moreover, the expression of PDL1 can vary between the different metastatic sites (spatial heterogeneity),Citation18 and also after chemotherapy treatment (temporal heterogeneity).Citation19 Plasmatic biomarkers offer the advantage of being easily accessible, repeatable, and able to reflect tumor heterogeneity. Several studies have suggested that ctDNA could be useful to predict tumor response and prognosis with ICI. Cabel et al. analyzed 15 patients with various solid tumors (NSCLC, uveal melanoma or microsatellite-instable colorectal cancer) treated by nivolumab or pembrolizumab, and showed that the decrease of ctDNA at 8 weeks was associated with tumor response.Citation14 In melanoma, equivalent results were obtained in 76 patients, with a strong negative prognostic and predictive impact of the absence of decrease of ctDNA concentration at 12 weeks in patients treated with ICI with or without ipilimumab.Citation16 In NSCLC, a recent report in two patients with advanced NSCLC treated with nivolumab described the possibility of monitoring the Kras-mutated ctDNA fraction to distinguish pseudo-progression from true progression.Citation15 Our study is, to date, the largest analysis of ctDNA dedicated to NSCLC patients treated with nivolumab (n = 15). We chose to analyze ctDNA with NGS, allowing the detection and follow-up of a wide variety of cancer-related genes. As a comparison, previous studies only used digital-dropplet PCR (ddPCR)Citation15,Citation16 or various techniques (pyrosequencing, ddPCR or NGS) in the melanoma study.Citation14 ddPCR is a highly sensitive technique, but allows the follow-up of only one pre-specified mutation. We believe that NGS is the most appropriate technique for follow-up of NSCLC that present often a large variety of somatic mutations, and do not need to have a pre-analytic selection of one specific mutation. In our study, using a gene-panel of 22 genes, we were able to follow predominant mutations on 9 different genes (TP53, NOTCH1, FBXW7, KRAS, SMAD4, KDR, DDR2, BRAF, PTEN). Serial analyses of ctDNA using NGS has already proven to be a robust strategy to evaluate tumor response to targeted therapies, with possibility for performing longitudinal studies of ctDNA samples with multiple mutations.Citation13
Beyond the association to tumor response, we clearly showed that the evaluation of ctDNA allowed an early detection of the patients who will have a clinical benefit of nivolumab, as assessed by a treatment given more than 6 months. The evaluation of the clinical benefit seems more adapted for ICI treatment evaluation rather than tumor response, as several patients can experience significant clinical improvement with prolonged stable disease, or a tumor control after an initial augmentation of tumor volumes on CT-scan.Citation2,Citation3
In our population, we found an ORR at 33.3%, which is higher than ORR reported in the Checkmate trials (around 20.0%).Citation2,Citation3 Our rate of PD-L1 positive tumors (≥1% and ≥50%) was however comparable to those reported in the literature.Citation2-4 Because of the small number of patients (n = 15), this high ORR could be due to hazard of patients inclusion.
The monitoring of ctDNA in ICI treatment opens a wide range of possible applications. The possibility to detect as soon as the first tumor evaluation, those patients who will not have tumor response or clinical benefit with nivolumab could offer the possibility of alternative treatment strategies for these patients, such as ICI combinations. In addition, ctDNA monitoring could also be used for early detection of secondary progression, and maybe reveal the appearance of resistance mutation, as described in melanoma with JAK1/JAK2 and beta-2 microglobulin mutations.Citation20 Further studies are needed in NSCLC to confirm these hypotheses.
In conclusion, ctDNA is a useful biomarker during nivolumab treatment in advanced NSCLC. NGS allow the detection and the follow-up of a wide variety of somatic mutations. A high ctDNA concentration at 2 months (i.e., first tumor evaluation) and an increase in concentration compared with baseline are associated with poor response and no long-term clinical benefit. Further studies with larger population and longer follow-up are needed to confirm our results.
Patients and methods
Patients and plasmas
We analyzed all consecutive patients in our hospital's department of Thoracic Oncology who received nivolumab in second-line or later treatment for a stage IIIb-IV NSCLC, between June 2015 and October 2016. Demographical, pathological and treatment-related data were collected from a prospective database. Patients received nivolumab at 3 mg/kg intravenously every 2 weeks until progression or intolerable toxicity. Tumor evaluation was performed at baseline with brain, thoracic and abdominal CT-scan at 2 months (after 4 injections of nivolumab) and then every 2 months under nivolumab. Tumor response was assessed according to iRECIST.Citation21 Patients who did not have restaging due to clinical disease progression before 2 months, as assessed by their clinician, were classified as progressive disease, with plasma taken at the moment of the clinical progression and included in the analysis. Clinical benefit was defined as patients treated with nivolumab for more than 6 months. Tumor burden was calculated by the sum of the largest diameter of all evaluable tumor lesions (more than 1 cm), at baseline and at the first tumor evaluation.
Plasma samples were taken at diagnosis, just before the first injection of nivolumab (C1), and at the first tumor evaluation (at 2 months, M2).
ctDNA analyses
Two 10 ml-EDTA tubes of peripheral blood were taken, and plasma was isolated within one hour after and immediately conserved at −80°C.
DNA Extraction from Tumor and Plasma Samples
DNAs were extracted on a Maxwell® 16 Forensic Instrument (Promega, France) using Maxwell® 16 FFPE Plus LEV DNA Purification Kit (Promega, France) for FFPE samples and the Maxwell® RSC ccfDNA Plasma Kit (Promega, France) for circulating cell-free DNAs. Quantification was done by Qubit Fluorometric Quantitation using the Qubit dsDNA HS Assay Kit and the Qubit dsDNA BR Assay Kit (Life Technologies–Thermo Fisher Scientific, Saint Aubin, France) for circulating DNA and tumor DNA respectively. DNAs were stored at −20°C before use.
Sequencing
Colon and Lung Cancer Panel V2 libraries were prepared using the Ion Ampliseq library preparation kit v2 from 30 ng of tumor DNA or 6 ul of cell free DNAs. Libraries were normalized (Ion Library Equalizer™ Kit), pooled, processed on a Ion Chef™ System for template preparation and chip loading (Ion PI™ Hi-Q™ Chef Kit, Ion PI™ Chip Kit v3, Thermo Fisher Scientific), and sequenced on a Ion Proton™ System.
Analysis
The FASTQs sequencing data were aligned to the human genome (hg19) and processed by the IonTorrent Suite V5.0.4.0. VCF files were generated using the built-in “Somatic – low stringency” parameters to automatically call variants with allele frequencies (AF) > 2%. Annotation pipeline was developed internally on a galaxy platform that uses SAFIR 2.4 report tool. In parallel, samples were analyzed by the BPER method after BAM recalibration, a specific algorithm developed to detect AF < 2%.Citation22 It is publicly available at https://cran.r-project.org/package=PlasmaMutationDetector. Results provided by BPER method and annotated VCFs were registered blindly from clinical data.
In case of multiple mutations, we chose for follow-up the predominant mutation found in NGS, according to the concentration of each mutated allele, based on the allelic ratio and circulating DNA concentrations.
IHC technique
IHC was performed using an automated method (Leica) and the E13LN anti-PD-L1 antibody (Cell signalling Technology) diluted to the 1/80th on 4 µm-slides from the treatment-naïve diagnostic samples. The assay was performed using human amygdala as positive control, and IgG as isotype negative control. The IHC was considered as being positive if at least one tumour cell out of 100 analysed tumour cells was positively stained.
Ethical considerations
All patients signed a consent form prior to plasma collection. The plasma collection and analyses were authorized by the Institutional Review Board CPP IDF n°8 (ID CRB 2014-A00187-40). The retrieval of data from our prospectively collected database conformed to the rules of the Commission Nationale de l'Informatique et des Libertés.
Statistical analyses
Clinical and pathological data were compared between patients with tumor response (overall response, OR) at the first evaluation versus no tumor response, and with clinical benefit of nivolumab versus no clinical benefit of nivolumab. Associations with qualitative variables were assessed using the χ2, whereas comparisons of continuous variables were assessed using the Student t test (variables with normal distribution) or Mann–Whitney test (variables without normal distribution). Analyses of correlation between tumor burden and ctDNA concentrations were made using the Spearman test. ROC curves were used to determine positivity thresholds to detect OR and clinical benefit, with expression of related sensitivity, specificity, positive predictive value, negative predictive value, and area under the curve (AUC) value. PFS and OS were estimated using the Kaplan–Meier method (p-value calculated using log-rank test). The censoring date was 18/07/2017. Continuous variables are expressed as mean (± standard deviation) if normally distributed or median (interquartile range, IQR) if non-normally distributed. Results were considered significant if the p-value was < 0.05. Statistical tests were performed with Xlstat 2017 software (Addinsoft).
Conflict of interest statement
EGL received research grants from Bristol-Myers-Squibb. Other authors don't have conflict of interest.
Financial support
This study was realized with the support of Legs Poix 2016 and Fondation de l'Avenir (Paris, France) 2016 n° AP-UMC-2016-001.
2017ONCOIMM0941R-f06-z-bw.pptx
Download MS Power Point (56.7 KB)Additional information
Funding
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi:10.3322/caac.21332.
- Brahmer J, Reckamp KL, Baas P, Crinò L, Eberhardt WEE, Poddubskaya E, Antonia S, Pluzanski A, Vokes EE, Holgado E, et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. N Engl J Med. 2015;373:123–35. doi:10.1056/NEJMoa1504627. PMID:26028407.
- Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, Chow LQ, Vokes EE, Felip E, Holgado E, et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N Engl J Med. 2015;373:1627–39. doi:10.1056/NEJMoa1507643. PMID:26412456.
- Reck M, Rodríguez-Abreu D, Robinson AG, Hui R, Csőszi T, Fülöp A, Gottfried M, Peled N, Tafreshi A, Cuffe S, et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med. 2016;375(19):1823–1833. doi:10.1056/NEJMoa1606774. PMID:27718847.
- Tissot C, Toffart A-C, Villar S, Souquet P-J, Merle P, Moro-Sibilot D, Pérol M, Zavadil J, Brambilla C, Olivier M, et al. Circulating free DNA concentration is an independent prognostic biomarker in lung cancer. Eur Respir J. 2015;46:1773–80. doi:10.1183/13993003.00676-2015. PMID:26493785.
- Li BT, Drilon A, Johnson ML, Hsu M, Sima CS, McGinn C, Sugita H, Kris MG, Azzoli CG. A prospective study of total plasma cell-free DNA as a predictive biomarker for response to systemic therapy in patients with advanced non-small-cell lung cancers. Ann Oncol. 2016;27:154–9. doi:10.1093/annonc/mdv498. PMID:26487589.
- Zhang H, Liu D, Li S, Zheng Y, Yang X, Li X, Zhang Q, Qin N, Lu J, Ren-Heidenreich L, et al. Comparison of EGFR signaling pathway somatic DNA mutations derived from peripheral blood and corresponding tumor tissue of patients with advanced non-small-cell lung cancer using liquidchip technology. J Mol Diagn. 2013;15:819–26. doi:10.1016/j.jmoldx.2013.06.006. PMID:23988622.
- Sorensen BS, Wu L, Wei W, Tsai J, Weber B, Nexo E, Meldgaard P. Monitoring of epidermal growth factor receptor tyrosine kinase inhibitor-sensitizing and resistance mutations in the plasma DNA of patients with advanced non-small cell lung cancer during treatment with erlotinib. Cancer. 2014;120:3896–901. doi:10.1002/cncr.28964. PMID:25103305.
- Ishii H, Azuma K, Sakai K, Kawahara A, Yamada K, Tokito T, Okamoto I, Nishio K, Hoshino T. Digital PCR analysis of plasma cell-free DNA for non-invasive detection of drug resistance mechanisms in EGFR mutant NSCLC: Correlation with paired tumor samples. Oncotarget. 2015;6:30850–8. doi:10.18632/oncotarget.5068. PMID:26334838.
- Sakai K, Horiike A, Irwin DL, Kudo K, Fujita Y, Tanimoto A, Sakatani T, Saito R, Kaburaki K, Yanagitani N, et al. Detection of epidermal growth factor receptor T790M mutation in plasma DNA from patients refractory to epidermal growth factor receptor tyrosine kinase inhibitor. Cancer Sci. 2013;104:1198–204. doi:10.1111/cas.12211. PMID:23721103.
- Marchetti A, Palma JF, Felicioni L, De Pas TM, Chiari R, Del Grammastro M, Filice G, Ludovini V, Brandes AA, Chella A, et al. Early Prediction of Response to Tyrosine Kinase Inhibitors by Quantification of EGFR Mutations in Plasma of NSCLC Patients. J Thorac Oncol. 2015;10:1437–43. doi:10.1097/JTO.0000000000000643. PMID:26295376.
- Oxnard GR, Thress KS, Alden RS, Lawrance R, Paweletz CP, Cantarini M, Yang JC-H, Barrett JC, Jänne PA. Association Between Plasma Genotyping and Outcomes of Treatment With Osimertinib (AZD9291) in Advanced Non-Small-Cell Lung Cancer. J Clin Oncol. 2016;34:3375–82. doi:10.1200/JCO.2016.66.7162. PMID:27354477.
- Frenel JS, Carreira S, Goodall J, Roda D, Perez-Lopez R, Tunariu N, Riisnaes R, Miranda S, Figueiredo I, Nava-Rodrigues D, et al. Serial Next-Generation Sequencing of Circulating Cell-Free DNA Evaluating Tumor Clone Response To Molecularly Targeted Drug Administration. Clin Cancer Res. 2015;21:4586–96. doi:10.1158/1078-0432.CCR-15-0584. PMID:26085511.
- Cabel L, Riva F, Servois V, Livartowski A, Daniel C, Rampanou A, Lantz O, Romano E, Milder M, Buecher B, et al. Circulating tumor DNA changes for early monitoring of anti-PD1 immunotherapy: a proof-of-concept study. Ann Oncol. 2017;28(8):1996–2001. doi:10.1093/annonc/mdx212. PMID:28459943.
- Guibert N, Mazieres J, Delaunay M, Casanova A, Farella M, Keller L, Favre G, Pradines A. Monitoring of KRAS-mutated ctDNA to discriminate pseudo-progression from true progression during anti-PD-1 treatment of lung adenocarcinoma. Oncotarget. 2017;8:38056–60. PMID:28445137.
- Lee JH, Long GV, Boyd S, Lo S, Menzies AM, Tembe V, Guminski A, Jakrot V, Scolyer RA, Mann GJ, et al. Circulating tumour DNA predicts response to anti-PD1 antibodies in metastatic melanoma. Annals of Oncology. 2017;28:1130–6. doi:10.1093/annonc/mdx026. PMID:28327969.
- Ilie M, Long-Mira E, Bence C, Butori C, Lassalle S, Bouhlel L, Fazzalari L, Zahaf K, Lalvée S, Washetine K, et al. Comparative study of the PD-L1 status between surgically resected specimens and matched biopsies of NSCLC patients reveal major discordances: a potential issue for anti-PD-L1 therapeutic strategies. Ann Oncol. 2016;27:147–53. doi:10.1093/annonc/mdv489. PMID:26483045.
- Mansfield AS, Aubry MC, Moser JC, Harrington SM, Dronca RS, Park SS, Dong H. Temporal and spatial discordance of programmed cell death-ligand 1 expression and lymphocyte tumor infiltration between paired primary lesions and brain metastases in lung cancer. Ann Oncol. 2016;27:1953–8. doi:10.1093/annonc/mdw289. PMID:27502709.
- Sheng J, Fang W, Yu J, Chen N, Zhan J, Ma Y, Yang Y, Huang Y, Yanhuang null, Zhao H, et al. Expression of programmed death ligand-1 on tumor cells varies pre and post chemotherapy in non-small cell lung cancer. Sci Rep. 2016;6:20090. doi:10.1038/srep20090. PMID:26822379.
- Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, Torrejon DY, Abril-Rodriguez G, Sandoval S, Barthly L, et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N Engl J Med. 2016;375:819–29. doi:10.1056/NEJMoa1604958. PMID:27433843.
- Seymour L, Bogaerts J, Perrone A, Ford R, Schwartz LH, Mandrekar S, Lin NU, Litière S, Dancey J, Chen A, et al. iRECIST: guidelines for response criteria for use in trials testing immunotherapeutics. Lancet Oncol. 2017;18:e143–52. doi:10.1016/S1470-2045(17)30074-8. PMID:28271869.
- Pécuchet N, Rozenholc Y, Zonta E, Pietraz D, Didelot A, Combe P, Gibault L, Bachet J-B, Taly V, Fabre E, et al. Analysis of Base-Position Error Rate of Next-Generation Sequencing to Detect Tumor Mutations in Circulating DNA. Clin Chem. 2016;62:1492–503. doi:10.1373/clinchem.2016.258236. PMID:27624137.