ABSTRACT
Analysis of the T cell receptor (TR) repertoire of chronic lymphocytic leukemia-like monoclonal B cell lymphocytosis (CLL-like MBL) and early stage CLL is relevant for understanding the dynamic interaction of expanded B cell clones with bystander T cells. Here we profiled the T cell receptor β chain (TRB) repertoire of the CD4+ and CD8+ T cell fractions from 16 CLL-like MBL and 13 untreated, Binet stage A/Rai stage 0 CLL patients using subcloning analysis followed by Sanger sequencing. The T cell subpopulations of both MBL and early stage CLL harbored restricted TRB gene repertoire, with CD4+ T cell clonal expansions whose frequency followed the numerical increase of clonal B cells. Longitudinal analysis in MBL cases revealed clonal persistence, alluding to persistent antigen stimulation. In addition, the identification of shared clonotypes among different MBL/early stage CLL cases pointed towards selection of the T cell clones by common antigenic elements. T cell clonotypes previously described in viral infections and immune disorders were also detected. Altogether, our findings evidence that antigen-mediated TR restriction occurs early in clonal evolution leading to CLL and may further increase together with B cell clonal expansion, possibly suggesting that the T cell selecting antigens are tumor-related.
Introduction
Clinical CLL-like monoclonal B cell lymphocytosis (MBL) is characterized by the presence of a clonal population of B lymphocytes in the peripheral blood (0.5 to <5 × 109/L) having a phenotype consistent with chronic lymphocytic leukemia (CLL). It is an asymptomatic condition, yet considered to be a premalignant precursor of CLL, with a progression rate of 1.1% per year to CLL requiring therapy. In fact, the great majority of CLL are preceded by an MBL stage.Citation1–3 Therefore, the study of MBL is critical to understand CLL ontogenesis and clinical evolution.
Several lines of evidence suggest selection of CLL clones by a restricted set of antigenic epitopes; perhaps the strongest argument is the remarkable restriction of the immunoglobulins (IG) expressed by the clonotypic B cell receptors (BcR),Citation4 including the existence of subsets with quasi-identical, stereotyped BcR IGs.Citation5–7 Recently, the role of antigens in shaping the T cell receptor (TR) repertoire in CLL has been also demonstrated, further corroborating the implication of antigen selection in the natural history of the disease.Citation8,Citation9
Interactions between CLL cells and tumor microenvironment, including other cells and soluble factors like cytokines, are crucial for disease development. Previous investigations in CLL reported (i) altered cytokine patterns,Citation10 (ii) dysfunctional T cells regardless of the presence of elevated absolute counts and oligoclonal expansionsCitation11–13 and (iii) crosstalk between tumor B cells and autologous T cells.Citation14 Importantly, T cell tolerance induced by CLL seems to be critical for CLL clonal expansion and may be reverted by immunomodulating drugs.Citation15
Studies concerning the microenvironment in MBL are scarce. While CLL is characterized by immune suppression and tolerized behavior of autologous T cells that hampers anti-tumor immunity,Citation16 prior investigations showed that the effector function of T cells in MBL is only slightly deviated.Citation17 On these grounds, the antigen mediated interactions of the aberrant B cells with autologous T cells and, subsequently, the architecture of the TR repertoire may differ between MBL and CLL at early stages compared to advanced disease, possibly reflecting more effective T cell immune surveillance. That notwithstanding, the implication of the antigenic elements both in the selection and survival of CLL/MBL cells, as well as the induction of autologous T cell tolerance, still remains unclear. Here, we extensively characterized the TR repertoire of the CD4+ and CD8+ T cell subpopulations in subjects with MBL and early stage CLL in order to assess the molecular characteristics and the dynamics of T cell clonal expansions in relation to B cell clonal evolution, so as to gain more insight into the potential role of the autologous T cells in the ontogeny and control of the emerging CLL clone.
Results
The T cell repertoire of MBL and early stage CLL is restricted
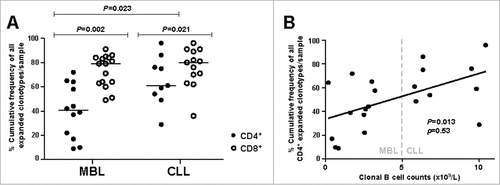
In all, 2,567 productive rearrangements were obtained (average: 39 rearrangements/sample, range: 21–62), corresponding to 1,337 distinct clonotypes (887 from MBL, 449 from CLL and one shared by MBL/CLL). Expanded clonotypes were detected in all 65 samples analyzed. The number of expanded clonotypes/sample ranged from 1–13 (median: 6) whereas the cumulative frequency of all expanded clonotypes ranged from 9–97% (median: 64%). A comparative analysis between groups and cell types restricted to samples with purity ≥90% is shown in .
Table 1. Clonality analysis for the CD4+ and the CD8+ T cell fractions. Values are given as median (range). Data from longitudinal samples of the same patients is not included. NS: not significant. #Only samples with purity ≥90%.
Concerning CD4+ T cells, the MBL group exhibited a significantly lower cumulative frequency of all expanded clonotypes compared to CLL (median: 40.4% vs. 61.0%, P = 0.023) (, ). In line with these observations, a significant positive correlation between the absolute count of clonal B cells and the cumulative frequency of all expanded CD4+ T cell clonotypes was noted (P = 0.013, ρ = 0.53) (). No differences in clonality between MBL and CLL neither correlation with clonal B cell counts were identified for the CD8+ T cell compartment. CD8+ T cell samples showed a significantly higher cumulative frequency of all expanded clonotypes than CD4+ T cell samples both in MBL (median: 79.2% vs. 40.4%, P = 0.002) and CLL (median: 79.6% vs. 61.0%, P = 0.021) (, ). When the cumulative frequencies of all expanded CD4+ and CD8+ T cell clonotypes were compared, a significant positive correlation was observed for CLL patients (P = 0.050, ρ = 0.67), but no significant correlation was detected in MBL (P = 0.145, ρ = 0.45) (Fig. S1).
Figure 1. Clonality analysis. A, Percentage cumulative frequency of all expanded CD4+ and CD8+ T cell clonotypes in MBL subjects and CLL-A(0) patients. Horizontal lines correspond to the median value for each case. B, Correlation between the absolute count of malignant B cells and the percentage cumulative frequency of all expanded CD4+ T cell clonotypes per sample. ρ: Spearman's rho correlation coefficient.
As for the TRBV gene repertoire of the CD4+ T cell fraction, 32 functional genes were identified (Table S1). A remarkable bias in the TRBV gene usage was observed both for MBL and CLL, with only six genes (TRBV10-3, TRBV6-1, TRBV28, TRBV19, TRBV27 and TRBV20-1) accounting for more than 50% of the entire repertoire in each group separately. Notably, when expanded clonotypes were considered, the frequencies of certain TRBV genes differed among groups (, Table S1). In detail, the TRBV6-2 or 6–3 gene was overrepresented in MBL compared to CLL (frequencies: 8.2% vs. 0% respectively, P = 0.032) whereas TRBV20-1 was less frequent in MBL than in CLL (frequencies: 3.3% vs. 13.4% respectively, P = 0.023).
Figure 2. TRBV gene repertoire analysis of the CD4+ (A) and CD8+ (B) expanded T cell clonotypes in the MBL and CLL groups. The 15 most frequently detected genes within the expanded clonotypes of the MBL group are detailed in a decreasing order in the x-axis. Significant differences (P < 0.05) are shown with #. Variation of the data (range) is detailed in Tables S1 and S2.
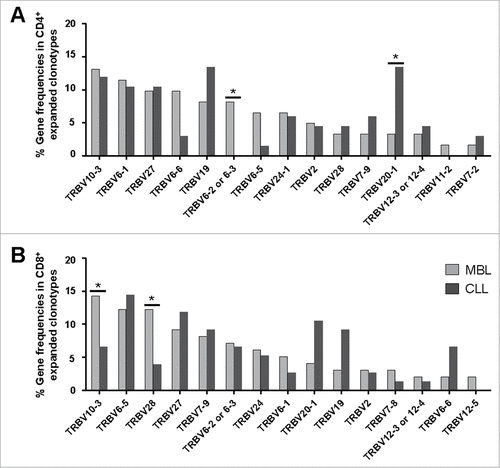
The TRBV gene repertoire of the CD8+ T cell compartment was also skewed. A total of 30 functional genes were identified (Table S2). Similarly to the CD4+ T cell fraction, only a few genes (TRBV6-5, TRBV10-3, TRBV28, TRBV6-2 or 6–3, TRBV27 and TRBV19) amounted for almost half of all clonotypic rearrangements in both MBL and CLL groups. Indeed, when focusing on the expanded clonotypes, the frequencies of some TRBV genes were also different between the two groups. The main differences concerned higher frequencies of the TRBV10-3 and TRBV28 genes in MBL compared to CLL (frequencies: 14.3% vs. 6.6%, P = 0.030 and 12.2% vs. 4.0%, P = 0.024, respectively) (, Table S2).
Interestingly, the expanded clonotype repertoire also exhibited differences in the TRBV gene usage between the CD4+ and the CD8+ T cell fractions (Fig. S1, Tables S1 and S2). In particular, the CD4+ T cell fraction of MBL cases displayed lower TRBV6-5 and TRBV28 gene frequencies compared to the respective CD8+ T cell fraction (frequency: 6.6% vs. 12.2%, P = 0.035 and 3.3% vs. 12.2%, P = 0.012, respectively). Within the CLL group, significant differences were observed concerning TRBV6-5 gene frequencies (CD4+ cells: 1.5%, CD8+ cells: 14.5%, P = 0.045).
When non-progressive MBL subjects (n = 12) and those that had progressed to CLL in the last follow-up (n = 4) were compared, no significant differences in terms of CD4+ and CD8+ T cell clonality or TRBV gene frequencies were detected.
Sequential analysis in MBL cases reveals T cell repertoire drift but also persisting clones
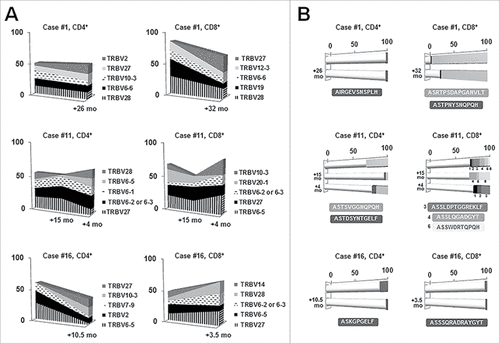
We studied longitudinal samples from three MBL cases to investigate whether the small-sized MBL clones (<5 × 109 cells/L) would persistently affect T cell clonal dynamics. CD4+ and CD8+ T cell samples were analyzed over two sequential time points (median follow-up: 18 months) for two MBL cases and over three sequential time points for another MBL case, after 15 and 19 months.
With the exception of one case where the CD4+ T cell fraction exhibited a relative stable TRBV gene repertoire, fluctuation in TRBV frequencies were detected over time in both CD4+ and CD8+ compartments, suggesting clonal drift (). Regarding clonotype distribution, except for one case, a pronounced clonal drift was observed (). Interestingly, in case #1, a highly expanded immunodominant clonotype of the CD8+ T cell fraction persisted after 32 months (detected frequency at the first and second time point: 85% and 66% respectively). In case #11, the immunodominant clonotype of the CD4+ T cell compartment at the first time point was detected in the other two sequential samples as well, being also the predominant clonotype after 19 months. In the remaining cases, the immunodominant clonotype at each time point differed, although at least one clonotype persisted over time ().
Figure 3. Longitudinal analysis in three selected MBL cases for the CD4+ and CD8+ T cell fractions. Gene frequencies (A) were assessed considering clonotypes whereas clonality (B) was measured considering rearrangements. A, TRBV gene repertoire dynamics over time. Only the five most frequent genes are represented. Sequential time points are indicated in the x-axis whereas the frequency (%) of each gene is shown in the y-axis. B, Clonal fluctuations over time. Each horizontal bar illustrates a different time point. White cylindrical parts of the bars account for the different clonotypes among the distinct time points whereas darker cubic parts represent persistent clonotypes. The frequency (%) of each clonotype is shown along the x-axis. Clonotypes shared by different time points, as well as their CDR3 amino acid sequence, are depicted in the same color.
Shared clonotypes and CDR3 regions between distinct cases, mostly MBL/CLL-specific
We next compared all the obtained TRB CDR3 amino acid sequences across all the MBL and CLL subjects included in the study. Notably, we found two identical TRB CDR3 used by pairs of cases and another one shared by three MBL individuals (). In all but one case, shared CDR3 were also accompanied by identical TRBV genes, thereby consisting of “public” (“stereotyped”) clonotypes. Of note, the nucleotide sequences coding for two of the three shared CDR3 amino acid sequences were different, excluding the possibility of cross-contamination and highlighting the role of antigenic selection at the CDR3 amino acid level. Restricted HLA usage was confirmed between all cases sharing the same CDR3 regions ().
Table 2. Groups of cases that displayed common CDR3 amino acid sequences. Three distinct clonotypes (identical TRBV-TRBJ genes and CDR3 amino acid sequence) were shared between cases #6 and #16, #12 and #22 and #7 and #16. Case #1 showed the same CDR3 but different TRBV gene than cases #7 and #16. A remarkable HLA restriction was observed between cases harboring identical CDR3; similarities are highlighted in bold. #Purity of CD4+ cells: 69%.
Finally, in order to obtain insight into the nature of the selecting antigens, we cross-compared all the clonotypes from our MBL and CLL cases (n = 1,337) and a panel of 5,264 unique and productive TRBV-TRBD-TRBJ rearrangements from several entities obtained from the IMGT/LIGM-DB sequence database or available to the groups involved in the study. Nine hits sharing 100% CDR3 amino acid sequence identity were identified (): (i) a match between one MBL case and a CLL patient belonging to subset #25, both cases displaying the same TRBV gene, thus carrying a shared clonotype; (ii) a CLL case matched with an Epstein-Barr virus-specific T cell clone; (iii) a MBL case matched with a hepatitis C virus specific T cell clone; (iv) the aforementioned MBL case also showed a match with a T cell large granular lymphocyte leukemia patient, although with distinct TRBV genes. The remaining hits, mainly related to immunological disorders, as well as detailed information about CDR3 sequences and TRBV-TRBD-TRBJ genes, are summarized in .
Table 3. Matches of the identified CDR3 regions with other entities. For each case, the first row represents the match identified in the IMGT/LIGM-DB or in the database available to the groups involved in the study and the second row corresponds to the MBL or CLL case from the studied cohort. ND: not detected. #Subsets definitions can be found in the study by Agathangelidis et alCitation5.
When a less restrictive threshold (>85%) for CDR3 amino acid sequence identity was applied, various additional matches were identified, including hits with CDR3 sequences of specific T cell clones in the context of herpes simplex virus-2 infection, multiple sclerosis and cervical intraepithelial neoplasia ().
Discussion
Molecular characterization of the TR confers a powerful tool for the detection of T cell clones potentially involved in immune surveillance and leukemogenesis. Although the T cell compartment was demonstrated to be dysfunctional in CLL,Citation11,Citation12 allowing tumor expansion and disease progression, the indolent clinical courses observed in MBL and early stage CLL may reflect different molecular mechanisms related to a distinct T cell behavior, with potential therapeutic relevance. That said, MBL may represent an early stage of CLL ontogeny, most likely concealing important clues about leukemogenesis. In the present study, we analyzed the T cell repertoire, separately for CD4+ and CD8+ T cell subpopulations, in the largest cohort of MBL and early stage CLL patients in order to gain insight into the role of T cells in CLL evolution.
The CD4+ T cell fraction in CLL exhibited a significantly higher clonality than in MBL. In line with this, CD4+ T cell clonal expansions followed the numerical increase of clonal B cells. These findings suggest that CD4+ T cell repertoire restriction may be influenced by the extent of B cell clonal expansions, occurring early in clonal evolution and increasing concurrently with tumor progression, which possibly alludes to tumor-related selecting antigens. Prior investigations in CLL showed increasing frequencies of CD4+ T cells with regulatory properties (Treg) accompanying tumor development,Citation16,Citation18 contributing to the immunosuppressive microenvironment that allows the neoplastic B cells to proliferate. Interestingly, increased numbers of Treg have been detected in MBL as well, albeit to a lesser extent.Citation17,Citation19 Although the present study had not been designed so as to answer the question whether the progressive increase of CD4+ T cell clonality could be due to Treg expansion, we plan to investigate this by employing high-throughput next-generation sequencing techniques. On the other hand, the fact that CLL cases showed a higher restriction in their T cell repertoire could also reflect the loss of effector T cell clones restraining CLL clonal expansions. In line with this, a recent investigation showed that the Bruton's tyrosine kinase inhibitor ibrutinib, a highly effective new therapy for CLL, increased T cell repertoire diversity.Citation20 This could suggest that eradication of the malignant cells allows T cell immune reconstitution. Whether this reconstitution is accompanied by changes in T cell function, perhaps abrogating tolerance and allowing them to mount cytotoxic responses against the tumor, is not yet clarified. Yet, it would be highly relevant for designing combinations of drugs with the aim of boosting immune responses for sustained tumor growth control.
Concerning the CD8+ T cell fraction, differences in clonality were not detected. However, this could be possibly attributed to the increased extent of clonality in the CD8+ T cell compartment, which may not allow the discrimination of small variations when performing the methodology employed in this study.
Notably, a previous study employing the same methodology did not show T cell oligoclonality in age-matched healthy controls (median cumulative frequency of all expanded T cell clonotypes: 5%),Citation8 which further supports that clonal CD4+ and CD8+ T cell expansions may occur in the presence of clonal B cells and are already detectable in MBL.
Differences in the TRBV gene usage between MBL and CLL groups were also identified, including significantly increased TRBV10-3 and TRBV28 frequencies in the expanded CD8+ T cell repertoire of MBL cases compared to CLL. Along this line, a previous subcloning study found that TRBV10-3 and TRBV28 genes were underrepresented in CLL in comparison to healthy subjects.Citation8 Altogether, these findings provide evidence of progressive modifications in the architecture of the T cell compartment that may occur following clonal B cell expansions.
When CD4+ and CD8+ T cells were compared, the correlation in the CLL group was higher than in MBL. In line with this, prior investigations in healthy subjects demonstrated that CD4+ and CD8+ TRBV diversity is highly individualized but comparable between both T cell fractions for a given person, which suggests that the TRBV repertoire of CD4+ and CD8+ T cells may be shaped in parallel.Citation21 In the context of CLL, this could imply the simultaneous skewing of CD4+ and CD8+ T cells by particular antigenic elements, potentially CLL-specific. The fact that similarities between both T cell fractions were higher in CLL compared to MBL could reflect an increased effect of antigen restriction with the expansion of clonal B cells, which alludes to tumor-related antigens.
We also demonstrated the persistence of some T cell clones over time in all MBL samples longitudinally analyzed. This is in agreement with prior investigations in CLLCitation8,Citation9 and extends the findings to MBL, suggesting that the selecting antigens may persist even from the early stages of CLL ontogeny.
Another important finding was the identification of shared (“public”) clonotypes between different MBL and/or CLL cases. Relevant to mention, the possibility of cross-contamination was essentially discarded due to several reasons, including subcloning experiments performed in different times or laboratories and distinct nucleotide sequences of shared amino acid clonotypes. Since the possibility that unrelated individuals harbor shared clonotypes by chance is extremely low, these public clonotypes may be associated with the pathophysiology of MBL/CLL, either due to recognizing tumor-specific antigens or within the context of immune responses against as yet unidentified infectious agents with a potential relevance in the development of the disease.Citation22,Citation23 These antigenic triggers likely occur very early in the natural history of CLL and may persist along with B cell clonal expansions, which was demonstrated herein by those shared clonotypes between MBL subjects but also between MBL and CLL cases. Of note, these public clonotypes were not detected in public databases, alluding to common antigenic stimulation that may be MBL/CLL-specific. Whether they correspond to the same antigens that are implicated in the selection of the malignant clone remains to be elucidated. Unexpectedly, some of these CDR3 sequences shared between MBL/CLL cases were detected in different T cell fractions. Several explanations can be inferred, such as: (i) the presence of CD4+ cells in the CD8+ cell fraction of the same patient, or vice versa, (ii) double-positive CD4+CD8+ T cells, which are increased in the peripheral blood of the elderlyCitation24 or (iii) potentially, different CD4+ and CD8+ T cells harboring TR with specificity for the same epitopes. In this sense, although it is generally accepted that CD4+ and CD8+ harbor TR that recognize different peptides on class II and class I MHC complexes, respectively, previous investigations suggested that some TR may be capable of reacting with both MHC class I- and class II-bound peptide ligands.Citation21,Citation25 Hence, the potential role of double-positive CD4+CD8+ T cells or switchable TR conformers in MBL/CLL should be further characterized.
As expected, HLA restrictions were identified between cases with shared clonotypes. A bias in HLA usage, mainly concerning the development of severe disease, was reported in CLL.Citation26–28 In addition, a very recent study associated HLA specificities with prognosis in MBL,Citation29 pointing to the existence of protective HLA-restricted T cell interactions involved in the control of tumor expansion. In line with this, two of our MBL cases with a shared TR CDR3 region also shared five of the eight analyzed HLA loci.
Several matches with infectious and immune disorders were also detected. Interestingly, a similar study performed with the same methodology in 58 CLL patients only found one match with a reactive CD8+ Epstein-Barr virus-specific T cell clone.Citation8 The fact that we identified considerably more matches within our MBL and early stage CLL cohort could reflect a potential role of infectious agents and immune alterations in the pathogenesis of the disease. Interestingly, we detected a clonotype in a MBL case that matched a T cell clone found in an early arthritis patient and that persisted after 32 months, which might also be relevant considering several reports that CLL BcR IGs often exhibit rheumatoid factor reactivity.Citation30–34 All these findings suggest that chronic exposure to self or exogenous antigens could trigger immune reactions and processes leading to CLL-like clonal expansions.
Taken together, our results demonstrate T cell oligoclonality in MBL with persisting T cell clones over time and increasing clonality within the CD4+ T cell subpopulation concurrently with the expansion of neoplastic B cells. In addition, the identification of the same clonotypes in different MBL/early stage CLL cases points to selection of the T cell clones by common antigenic elements, very early in the clonal evolution process leading to CLL. The fact that these shared T cell clonotypes were not found in public databases alludes to common antigenic stimulation that may be potentially considered MBL/CLL-specific. Further investigations are necessary to clarify the exact role of these antigens in the pathogenesis of the disease.
Materials and methods
Patient groups
In total, 16 clinical CLL-like MBL (median age: 76, range: 60–83 years) were analyzed. Clinical CLL-like MBL was defined on the basis of an unbalanced κ/λ ratio (>3:1 or <1:3) within CD19+, CD5bright, CD23+ and CD20dim cells determined by flow cytometry and an absolute clonal B cell count of 0.5 to 5 × 109/L without other manifestations of CLL. The median white blood cell (WBC) count of clinical MBL cases was 10.2 × 109/L (range: 5–15.8 × 109/L). Besides, 13 untreated early stage CLL patients were also studied (Binet A/Rai 0; median WBC count: 16.3, range: 11.3–24 × 109/L; median age: 73, range: 59–88 years). Three MBL cases were analyzed over time, as follows: case #1, second CD4+ and CD8+ samples studied after 26 and 32 months, respectively; case #11, second CD4+ and CD8+ samples studied after 15 months, third CD4+ and CD8+ samples studied after 4 months from the second time point; case #16, second CD4+ and CD8+ samples studied after 10.5 and 3.5 months, respectively. In total, 65 peripheral blood samples were analyzed. No case had evidence of infection at sampling. All cases remained asymptomatic and none progressed to CLL requiring therapy (median follow-up: 38 months; range: 3–55 months). Four MBL cases had progressed to CLL (>5 × 109 clonal B cells/L) in the last follow-up. The study was performed in accordance with national and international guidelines (Professional Code of Conduct, Declaration of Helsinki) and approved by the Ethics Committee of Hospital del Mar, Barcelona (2011/4317/I).
PCR amplification of TRBV-TRBD-TRBJ gene rearrangements
In all cases, CD4+ and CD8+ cells were isolated from peripheral blood by positive selection using magnetic beads (Miltenyi Biotec). Median purity of CD4+ cells out of the total number of events analyzed by flow cytometry was 94.9% (range: 67.8%−99.1%), whereas median purity of CD8+ cells out of the total CD3+ cells was 99.4% (range: 97.7%−99.9%). Total cellular RNA was then extracted (RNeasy Plus Mini Kit, QIAGEN) and reverse transcribed to cDNA (Ovation® Pico WTA System V2, NUGEN). PCR of TRBV-TRBD-TRBJ rearrangements was performed according to the BIOMED2 protocol.Citation35 PCR products were gel-purified (QIAquick Gel Extraction Kit, QIAGEN) and then used for ligation into the pCR2.1 vector and transformation into chemically competent E.coli/TOP10F’ (Invitrogen Life Technologies). Colonies were randomly isolated and the insert of interest was subjected to Sanger sequencing using M13 primers.
Sequence analysis, definitions and interpretation
Subcloned sequences were introduced into the IMGT/V-QUEST tool (http://www.imgt.org) from the international ImMunoGeneTics information system for analysis. Only productive TRBV-TRBD-TRBJ rearrangements were considered (n = 2,567).
Clonotypes were defined as unique rearrangements carrying identical TRBV-TRBJ genes and complementary determining region 3 (CDR3) amino acid sequence and were considered expanded (in clusters) within a sample when corresponding to ≥2 sequences. The most expanded clonotype within each sample was referred to as the immunodominant (predominant) clonotype. The relative frequency of each clonotype within a sample was assessed as the number of rearrangements corresponding to that clonotype divided by the total number of productive rearrangements sequenced for that specific sample. For TRBV gene repertoire studies, clonotypes instead of single rearrangements were considered in order to avoid skewing due to T cell expansions after antigenic stimulation. For clonality and TRBV gene repertoire studies, only samples with purity ≥90% were considered.
Comparison to public data
Clonotype comparison across all MBL and CLL individuals was performed. A panel of 5,264 productive, unique and well-annotated TRBV-TRBD-TRBJ rearrangements from T cell clones of different entities (available to groups involved in the study, n = 1,262; or extracted from the IMGT/LIGM-DB sequence database, n = 4,002, http://www.imgt.org/IMGTindex/LGM.html) was also compared with all the clonotypes of this series.
HLA typing
Typing of the HLA-A, -B and -C loci for low/intermediate resolution [CWD (Common Well Defined) level] and HLA-DRB1 locus for high resolution was performed for all of the subjects included in the study by sequence-specific oligonucleotide-PCR (PCR-SSO) by microbeads array (Luminex Technology, One Lambda).
Statistical analysis
Counts, frequency distributions, means, medians and ranges were assessed for quantitative variables. Comparisons among independent groups (MBL vs. CLL) were performed employing the non-parametric Mann-Whitney test, whereas the results from the CD4+ and CD8+ T cell fractions (both belonging to the same subject) were compared using the Wilcoxon test. Simple linear regression analysis and Spearman correlation were calculated to analyze the relationship between T cell clonality (CD4+ or CD8+) and B cell clonal size, or CD4+ T cell clonality and CD8+ T cell clonality. Statistical analyses were performed using SPSS v.22 software (SPSS Inc., Chicago, IL, USA). P-values below 0.05 were considered statistically significant.
Abbreviations
| BcR | = | B cell receptor |
| CDR3 | = | complementary determining region 3 |
| CLL | = | chronic lymphocytic leukemia |
| HLA | = | human leukocyte antigen |
| IG | = | immunoglobulins |
| MBL | = | monoclonal B cell lymphocytosis |
| MHC | = | major histocompatibility complex |
| TR | = | T cell receptor |
| TRB | = | T cell receptor β chain |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
koni_a_1432328_sm9394.pdf
Download PDF (330.6 KB)Additional information
Funding
References
- Rawstron AC, Bennett FL, O'Connor SJ, Kwok M, Fenton JA, Plummer M, de Tute R, Owen RG, Richards SJ, Jack AS, Hillmen P. Monoclonal B lymphocytosis and chronic lymphocytic leukemia. N Engl J Med. 2008;359:575–83. doi:10.1056/NEJMoa075290. PMID:18687638.
- Vardi A, Dagklis A, Scarfò L, Jelinek D, Newton D, Bennett F, Almeida J, Rodriguez-Caballero A, Allgood S, Lanasa M, et al. Immunogenetics shows that not all MBL are equal: the larger the clone, the more similar to CLL. Blood. 2013;121:4521–8. doi:10.1182/blood-2012-12-471698. PMID:23596047.
- Strati P, Shanafelt TD. Monoclonal B cell lymphocytosis and early stage chronic lymphocytic leukemia: diagnosis, natural history, and risk stratification. Blood. 2015;126:454–62. doi:10.1182/blood-2015-02-585059. PMID:26065657.
- Fais F, Ghiotto F, Hashimoto S, Sellars B, Valetto A, Allen SL, Schulman P, Vinciguerra VP, Rai K, Rassenti LZ, et al. Chronic lymphocytic leukemia B cells express restricted sets of mutated and unmutated antigen receptors. J Clin Invest. 1998;102:1515–25. doi:10.1172/JCI3009. PMID:9788964.
- Agathangelidis A, Darzentas N, Hadzidimitriou A, Brochet X, Murray F, Yan XJ, Davis Z, van Gastel-Mol EJ, Tresoldi C, Chu CC, et al. Stereotyped B cell receptors in one-third of chronic lymphocytic leukemia: a molecular classification with implications for targeted therapies. Blood. 2012;119:4467–75. doi:10.1182/blood-2011-11-393694. PMID:22415752.
- Agathangelidis A, Vardi A, Baliakas P, Stamatopoulos K. Stereotyped B cell receptors in chronic lymphocytic leukemia. Leuk Lymphoma. 2014;55:2252–61. doi:10.3109/10428194.2013.879715. PMID:24397617.
- Stamatopoulos K, Agathangelidis A, Rosenquist R, Ghia P. Antigen receptor stereotypy in chronic lymphocytic leukemia. Leukemia. 2017;31:282–291. doi:10.1038/leu.2016.322. PMID:27811850.
- Vardi A, Agathangelidis A, Stalika E, Karypidou M, Siorenta A, Anagnostopoulos A, Rosenquist R, Hadzidimitriou A, Ghia P, Sutton LA, Stamatopoulos K. Antigen selection shapes the T cell repertoire in chronic lymphocytic leukemia. Clin Cancer Res. 2016;22:167–74. doi:10.1158/1078-0432.CCR-14-3017. PMID:26338994.
- Vardi A, Vlachonikola E, Karypidou M, Stalika E, Bikos V, Gemenetzi K, Maramis C, Siorenta A, Anagnostopoulos A, Pospisilova S, et al. Restrictions in the T cell repertoire of chronic lymphocytic leukemia: high-throughput immunoprofiling supports selection by shared antigenic elements. Leukemia. 2017;31(7):1555–1561. doi:10.1038/leu.2016.362. PMID:27904140.
- Yan XJ, Dozmorov I, Li W, Yancopoulos S, Sison C, Centola M, Jain P, Allen SL, Kolitz JE, Rai KR, et al. Identification of outcome-correlated cytokine clusters in chronic lymphocytic leukemia. Blood. 2011;118:5201–10. doi:10.1182/blood-2011-03-342436. PMID:21911837.
- Görgün G, Holderried TA, Zahrieh D, Neuberg D, Gribben JG. Chronic lymphocytic leukemia cells induce changes in gene expression of CD4 and CD8 T cells. J Clin Invest. 2005;115:1797–805. doi:10.1172/JCI24176. PMID:15965501.
- Palma M, Gentilcore G, Heimersson K, Mozaffari F, Näsman-Glaser B, Young E, Rosenquist R, Hansson L, Österborg A, Mellstedt H. T cells in chronic lymphocytic leukemia display dysregulated expression of immune checkpoints and activation markers. Haematologica. 2017;102:562–572. doi:10.3324/haematol.2016.151100. PMID:27927767.
- Rezvany MR, Jeddi-Tehrani M, Osterborg A, Kimby E, Wigzell H, Mellstedt H. Oligoclonal TCRBV gene usage in B chronic lymphocytic leukemia: major perturbations are preferentially seen within the CD4 T cell subset. Blood. 1999;94:1063–9. PMID:10419899.
- Bagnara D, Kaufman MS, Calissano C, Marsilio S, Patten PE, Simone R, Chum P, Yan XJ, Allen SL, Kolitz JE, et al. A novel adoptive transfer model of chronic lymphocytic leukemia suggests a key role for T lymphocytes in the disease. Blood. 2011;117:5463–72. doi:10.1182/blood-2010-12-324210. PMID:21385850.
- Ramsay AG, Clear AJ, Fatah R, Gribben JG. Multiple inhibitory ligands induce impaired T cell immunologic synapse function in chronic lymphocytic leukemia that can be blocked with lenalidomide: establishing a reversible immune evasion mechanism in human cancer. Blood. 2012;120:1412–21. doi:10.1182/blood-2012-02-411678. PMID:22547582.
- Jadidi-Niaragh F, Yousefi M, Memarian A, Hojjat-Farsangi M, Khoshnoodi J, Razavi SM, Jeddi-Tehrani M, Shokri F. Increased frequency of CD8+ and CD4+ regulatory T cells in chronic lymphocytic leukemia: association with disease progression. Cancer Invest. 2013;31:121–31. doi:10.3109/07357907.2012.756110. PMID:23286587.
- Rissiek A, Schulze C, Bacher U, Schieferdecker A, Thiele B, Jacholkowski A, Flammiger A, Horn C, Haag F, Tiegs G, et al. Multidimensional scaling analysis identifies pathological and prognostically relevant profiles of circulating T cells in chronic lymphocytic leukemia. Int J Cancer. 2014;135:2370–9. doi:10.1002/ijc.28884. PMID:24723150.
- Dasgupta A, Mahapatra M, Saxena R. Flow cytometric immunophenotyping of regulatory T cells in chronic lymphocytic leukemia: comparative assessment of various markers and use of novel antibody panel with CD127 as alternative to transcription factor FoxP3. Leuk Lymphoma. 2013;54:778–89. doi:10.3109/10428194.2012.730614. PMID:22989355.
- D'Arena G, Rossi G, Minervini MM, Savino L, D'Auria F, Laurenti L, Del Principe MI, Deaglio S, Biagi A, De Martino L, et al. Circulating regulatory T cells in “clinical” monoclonal B lymphocytosis. Int J Immunopathol Pharmacol. 2011;24:915–23. doi:10.1177/039463201102400410. PMID:22230398.
- Yin Q, Sivina M, Robins H, Yusko E, Vignali M, O'Brien S, Keating MJ, Ferrajoli A, Estrov Z, Jain N, et al. Ibrutinib therapy increases T cell repertoire diversity in patients with chronic lymphocytic leukemia. J Immunol. 2017;198:1740–7. doi:10.4049/jimmunol.1601190. PMID:28077600.
- Li HM, Hiroi T, Zhang Y, Shi A, Chen G, De S, Metter EJ, Wood WH, 3rd, Sharov A, Milner JD, et al. TCRβ repertoire of CD4+ and CD8+ T cells is distinct in richness, distribution, and CDR3 amino acid composition. J Leukoc Biol. 2016;99:505–13. doi:10.1189/jlb.6A0215-071RR. PMID:26394815.
- Fazi C, Dagklis A, Cottini F, Scarfò L, Bertilaccio MT, Finazzi R, Memoli M, Ghia P. Monoclonal B cell lymphocytosis in hepatitis C virus infected individuals. Cytometry B Clin Cytom. 2010;78 Suppl 1:S61–8. doi:10.1002/cyto.b.20545. PMID:20839339.
- Criado I, Muñoz-Criado S, Rodríguez-Caballero A, Nieto WG, Romero A, Fernández-Navarro P, Alcoceba M, Contreras T, González M, Orfao A, Almeida J, Primary Health Care Group of Salamanca for the Study of MBL. Host virus and pneumococcus-specific immune responses in high-count monoclonal B lymphocytosis and chronic lymphocytic leukemia: implications for disease progression. Haematologica. 2017;102:1238–46. doi:10.3324/haematol.2016.159012. PMID:28385786.
- Ghia P, Prato G, Stella S, Scielzo C, Geuna M, Caligaris-Cappio F. Age-dependent accumulation of monoclonal CD4+CD8+ double positive T lymphocytes in the peripheral blood of the elderly. Br J Haematol. 2007;139:780–90. doi:10.1111/j.1365-2141.2007.06867.x. PMID:18021092.
- Yin L, Huseby E, Scott-Browne J, Rubtsova K, Pinilla C, Crawford F, Marrack P, Dai S, Kappler JW. A single T cell receptor bound to major histocompatibility complex class I and class II glycoproteins reveals switchable TCR conformers. Immunity. 2011;35:23–33. doi:10.1016/j.immuni.2011.04.017. PMID:21683626.
- Gragert L, Fingerson S, Albrecht M, Maiers M, Kalaycio M, Hill BT. Fine-mapping of HLA associations with chronic lymphocytic leukemia in US populations. Blood. 2014;124:2657–65. doi:10.1182/blood-2014-02-558767. PMID:25232063.
- Shah N, Decker WK, Lapushin R, Xing D, Robinson SN, Yang H, Parmar S, Tung SS, O'Brien S, Fernandez-Viña M, et al. HLA homozygosity and haplotype bias among patients with chronic lymphocytic leukemia: implications for disease control by physiological immune surveillance. Leukemia. 2011;25:1036–9. doi:10.1038/leu.2011.30. PMID:21350559.
- Di Bernardo MC, Broderick P, Harris S, Dyer MJ, Matutes E, Dearden C, Catovsky D, Houlston RS. Risk of developing chronic lymphocytic leukemia is influenced by HLA-A class I variation. Leukemia. 2013;27:255–8. doi:10.1038/leu.2012.173. PMID:22814293.
- García-Álvarez M, Alcoceba M, López-Parra M, Puig N, Antón A, Balanzategui A, Prieto-Conde I, Jiménez C, Sarasquete ME, Chillón MC, et al. HLA specificities are associated with prognosis in IGHV-mutated CLL-like high-count monoclonal B cell lymphocytosis. PLoS One. 2017;12:e0172978. doi:10.1371/journal.pone.0172978. PMID:28249016.
- Kostareli E, Gounari M, Janus A, Murray F, Brochet X, Giudicelli V, Pospisilova S, Oscier D, Foroni L, di Celle PF, et al. Antigen receptor stereotypy across B cell lymphoproliferations: the case of IGHV4-59/IGKV3-20 receptors with rheumatoid factor activity. Leukemia. 2012;26:1127–31. doi:10.1038/leu.2011.311. PMID:22051533.
- Lanemo Myhrinder A, Hellqvist E, Sidorova E, Söderberg A, Baxendale H, Dahle C, Willander K, Tobin G, Bäckman E, Söderberg O, et al. A new perspective: molecular motifs on oxidized LDL, apoptotic cells, and bacteria are targets for chronic lymphocytic leukemia antibodies. Blood. 2008;111:3838–48. doi:10.1182/blood-2007-11-125450. PMID:18223168.
- Chu CC, Catera R, Hatzi K, Yan XJ, Zhang L, Wang XB, Fales HM, Allen SL, Kolitz JE, Rai KR, Chiorazzi N. Chronic lymphocytic leukemia antibodies with a common stereotypic rearrangement recognize nonmuscle myosin heavy chain IIA. Blood. 2008;112:5122–9. doi:10.1182/blood-2008-06-162024. PMID:18812466.
- Borche L, Lim A, Binet JL, Dighiero G. Evidence that chronic lymphocytic leukemia B lymphocytes are frequently committed to production of natural autoantibodies. Blood. 1990;76:562–9. PMID:2378986.
- Sthoeger ZM, Wakai M, Tse DB, Vinciguerra VP, Allen SL, Budman DR, Lichtman SM, Schulman P, Weiselberg LR, Chiorazzi N. Production of autoantibodies by CD5-expressing B lymphocytes from patients with chronic lymphocytic leukemia. J Exp Med. 1989;169:255–68. doi:10.1084/jem.169.1.255. PMID:2462608.
- van Dongen JJ, Langerak AW, Brüggemann M, Evans PA, Hummel M, Lavender FL, Delabesse E, Davi F, Schuuring E, García-Sanz R, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17:2257–317. doi:10.1038/sj.leu.2403202. PMID:14671650
