ABSTRACT
Inhibitors targeting the PDCD1 (programmed cell death 1, PD-1) immune checkpoint pathway have revolutionized cancer treatment strategies. The TIME (Tumor Immunity in the MicroEnvironment) classification based on tumor CD274 (PDCD1 ligand 1, PD-L1) expression and tumor-infiltrating lymphocytes (TIL) has been proposed to predict response to immunotherapy. It remains to be determined clinical, pathological, and molecular features of TIME subtypes of colorectal cancer. Using 812 colon and rectal carcinoma cases from the Nurses' Health Study and Health Professionals Follow-up Study, we examined the association of tumor characteristics and survival outcomes with four TIME subtypes (TIME 1, CD274low/TILabsent; TIME 2, CD274high/TILpresent; TIME 3, CD274low/TILpresent; and TIME 4, CD274high/TILabsent). In survival analyses, Cox proportional hazards models were adjusted for potential confounders, including microsatellite instability (MSI) status, CpG island methylator phenotype (CIMP) status, LINE-1 methylation level, and KRAS, BRAF, and PIK3CA mutation status. TIME subtypes 1, 2, 3 and 4 had 218 (27%), 117 (14%), 103 (13%), and 374 (46%) colorectal cancer cases, respectively. Compared with TIL-absent subtypes (TIME 1 and 4), TIL-present subtypes (TIME 2 and 3) were associated with high-level MSI, high-degree CIMP, BRAF mutation, and higher amounts of neoantigens (p < 0.001). TIME subtypes were not significantly associated with colorectal cancer-specific or overall survival. In conclusion, TIL-present TIME subtypes of colorectal cancer are associated with high levels of MSI and neoantigen load, supporting better responsiveness to cancer immunotherapy. Further studies examining tumor molecular alterations and additional factors in the tumor microenvironment may inform development of immunoprevention and immunotherapy strategies.
Introduction
Immunotherapy has emerged in recent years as an attractive therapeutic modality in cancer management.Citation1-Citation3 In particular, immune checkpoint inhibitors that block the PDCD1 (programmed cell death 1, PD-1) or CD274 (PDCD1 ligand 1, PD-L1) protein have shown great promise in treating various malignancies with durable clinical remissions.Citation4-Citation8 How to effectively identify patients who would derive clinical benefits from immune checkpoint blockade therapy has therefore become a clinical question of paramount importance.
The TIME (Tumor Immunity in the MicroEnvironment) classification system has been proposed as a first-step framework to predict immunotherapeutic response based on cross-classified levels of tumor CD274 (PD-L1) expression (low vs. high) and tumor-infiltrating lymphocytes (TIL, absent vs. present).Citation9-Citation12 However, how this proposed scheme may correlate with tumor molecular features and clinical outcomes in general and in colorectal cancer remains to be determined. The CD274 protein expressed on cancer cells impairs T cell-mediated tumor-specific immune response by binding to PDCD1 (PD-1) on T cells.Citation5,Citation7 In colorectal cancer, tumor CD274 expression has been inversely correlated with FOXP3+ regulatory T cells in tumor, suggesting mutually exclusive immunosuppressive mechanisms.Citation13 Immune reaction plays a key role in suppressing tumor development and progression,Citation14-Citation16 and high-level infiltrates of lymphocytes in colorectal cancer have been associated with better clinical outcomes.Citation17-Citation22 Four tumor subtypes can be created based on the TIME framework,Citation9-Citation12 including the CD274high/TILpresent subtype (TIME 2) which generally responds to immunotherapy, the CD274low/TILpresent subtype (TIME 3) which suggests presence of other suppressor pathways in immune tolerance, the CD274high/TILabsent subtype (TIME 4) which indicates intrinsic induction of the CD274 pathway, and the CD274low/TILabsent subtype (TIME 1) which reflects immune “ignorance.”
The TIME model has been developed based on the findings in melanoma,Citation9 and has been applied to other tumor types.Citation23-Citation26 The prevalence and clinical implications of different TIME subtypes likely vary by tumor-specific genetic aberrations and oncogenic drivers as well as other factors defining the tumor microenvironment.Citation26,Citation27 In colorectal cancer, it is known that high-level microsatellite instability (MSI) and resultant frameshift mutations are associated with abundant immunogenic peptides (“neoantigens”), leading to increased likelihood of clinical benefits from immune checkpoint blockade.Citation8,Citation28-Citation30 The carcinogenesis of colorectal carcinoma, however, is by itself a fairly heterogeneous process which involves stepwise accumulation of multiple genetic and epigenetic aberrations, as well as complex interactions with environmental exposures and host features.Citation31-Citation34 Phenotypic profiling of colorectal carcinomas that would respond to immunotherapy would therefore likely require consideration of multiple factors in association with TIME categories to precisely predict tumor immune resistance.Citation35,Citation36
Using a molecular pathological epidemiology database derived from two large prospective cohort studies in the U.S., we examined the association of TIME subtypes of colorectal cancer with clinical, pathological, and molecular characteristics, and patient survival.
Results
Among 812 colorectal carcinoma cases with available data on tumor CD274 (PD-L1) expression status and TIL, TIME subtypes 1, 2, 3 and 4 had 218 (27%), 117 (14%), 103 (13%), and 374 (46%) cases, respectively. summarizes clinical, pathological, and molecular characteristics of colorectal cancer cases according to TIME subtypes. Compared with TIL-absent subtypes (TIME 1 and 4), TIL-present subtypes (TIME 2 and 3) were statistically significantly associated with tumor location at the proximal colon, high-level MSI, high-degree CpG island methylator phenotype (CIMP), high-level long interspersed nucleotide element-1 (LINE-1) methylation, BRAF mutation, negative nuclear CTNNB1 expression, and high neoantigen load (p < 0.001 with adjusted α level of 0.003). Interestingly, TIL-present subtypes were more likely to have more poorly-differentiated tumors but lower disease stage (p < 0.003). There were no significant differences in characteristics by tumor CD274 expression status in strata of levels of TIL (p > 0.01 with adjusted α level of 0.003).
Table 1. Clinical, pathological, and molecular characteristics of colorectal cancer cases according to TIME (Tumor Immunity in the MicroEnvironment) subtypes based on tumor CD274 (PD-L1) expression status and tumor-infiltrating lymphocytes (TIL).
We also examined the association of TIME subtypes with colorectal cancer survival. During the median follow-up time of 12.1 years (interquartile range, 8.4–16.1 years) for all censored cases, there were 479 all-cause deaths, including 247 colorectal cancer-specific deaths. shows Kaplan-Meier survival curves of colorectal cancer cases by TIME subtypes. In multivariable Cox regression analyses, TIME subtypes were not statistically significantly associated with colorectal cancer-specific or overall survival ( and Table S1). Compared with TIME subtype 1, multivariable-adjusted hazard ratios for colorectal cancer-specific mortality were 0.61 [95% confidence interval (CI), 0.37–1.01] for subtype 2, 0.76 (95% CI, 0.46–1.25) for subtype 3, and 0.93 (95% CI, 0.69–1.26) for subtype 4. We did not observe a statistically significant interaction between tumor CD274 expression status and TIL in relation to cancer-specific or overall survival (p interaction > 0.67).
Figure 1. Kaplan-Meier survival curves of colorectal cancer patients according to TIME (Tumor Immunity in the MicroEnvironment) subtypes based on tumor CD274 (PD-L1) expression status and tumor-infiltrating lymphocytes (TIL). The p values were calculated using the log-rank test (two-sided). (A), colorectal cancer-specific survival; (B), overall survival.
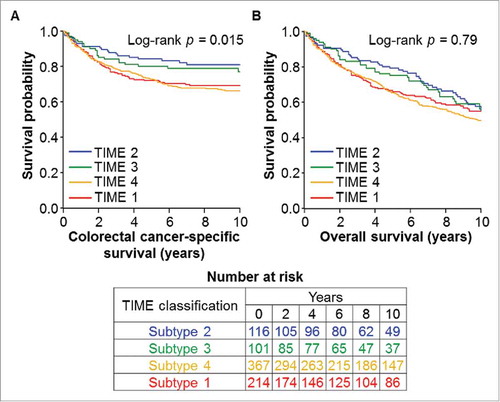
Table 2. Colorectal cancer survival according to TIME (Tumor Immunity in the MicroEnvironment) subtypes based on tumor CD274 (PD-L1) expression status and tumor-infiltrating lymphocytes (TIL).
In exploratory secondary analyses, characteristics and survival of colorectal carcinoma cases were evaluated according to a modified TIME classification scheme defined by tumor CD274 expression status and intratumoral periglandular reaction. The analyses did not yield significant differences in associations (Tables S2 and S3). When modified TIME subtypes were defined by tumor CD274 expression status and density of CD3+ cells in tumor tissue (Tables S4 and S5), no significant differences across TIME subtypes in association with colorectal cancer characteristics or survival were observed.
Discussion
We conducted this study based on two large prospective cohorts to examine the association of TIME subtypes with characteristics and survival outcomes of colorectal cancer. Our findings suggest that TIL-present TIME subtypes are more likely to present with high levels of MSI, CIMP, and neoantigens, and less likely to present with LINE-1 hypomethylation and nuclear CTNNB1 expression when compared with TIL-absent subtypes. There were no significant differences in colorectal cancer-specific or overall survival by TIME subtypes, which may be supported by our previous findings of the lack of a prognostic role of tumor CD274 (PD-L1) expression status in colorectal cancer.Citation13 No statistically significant interaction was noted between tumor CD274 expression and TIL in relation to patient survival.
TIL has been demonstrated in multiple tumor types to reflect local immune effector response, and, along with regulatory T cells, has generally been associated with improved survival in colorectal carcinomas.Citation17,Citation37,Citation38 Tumor CD274 (PD-L1) expression was previously shown to inversely associate with FOXP3+ regulatory T cells, but not with CD3+ pan-T cells or CD8+ cytotoxic T cells in colorectal cancer,Citation13 suggesting the potential influence of CD274-expressing cells on the tumor microenvironment via immune regulation. The TIME classification framework based on tumor CD274 expression status and TIL was hence proposed as a pragmatic approach to predict the immune resistance of tumors.Citation9-Citation12 Evidence suggests that the CD274 protein can be selectively expressed on tumor cells in response to proinflammatory cytokine IFNG (interferon-gamma) released from activated antigen-specific CD8+ T cells,Citation6,Citation9,Citation36 rather than through oncogenic pathways. In TIME 2 (CD274high/TILpresent) tumors containing abundant TIL, the CD274-PDCD1 pathway is adaptively activated as negative feedback to help cancer cells evade immune attack; therefore, these tumors are believed to represent the group that would largely benefit from immune checkpoint blockade therapy. TIME 3 (CD274low/TILpresent) tumors, while harboring high-level TIL, utilize tumor suppressive pathways other than the CD274-PDCD1 axis, and hence, might better respond to non-CD274 immune checkpoint inhibitors. TIME 1 and 4 subtypes lack intrinsic TIL and may therefore require complementary therapeutic strategies to recruit lymphocytes into local tumors to increase efficacy of immune checkpoint inhibitors, such as via combination CTLA4 blockade or radiotherapy to induce T cell response.Citation39-Citation41 Application of the TIME classification needs to be interpreted in the context of other variables defining the tumor microenvironment for precise disease management and response prediction in the era of precision medicine. In this study, we sought to evaluate the implications of TIME subtypes on colorectal cancer in association with patient survival as well as tumor histopathologic and molecular features.
Integrated analyses of tumor, immunity, and microenvironment including the microbiota are important.Citation16,Citation42-Citation45 While the TIME model does not correlate significantly with survival outcomes, our findings highlight a distinct molecular profile correlated with TIL-present TIME subtypes regardless of tumor CD274 expression status. TIL-present TIME subtypes were shown to be associated with presence of BRAF mutation, as well as high levels of MSI and neoantigens, which have been among the most well-documented predictive factors for tumor CD274 expression and/or response to immune checkpoint inhibitors regardless of primary organ site.Citation8,Citation30,Citation46 Of note, a smaller proportion of CD274high/TILpresent subtype showed positive nuclear CTNNB1 (beta-catenin) expression when compared to CD274low/TILpresent subtype, although both subtypes were associated with high-level MSI. Nuclear expression or accumulation of CTNNB1 has been associated with more aggressive cancer behavior and suppression of anti-tumor immune response.Citation47,Citation48 Its presence in a greater proportion of CD274low/TILpresent TIME subtype could reflect potential mutations in the WNT signaling pathway or tumor-intrinsic activated WNT signaling which could contribute in part to no response to immunotherapy due to immune resistance and/or carcinogenic effects.Citation47,Citation48 Another observation of interest is that PTGS2 (cyclooxygenase-2) overexpression was commonly observed across TIME subtypes. PTGS2 produces inflammatory mediator prostaglandin E2, which contributes to immunosuppressive tumor microenvironment via recruitment of myeloid-derived suppressor cells and suppression of tumor-specific T cells.Citation49,Citation50 Intriguingly, experimental and clinical evidence supports a synergistic effect of PTGS suppression and immune checkpoint blockade on stimulating T cell-mediated anti-tumor immune response,Citation49,Citation51 suggesting potential benefit of considering PTGS suppression combined with CD274-PDCD1 immune checkpoint blockade.
Our findings would inform further studies to elucidate the associations between TIME subtypes and other parameters within the tumor microenvironment to better tailor combination immunotherapies. Additional analyses with other local immune effector cells (e.g., memory and regulatory T cells, tumor-associated macrophagesCitation52), as well as their densities and spatial distribution in relation to tumor invasive fronts, would enrich the TIME model in predicting tumor immune deficits and resistance. Sequencing-based assessment of intratumoral genetic heterogeneity of tumors at primary and metastatic sites in relation to CD274 expression and TIL would also facilitate understanding of the tumor microenvironment and its implications on therapeutic options. Other factors that could influence tumor recruitment and extravasation of immune effector cells to tumor sites, such as the gut microbiome, tumor vasculature, and related expression of adhesion molecules, would also warrant further investigations to correlate with TIME subtypes and their prognosis.Citation15
Our study has notable strengths including the use of a molecular pathological epidemiology database derived from two U.S. prospective cohort studies with long duration of follow-up.Citation53,Citation54 Integrated data on tumor molecular characteristics and pathological findings allowed us to comprehensively characterize TIME subtypes of colorectal cancer. Of note, our study population was derived from a large number of cases from hospitals located throughout the U.S., contributing to increased generalizability of our findings.
Limitations should be considered in our study. While we cannot entirely exclude the possibility of potential unmeasured or residual confounding in survival analyses, we collected detailed data on a comprehensive panel of colorectal cancer characteristics and evaluated them by multivariable models to control for potential confounding. Our study was also limited in information on cancer treatments. This lack of data, however, was unlikely to differ substantially by tumor CD274 expression or TIL levels as such information would not have been available for decision-making in management upfront. Our study was also based on relatively selected populations as most participants were non-Hispanic health professionals; therefore, our findings would need to be validated in independent cohorts. In addition, due to cellular structural changes caused by tissue processing and lack of standardized antibodies for CD274,Citation10,Citation55 assessment of tumor CD274 expression status and TIL in tissue samples could pose challenges that might have resulted in potential misclassifications. Intratumoral spatial heterogeneity and inter-observer variability in evaluating tumor CD274 expression and TIL might also lead to misclassifications.Citation10,Citation55 Our previous studies on that regard, however, did not reveal considerable intratumoral heterogeneity in CD274 expression in whole tissue sections,Citation13 and showed reasonable agreement between two pathologists in the assessment of tumor CD274 expression status and TIL.Citation13,Citation17 Finally, tumor CD274 expression and TIL were scored on only one sample per participant in this study. As immunotherapies are commonly used to treat refractory advanced tumors, changes in tumor CD274 expression and TIL along the clinical course might occurCitation14,Citation55; as such, assessment of TIME subtypes with biopsies at sites of progression in relation to clinical course may be relevant to update subtype designation in guiding immunotherapeutic treatments.
In summary, our findings suggest distinctive pathologic and molecular characteristics of colorectal cancer associated with subtypes defined by the TIME classification. Consistent with prior literature, our data support the role of TIL as an important effector in tumor-immune interactions. Our findings would likely inform future studies to better understand tumor-immune microenvironment of colorectal carcinomas in the era of immunotherapy.
Patients and methods
Study population
We utilized two prospective cohort studies in the U.S., the Nurses' Health Study (NHS, 121,701 women aged 30–55 years followed since 1976) and the Health Professionals Follow-up Study (HPFS, 51,529 men aged 40–75 years followed since 1986).Citation56 Participants are followed with biennial questionnaires on lifestyle factors and newly-diagnosed diseases including colorectal cancer. The response rate has been more than 90% for each follow-up questionnaire in both cohorts. In both studies, the National Death Index was used to ascertain deaths of participants and to identify unreported lethal colorectal cancer cases. Study physicians, who were blinded to exposure data, reviewed medical records of identified colorectal cancer cases to confirm the disease diagnosis and to collect data on tumor clinical characteristics including tumor size, anatomical location, and disease stage.
Among participants diagnosed with colorectal cancer until 2012, we analyzed 812 cases with available data on tumor CD274 (PD-L1) expression status and TIL in tissue samples. We included both colon and rectal carcinomas based on the colorectal continuum model.Citation57,Citation58 Participants with a history of inflammatory bowel disease or cancer (except for non-melanoma skin cancer) were excluded from this study. Participants were followed until death or the end of follow-up (30 June 2014 for the NHS; and 1 January 2014 for the HPFS), whichever came first.
Informed consent was obtained from all participants at enrollment. This study was approved by the institutional review boards at Harvard T.H. Chan School of Public Health and Brigham and Women's Hospital (Boston, MA, USA).
Histopathologic evaluation of colorectal cancer
Formalin-fixed paraffin-embedded tumor tissue blocks were collected from hospitals throughout the U.S. where colorectal cancer patients had undergone surgical resection. Hematoxylin and eosin-stained tissue sections were examined by a pathologist (S.O.) who was blinded to other data. Tumor differentiation was categorized as well to moderate vs. poor (> 50% vs. ≤ 50% gland formation, respectively). Lymphocytic reaction to tumor was histopathologically evaluated, as previously described.Citation17 TIL was defined as lymphocytes on top of cancer cells (Fig. S1). Intratumoral periglandular reaction was defined as lymphocytic reaction in intratumoral stroma. Each lymphocytic reaction pattern was graded as negative/low, intermediate, or high. Lymphocytic reaction patterns in a subset of cases were independently reviewed by a second pathologist (J.N.G.) with a good inter-observer correlation, as previously described.Citation17 In the present study, TIL was categorized into absent (negative/low) vs. present (intermediate to high), and intratumoral periglandular reaction was categorized into low (negative/low to intermediate) vs. high.
Immunohistochemical evaluation
We constructed tissue microarrays of colorectal cancer cases with sufficient tissue materials, including up to four tumor cores approximately 600 µm in diameter from each case in one tissue microarray block.Citation59 Immunohistochemical study for CD274 (PD-L1) was performed using an anti-CD274 antibody (dilution, 1:50; eBioscience, San Diego, CA, USA; Fig. S2). As previously described,Citation13,Citation51 a single pathologist (Y.M.) scored overall tumor CD274 expression level as an ordinal scale of 0–4 by summing cytoplasmic intensity score [absent (0), weak (1), moderate (2), or strong (3)] and membrane expression score [absent (0) or present (1; if distinct membrane staining above cytoplasmic expression level existed)]. When the staining intensity was different across tumor cores in the same case, predominant staining pattern in tumor cells was recorded. CD274 expression in selected tumors (n = 148) was independently examined by a second pathologist (A.dS.), and the concordance between the two observers was reasonable with a weighted κ of 0.65 (95% CI, 0.57–0.73).Citation13 We categorized CD274 levels as low (scale of 0 to 1) vs. high (scale of 2 to 4), as consistent with our previous study.Citation51
As previously described,Citation59,Citation60 immunohistochemical analyses for PTGS2 (cyclooxygenase-2) and nuclear CTNNB1 (beta-catenin) expression were performed using an anti-PTGS2 antibody (dilution, 1:300; Cayman Chemical, Ann Arbor, MI, USA) and anti-CTNNB1 antibody (dilution, 1:400; BD Transduction Laboratories, Franklin Lakes, NJ, USA), respectively. We measured densities (cells/mm2) of CD3+ cells in colorectal cancer tissue, based on immunohistochemistry using an anti-CD3 antibody (dilution, 1:250; Dako Cytomation, Carpinteria, CA, USA) and image analysis using an automated scanning microscope and the Ariol image analysis system (Genetix, San Jose, CA, USA), as previously described.Citation18
Evaluation of tumor molecular characteristics
DNA was extracted from colorectal cancer tissue in archival formalin-fixed paraffin-embedded tissue sections using QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany). MSI status was analyzed using 10 microsatellite markers (D2S123, D5S346, D17S250, BAT25, BAT26, BAT40, D18S55, D18S56, D18S67, and D18S487). MSI-high was defined as presence of instability in ≥ 30% of the markers, and non-MSI-high as instability in < 30% of the markers, as previously described.Citation61 Using bisulfite-treated DNA, methylation status of eight CIMP-specific promoters (CACNA1G, CDKN2 A, CRABP1, IGF2, MLH1, NEUROG1, RUNX3, and SOCS1) and LINE-1 was determined, as previously described.Citation40,Citation41 CIMP-high was defined as ≥ 6 methylated promoters of eight promoters, and CIMP-low/negative as 0–5 methylated promoters, as previously described.Citation62 Polymerase chain reaction and pyrosequencing were performed for KRAS (codons 12, 13, 61, and 146),Citation63 BRAF (codon 600),Citation61 and PIK3CA (exons 9 and 20).Citation64 Neoantigen load, the number of immunogenic peptides, was predicted by using a neoantigen prediction pipeline for somatic mutations based on whole-exome sequencing and identifying peptides that bind to personal HLA molecules with high affinity (< 500 nM), as previously described.Citation29 Using NetMHCpan (version 2.4),Citation65 we predicted the binding affinities of all possible 9- and 10-mer mutant peptides to the corresponding HLA alleles inferred by the POLYSOLVER algorithm.
Definitions of TIME (Tumor Immunity in the MicroEnvironment) subtypes
TIME subtypes of colorectal carcinoma were assessed as the primary outcome in cross-sectional analyses with tumor characteristics, and as the primary exposure in survival analyses with colorectal cancer survival outcomes. As described previously,Citation9-Citation12 TIME subtypes were defined based on tumor CD274 (PD-L1) expression status (low vs. high) and TIL (absent vs. present): TIME 1, CD274 expression-low and TIL-absent; TIME 2, CD274 expression-high and TIL-present; TIME 3, CD274 expression-low and TIL-present; and TIME 4, CD274 expression-high and TIL-absent.
Exploratory secondary analyses with modified TIME classification schemes were also performed with assessment of intratumoral periglandular reaction (low vs. high), as well as density of CD3+ cells (dichotomized by median value), instead of TIL as markers for tumor immune status.
Statistical analyses
All statistical analyses were performed using SAS software (version 9.4; SAS Institute, Cary, NC, USA), and all p values were two-sided.
We used the chi-square test for categorical variables, and analysis of variance for continuous variables to compare tumor characteristic across TIME subtypes. The α level was set at 0.003 (≈ 0.05/17) with Bonferroni correction to adjust for multiple hypothesis testing. In subgroup analyses, results were interpreted cautiously in addition to the use of adjusted α level of 0.003.
Cumulative survival probabilities were estimated using the Kaplan-Meier method and compared using the log-rank test. Deaths from other causes were dealt as censored. Univariable and multivariable Cox proportional hazards models were used to evaluate the association of TIME subtypes with colorectal cancer-specific and overall mortality. Covariates assessed as potential confounders included sex (female vs. male), age at diagnosis (continuous variable), year of diagnosis (continuous variable), family history of colorectal cancer (absent vs. present), prediagnosis body mass index (< 25 kg/m2 vs. 25–29.9 kg/m2 vs. ≥ 30 kg/m2), tumor location (proximal colon vs. distal colon vs. rectum), tumor differentiation (well to moderate vs. poor), disease stage (I/II vs. III/IV), MSI status (high vs. non-high), CIMP status (high vs. low/negative), LINE-1 methylation level (continuous variable), KRAS mutation (wild-type vs. mutant), BRAF mutation (wild-type vs. mutant), PIK3CA mutation (wild-type vs. mutant), PTGS2 expression (negative vs. positive), and nuclear CTNNB1 expression (negative vs. positive). Backward eliminations with a threshold p of 0.05 were used to determine the most parsimonious final multivariable models. The two-sided α level in survival analyses was adjusted to 0.01 to account for multiple hypothesis testing. Cases with missing data were assigned to the majority category of a given categorical covariate to limit the degrees of freedom of the models: family history of colorectal cancer (1.1%), prediagnosis body mass index (0.7%), tumor location (0.5%), tumor differentiation (0.3%), MSI status (3.0%), CIMP status (8.4%), KRAS mutation (3.3%), BRAF mutation (2.5%), PIK3CA mutation (8.7%), PTGS2 expression (2.2%), and nuclear CTNNB1 expression (4.1%). For cases with missing data on LINE-1 methylation level (2.8%), a separate indicator variable was used. Excluding cases with missing data on any of the covariates did not yield substantial differences in our results (data not shown). Statistical interaction between tumor CD274 expression status (low vs. high) and TIL (absent vs. present) was evaluated using the Wald test on the cross-product. The assumption of proportional hazards was validated using a time-varying covariate in the models; i.e., cross-product of TIME subtype and survival time (p > 0.06).
Abbreviations
| CI | = | confidence interval |
| CIMP | = | CpG island methylator phenotype |
| HPFS | = | Health Professionals Follow-up Study |
| LINE-1 | = | long interspersed nucleotide element-1 |
| MSI | = | microsatellite instability |
| NHS | = | Nurses' Health Study |
| TIL | = | tumor-infiltrating lymphocytes |
| TIME | = | tumor immunity in the microenvironment |
Disclosure of potential conflict of interest
A.T.C. previously served as a consultant for Bayer Healthcare, Pfizer Inc., and Aralez Pharmaceuticals. This study was not funded by Bayer Healthcare, Pfizer Inc., or Aralez Pharmaceuticals. No other conflicts of interest exist. The other authors declare that they have no conflicts of interest.
Use of standardized official symbols: We use HUGO (Human Genome Organisation)-approved official symbols (or root symbols) for genes and gene products, including BRAF, CACNA1G, CD3, CD8, CD274, CDKN2 A, CRABP1, CTLA4, CTNNB1, FOXP3, HLA, IFNG, IGF2, KRAS, MLH1, NEUROG1, PDCD1, PIK3CA, PTGS2, RUNX3, SOCS1, and WNT; all of which are described at www.genenames.org. The official symbols are italicized to differentiate from non-italicized colloquial names that are used along with the official symbols. This format enables readers to familiarize themselves with the official symbols for genes and gene products together with common colloquial names.
2017ONCOIMM1013R-s03.docx
Download MS Word (924.1 KB)Acknowledgments
We would like to thank the participants and staff of the Nurses' Health Study and the Health Professionals Follow-up Study for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. The authors assume full responsibility for analyses and interpretation of these data.
Additional information
Funding
References
- Fesnak AD, June CH, Levine BL. Engineered T cells: the promise and challenges of cancer immunotherapy. Nat Rev Cancer. 2016;16:566–81. doi:10.1038/nrc.2016.97. PMID:27550819.
- Wang J, Reiss KA, Khatri R, Jaffee E, Laheru D. Immune Therapy in GI Malignancies: A Review. J Clin Oncol. 2015;33:1745–53. doi:10.1200/jco.2015.60.7879.
- Basile D, Garattini SK, Bonotto M, Ongaro E, Casagrande M, Cattaneo M, Fanotto V, De Carlo E, Loupakis F, Urbano F, et al. Immunotherapy for colorectal cancer: where are we heading? Expert Opin Biol Ther. 2017;17:709–21. doi:10.1080/14712598.2017.1315405. PMID:28375039.
- Gentzler R, Hall R, Kunk PR, Gaughan E, Dillon P, Slingluff CL, Jr., Rahma OE. Beyond melanoma: inhibiting the PD-1/PD-L1 pathway in solid tumors. Immunotherapy. 2016;8:583–600. doi:10.2217/imt-2015-0029. PMID:27140411.
- Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27:450–61. doi:10.1016/j.ccell.2015.03.001.
- Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–71. doi:10.1038/nature13954. PMID:25428505.
- Boussiotis VA. Molecular and Biochemical Aspects of the PD-1 Checkpoint Pathway. N Engl J Med. 2016;375:1767–78. doi:10.1056/NEJMra1514296. PMID:27806234.
- Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, Lu S, Kemberling H, Wilt C, Luber BS, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409–13. doi:10.1126/science.aan6733. PMID:28596308.
- Taube JM, Anders RA, Young GD, Xu H, Sharma R, McMiller TL, Chen S, Klein AP, Pardoll DM, Topalian SL, et al. Colocalization of inflammatory response with B7-h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci Transl Med. 2012;4:127ra37. doi:10.1126/scitranslmed.l3003689.
- Sanmamed MF, Chen L. Inducible expression of B7-H1 (PD-L1) and its selective role in tumor site immune modulation. Cancer J. 2014;20:256–61. doi:10.1097/PPO.0000000000000061. PMID:25098285.
- Zhang Y, Chen L. Classification of Advanced Human Cancers Based on Tumor Immunity in the MicroEnvironment (TIME) for Cancer Immunotherapy. JAMA Oncol. 2016;2:1403–4. doi:10.1001/jamaoncol.2016.2450. PMID:27490017.
- Teng MW, Ngiow SF, Ribas A, Smyth MJ. Classifying Cancers Based on T-cell Infiltration and PD-L1. Cancer Res. 2015;75:2139–45. doi:10.1158/0008-5472.CAN-15-0255. PMID:25977340.
- Masugi Y, Nishihara R, Yang J, Mima K, da Silva A, Shi Y, Inamura K, Cao Y, Song M, Nowak JA, et al. Tumour CD274 (PD-L1) expression and T cells in colorectal cancer. Gut. 2017;66:1463–73. doi:10.1136/gutjnl-2016-311421. PMID:27196573.
- Fridman WH, Zitvogel L, Sautes-Fridman C, Kroemer G. The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol. 2017;14:717–34. doi:10.1038/nrclinonc.2017.101. PMID:28741618.
- Ogino S, Nowak JA, Hamada T, Phipps AI, Peters U, Milner DA, Jr., Giovannucci EL, Nishihara R, Giannakis M, Garrett WS, et al. Integrative analysis of exogenous, endogenous, tumour, and immune factors for precision medicine. Gut 2018 in press.
- Fletcher R, Wang YJ, Schoen RE, Finn OJ, Yu J, Zhang L. Colorectal cancer prevention: Immune modulation taking the stage. Biochim Biophys Acta. 2018;1869:138–48. doi:10.1016/j.bbcan.2017.12.002. PMID:29391185.
- Ogino S, Nosho K, Irahara N, Meyerhardt JA, Baba Y, Shima K, Glickman JN, Ferrone CR, Mino-Kenudson M, Tanaka N, et al. Lymphocytic reaction to colorectal cancer is associated with longer survival, independent of lymph node count, microsatellite instability, and CpG island methylator phenotype. Clin Cancer Res. 2009;15:6412–20. doi:10.1158/1078-0432.CCR-09-1438. PMID:19825961.
- Nosho K, Baba Y, Tanaka N, Shima K, Hayashi M, Meyerhardt JA, Giovannucci E, Dranoff G, Fuchs CS, Ogino S. Tumour-infiltrating T-cell subsets, molecular changes in colorectal cancer, and prognosis: cohort study and literature review. J Pathol. 2010;222:350–66. doi:10.1002/path.2774. PMID:20927778.
- Mlecnik B, Bindea G, Angell HK, Maby P, Angelova M, Tougeron D, Church SE, Lafontaine L, Fischer M, Fredriksen T, et al. Integrative Analyses of Colorectal Cancer Show Immunoscore Is a Stronger Predictor of Patient Survival Than Microsatellite Instability. Immunity. 2016;44:698–711. doi:10.1016/j.immuni.2016.02.025. PMID:26982367.
- Rozek LS, Schmit SL, Greenson JK, Tomsho LP, Rennert HS, Rennert G, Gruber SB. Tumor-Infiltrating Lymphocytes, Crohn's-Like Lymphoid Reaction, and Survival From Colorectal Cancer. J Natl Cancer Inst. 2016;108:djw027. doi:10.1093/jnci/djw027.
- Williams DS, Mouradov D, Jorissen RN, Newman MR, Amini E, Nickless DK, Teague JA, Fang CG, Palmieri M, Parsons MJ, et al. Lymphocytic response to tumour and deficient DNA mismatch repair identify subtypes of stage II/III colorectal cancer associated with patient outcomes. Gut. 2018 in press. doi:10.1136/gutjnl-2017-315664. PMID:29382774
- Grizzi F, Basso G, Borroni EM, Cavalleri T, Bianchi P, Stifter S, Chiriva-Internati M, Malesci A, Laghi L. Evolving notions on immune response in colorectal cancer and their implications for biomarker development. Inflamm Res. 2018 in press. doi:10.1007/s00011-017-1128-1. PMID:29322204
- Yagi T, Baba Y, Ishimoto T, Iwatsuki M, Miyamoto Y, Yoshida N, Watanabe M, Baba H. PD-L1 Expression, Tumor-infiltrating Lymphocytes, and Clinical Outcome in Patients With Surgically Resected Esophageal Cancer. Ann Surg. 2017 in press. doi:10.1097/SLA.0000000000002616.
- `Koh J, Ock CY, Kim JW, Nam SK, Kwak Y, Yun S, Ahn SH, Park DJ, Kim HH, Kim WH, et al. Clinicopathologic implications of immune classification by PD-L1 expression and CD8-positive tumor-infiltrating lymphocytes in stage II and III gastric cancer patients. Oncotarget. 2017;8:26356–67. doi:10.18632/oncotarget.15465. PMID:28412752.
- Ono T, Azuma K, Kawahara A, Sasada T, Hattori S, Sato F, Shin B, Chitose SI, Akiba J, Hirohito U. Association between PD-L1 expression combined with tumor-infiltrating lymphocytes and the prognosis of patients with advanced hypopharyngeal squamous cell carcinoma. Oncotarget. 2017;8:92699–714. doi:10.18632/oncotarget.21564. PMID:29190949.
- Ock CY, Keam B, Kim S, Lee JS, Kim M, Kim TM, Jeon YK, Kim DW, Chung DH, Heo DS. Pan-Cancer Immunogenomic Perspective on the Tumor Microenvironment Based on PD-L1 and CD8 T-Cell Infiltration. Clin Cancer Res. 2016;22:2261–70. doi:10.1158/1078-0432.CCR-15-2834. PMID:26819449.
- Taube JM, Galon J, Sholl LM, Rodig SJ, Cottrell TR, Giraldo NA, Baras AS, Patel SS, Anders RA, Rimm DL, et al. Implications of the tumor immune microenvironment for staging and therapeutics. Mod Pathol. 2018;31:214–34. doi:10.1038/modpathol.2017.156. PMID:29192647.
- Li SK, Martin A. Mismatch Repair and Colon Cancer: Mechanisms and Therapies Explored. Trends Mol Med. 2016;22:274–89. doi:10.1016/j.molmed.2016.02.003.
- Giannakis M, Mu XJ, Shukla SA, Qian ZR, Cohen O, Nishihara R, Bahl S, Cao Y, Amin-Mansour A, Yamauchi M, et al. Genomic Correlates of Immune-Cell Infiltrates in Colorectal Carcinoma. Cell Rep. 2016;15:857–65. doi:10.1016/j.celrep.2016.03.075.
- Overman MJ, McDermott R, Leach JL, Lonardi S, Lenz HJ, Morse MA, Desai J, Hill A, Axelson M, Moss RA, et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol. 2017;18:1182–91. doi:10.1016/S1470-2045(17)30422-9. PMID:28734759.
- Ogino S, Chan AT, Fuchs CS, Giovannucci E. Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut. 2011;60:397–411. doi:10.1136/gut.2010.217182. PMID:21036793.
- Guinney J, Dienstmann R, Wang X, de Reynies A, Schlicker A, Soneson C, Marisa L, Roepman P, Nyamundanda G, Angelino P, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350–6. doi:10.1038/nm.3967. PMID:26457759.
- Punt CJ, Koopman M, Vermeulen L. From tumour heterogeneity to advances in precision treatment of colorectal cancer. Nat Rev Clin Oncol. 2017;14:235–46. doi:10.1038/nrclinonc.2016.171. PMID:27922044.
- Bijlsma MF, Sadanandam A, Tan P, Vermeulen L. Molecular subtypes in cancers of the gastrointestinal tract. Nat Rev Gastroenterol Hepatol. 2017;14:333–42. doi:10.1038/nrgastro.2017.33. PMID:28400627.
- Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, Berent-Maoz B, Pang J, Chmielowski B, Cherry G, et al. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell. 2016;165:35–44. doi:10.1016/j.cell.2016.02.065. PMID:26997480.
- Nishino M, Ramaiya NH, Hatabu H, Hodi FS. Monitoring immune-checkpoint blockade: response evaluation and biomarker development. Nat Rev Clin Oncol. 2017;14:655–68. doi:10.1038/nrclinonc.2017.88. PMID:28653677.
- Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, Tosolini M, Camus M, Berger A, Wind P, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–4. doi:10.1126/science.1129139. PMID:17008531.
- Shang B, Liu Y, Jiang SJ, Liu Y. Prognostic value of tumor-infiltrating FoxP3+ regulatory T cells in cancers: a systematic review and meta-analysis. Sci Rep. 2015;5:15179. doi:10.1038/srep15179. PMID:26462617.
- Bald T, Landsberg J, Lopez-Ramos D, Renn M, Glodde N, Jansen P, Gaffal E, Steitz J, Tolba R, Kalinke U, et al. Immune cell-poor melanomas benefit from PD-1 blockade after targeted type I IFN activation. Cancer Discov. 2014;4:674–87. doi:10.1158/2159-8290.CD-13-0458. PMID:24589924.
- Huang RR, Jalil J, Economou JS, Chmielowski B, Koya RC, Mok S, Sazegar H, Seja E, Villanueva A, Gomez-Navarro J, et al. CTLA4 blockade induces frequent tumor infiltration by activated lymphocytes regardless of clinical responses in humans. Clin Cancer Res. 2011;17:4101–9. doi:10.1158/1078-0432.CCR-11-0407. PMID:21558401.
- Kalbasi A, June CH, Haas N, Vapiwala N. Radiation and immunotherapy: a synergistic combination. J Clin Invest. 2013;123:2756–63. doi:10.1172/JCI69219. PMID:23863633.
- Rajpoot M, Sharma AK, Sharma A, Gupta GK. Understanding the Microbiome: Emerging Biomarkers for Exploiting the Microbiota for Personalized Medicine against Cancer. Semin Cancer Biol. 2018 in press. doi:10.1016/j.semcancer.2018.02.003. PMID:29425888
- Hughes LAE, Simons C, van den Brandt PA, van Engeland M, Weijenberg MP. Lifestyle, Diet, and Colorectal Cancer Risk According to (Epi)genetic Instability: Current Evidence and Future Directions of Molecular Pathological Epidemiology. Curr Colorectal Cancer Rep. 2017;13:455–69. doi:10.1007/s11888-017-0395-0. PMID:29249914.
- Rescigno T, Micolucci L, Tecce MF, Capasso A. Bioactive Nutrients and Nutrigenomics in Age-Related Diseases. Molecules. 2017;22. doi:10.3390/molecules22010105. PMID:28075340.
- Inamura K. Colorectal Cancers: An Update on Their Molecular Pathology. Cancers (Basel). 2018;10. doi:10.3390/cancers10010026. PMID:29361689.
- Rosenbaum MW, Bledsoe JR, Morales-Oyarvide V, Huynh TG, Mino-Kenudson M. PD-L1 expression in colorectal cancer is associated with microsatellite instability, BRAF mutation, medullary morphology and cytotoxic tumor-infiltrating lymphocytes. Mod Pathol. 2016;29:1104–12. doi:10.1038/modpathol.2016.95. PMID:27198569.
- Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature. 2015;523:231–5. doi:10.1038/nature14404. PMID:25970248.
- Pai SG, Carneiro BA, Mota JM, Costa R, Leite CA, Barroso-Sousa R, Kaplan JB, Chae YK, Giles FJ. Wnt/beta-catenin pathway: modulating anticancer immune response. J Hematol Oncol. 2017;10:101. doi:10.1186/s13045-017-0471-6. PMID:28476164.
- Zelenay S, van der Veen AG, Bottcher JP, Snelgrove KJ, Rogers N, Acton SE, Chakravarty P, Girotti MR, Marais R, Quezada SA, et al. Cyclooxygenase-Dependent Tumor Growth through Evasion of Immunity. Cell. 2015;162:1257–70. doi:10.1016/j.cell.2015.08.015. PMID:26343581.
- Wu AA, Drake V, Huang HS, Chiu S, Zheng L. Reprogramming the tumor microenvironment: tumor-induced immunosuppressive factors paralyze T cells. Oncoimmunology. 2015;4:e1016700. doi:10.1080/2162402X.2015.1016700. PMID:26140242.
- Hamada T, Cao Y, Qian ZR, Masugi Y, Nowak JA, Yang J, Song M, Mima K, Kosumi K, Liu L, et al. Aspirin Use and Colorectal Cancer Survival According to Tumor CD274 (Programmed Cell Death 1 Ligand 1) Expression Status. J Clin Oncol. 2017;35:1836–44. doi:10.1200/JCO.2016.70.7547. PMID:28406723.
- Nagorsen D, Voigt S, Berg E, Stein H, Thiel E, Loddenkemper C. Tumor-infiltrating macrophages and dendritic cells in human colorectal cancer: relation to local regulatory T cells, systemic T-cell response against tumor-associated antigens and survival. J Transl Med. 2007;5:62. doi:10.1186/1479-5876-5-62. PMID:18047662.
- Ogino S, Jhun I, Mata DA, Soong TR, Hamada T, Liu L, Nishihara R, Giannakis M, Cao Y, Manson JE, et al. Integration of pharmacology, molecular pathology, and population data science to support precision gastrointestinal oncology. NPJ Precis Oncol. 2017;1:40. doi:10.1038/s41698-017-0042-x.
- Hamada T, Keum N, Nishihara R, Ogino S. Molecular pathological epidemiology: new developing frontiers of big data science to study etiologies and pathogenesis. J Gastroenterol. 2017;52:265–75. doi:10.1007/s00535-016-1272-3. PMID:27738762.
- Topalian SL, Taube JM, Anders RA, Pardoll DM. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer. 2016;16:275–87. doi:10.1038/nrc.2016.36. PMID:27079802.
- Nishihara R, Wu K, Lochhead P, Morikawa T, Liao X, Qian ZR, Inamura K, Kim SA, Kuchiba A, Yamauchi M, et al. Long-term colorectal-cancer incidence and mortality after lower endoscopy. N Engl J Med. 2013;369:1095–105. doi:10.1056/NEJMoa1301969. PMID:24047059.
- Yamauchi M, Morikawa T, Kuchiba A, Imamura Y, Qian ZR, Nishihara R, Liao X, Waldron L, Hoshida Y, Huttenhower C, et al. Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut. 2012;61:847–54. doi:10.1136/gutjnl-2011-300865. PMID:22427238.
- Mima K, Cao Y, Chan AT, Qian ZR, Nowak JA, Masugi Y, Shi Y, Song M, da Silva A, Gu M, et al. Fusobacterium nucleatum in Colorectal Carcinoma Tissue According to Tumor Location. Clin Transl Gastroenterol. 2016;7:e200. doi:10.1038/ctg.2016.53. PMID:27811909.
- Chan AT, Ogino S, Fuchs CS. Aspirin and the risk of colorectal cancer in relation to the expression of COX-2. N Engl J Med. 2007;356:2131–42. doi:10.1056/NEJMoa067208. PMID:17522398.
- Morikawa T, Kuchiba A, Yamauchi M, Meyerhardt JA, Shima K, Nosho K, Chan AT, Giovannucci E, Fuchs CS, Ogino S. Association of CTNNB1 (beta-catenin) alterations, body mass index, and physical activity with survival in patients with colorectal cancer. JAMA. 2011;305:1685–94. doi:10.1001/jama.2011.513. PMID:21521850.
- Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Meyerhardt JA, Loda M, Giovannucci EL, Fuchs CS. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90–6. doi:10.1136/gut.2008.155473. PMID:18832519.
- Nosho K, Irahara N, Shima K, Kure S, Kirkner GJ, Schernhammer ES, Hazra A, Hunter DJ, Quackenbush J, Spiegelman D, et al. Comprehensive biostatistical analysis of CpG island methylator phenotype in colorectal cancer using a large population-based sample. PLoS One. 2008;3:e3698. doi:10.1371/journal.pone.0003698. PMID:19002263.
- Imamura Y, Lochhead P, Yamauchi M, Kuchiba A, Qian ZR, Liao X, Nishihara R, Jung S, Wu K, Nosho K, et al. Analyses of clinicopathological, molecular, and prognostic associations of KRAS codon 61 and codon 146 mutations in colorectal cancer: cohort study and literature review. Mol Cancer. 2014;13:135. doi:10.1186/1476-4598-13-135. PMID:24885062.
- Liao X, Lochhead P, Nishihara R, Morikawa T, Kuchiba A, Yamauchi M, Imamura Y, Qian ZR, Baba Y, Shima K, et al. Aspirin use, tumor PIK3CA mutation, and colorectal-cancer survival. N Engl J Med. 2012;367:1596–606. doi:10.1056/NEJMoa1207756. PMID:23094721.
- Nielsen M, Andreatta M. NetMHCpan-3.0; improved prediction of binding to MHC class I molecules integrating information from multiple receptor and peptide length datasets. Genome Med. 2016;8:33. doi:10.1186/s13073-016-0288-x. PMID:27029192.
