ABSTRACT
Mismatch repair (MMR)-deficient cancers accumulate multiple insertion/deletion mutations at coding microsatellites (cMS), which give rise to frameshift peptide neoantigens. The high mutational neoantigen load of MMR-deficient cancers is reflected by pronounced anti-tumoral immune responses of the host and high responsiveness towards immune checkpoint blockade. However, immune evasion mechanisms can interfere with the immune response against MMR-deficient tumors. We here performed a comprehensive analysis of immune evasion in MMR-deficient colorectal cancers, focusing on HLA class I-mediated antigen presentation. 72% of MMR-deficient colorectal cancers of the DFCI database harbored alterations affecting genes involved in HLA class I-mediated antigen presentation, and 54% of these mutations were predicted to abrogate function. Mutations affecting the HLA class I transactivator NLRC5 were observed as a potential new immune evasion mechanism in 26% (6% abrogating) of the analyzed tumors. NLRC5 mutations in MMR-deficient cancers were associated with decreased levels of HLA class I antigen expression. In summary, the majority of MMR-deficient cancers display mutations interfering with HLA class I antigen presentation that reflect active immune surveillance and immunoselection during tumor development. Clinical studies focusing on immune checkpoint blockade in MSI cancer should account for the broad variety of immune evasion mechanisms as potential biomarkers of therapy success.
Introduction
DNA mismatch repair (MMR) deficiency is responsible for around 15% of colorectal cancers. MMR-deficient cancers accumulate insertion and deletion mutations in short repetitive sequence stretches called microsatellites, leading to microsatellite instability (MSI) phenotype.Citation1 MSI tumors occur as sporadic cancer or in the context of Lynch syndrome, which is caused by germline mutations of the MMR genes.Citation2 MMR deficiency-induced insertion/deletion mutations in coding microsatellites (cMS) have been identified as the mechanism underlying the high immunogenicity of MSI cancers. These cMS mutations lead to a high load of frameshift peptide (FSP) neoantigens, which can encompass multiple HLA class I and II epitopes and elicit pronounced immune responses of the host.Citation3 These immune responses manifest for example through a dense infiltration with cytotoxic CD8T cells, which is the hallmark of MSI tumors.Citation4,Citation5 Clinically, the pronounced immunogenicity of MSI tumors is of high relevance, because MSI tumor patients respond particularly well to treatment with immune checkpoint blockade.Citation6-Citation8
The recent success of immune therapy against MSI cancer has also raised the question, which patients are likely to benefit from therapy and whether there are predictive markers that allow tailored application of immune checkpoint blockade. As one-third to half of MSI tumor patients do not show benefit from immune checkpoint blockade, the availability of predictive markers would significantly reduce unnecessary side effects and therapy costs.Citation9
One possible reason why MSI tumor patients do not respond to immune checkpoint blockade is tumor immune evasion. It has been known for a long time that MSI tumors frequently present with alterations that lead to a breakdown of HLA class I and HLA class II-mediated antigen presentation (For a review, see Citationref. 10). The most frequent and best characterized mechanism of immune evasion of MSI cancer cells are mutations of the Beta2-microglobulin (B2M) gene.Citation11-Citation13 Mutations in B2M lead to a complete loss of HLA class I antigen expression on the cell surface.Citation14 It has recently been demonstrated that B2M mutations particularly occur in MSI tumors growing out in an environment of activated T cells, supporting the hypothesis that B2M mutations develop as an immune evasion mechanism and a result of immune selection.Citation10,Citation15,Citation16 B2M mutation-induced loss of HLA class I-mediated antigen presentation has been discussed as a possible mechanism leading to secondary resistance of MSI cancers against immune therapy,Citation17,Citation18 in analogy to melanoma patients, where B2M mutations have been found in patients treated with adoptive T cell transfer or immune checkpoint blockade.Citation19,Citation20 However, B2M mutations only occur in about 30% of MSI cancers and cannot explain tumor outgrowth and immune evasion in all MSI cancer patients.Citation13,Citation17
In the present study, we therefore aimed at a more comprehensive characterization of immune evasion phenomena impairing HLA class I-mediated antigen presentation in MSI colorectal cancers. We started from mutation data deposited in public databases of colorectal cancers characterized for their MSI statusCitation21 and validated findings for specific mutations in tumor specimens from our own cohorts.
Results
Mutations of HLA class I-related genes in MSI cancer
In order to obtain a more complete picture of immune evasion mechanisms in MSI colorectal cancer, we gathered mutation data from the publicly available DFCI database using the cbioportal website.Citation21-Citation23 Specifically, mutations of B2M, MHC class I heavy chain-encoding genes HLA-A, HLA-B, HLA-C were analyzed in 91 MSI colorectal adenocarcinoma samples. Data were complemented by mutation data for transporters of antigen presentation TAP1, TAP2 and the recently described transactivator of MHC class I genes NLRC5 (, ). In total, 66 (72%) out of 91 analyzed MSI colorectal cancer specimens displayed mutations in at least one of the components tested (, ). The most common alteration detected was mutations of the B2M gene, with a frequency of 28 (30.8%) of 91 tumors, 27 (96.4%) of those being protein-truncating and therefore of likely functional relevance. Mutations of the HLA-B heavy chain were the second most common alteration with 19 (20.9%) of 91 MSI cancers affected, followed by mutations of HLA-C and HLA-B, which were present in 9 (9.9%) and 8 (8.8%) of the tumors, respectively. Two-thirds of the identified HLA-A and HLA-B heavy chain mutations were truncating (18 out of 27), supporting their functional significance. In contrast, only one (11.1%) of the observed HLA-C mutations were truncating.
Table 1. Mutual exclusivity of HLA class I-related genes Log odds ratio shows how strongly presence or absence of alterations in gene A are related with presence or absence of alterations in gene B. Log odds ratio of more than 0 corresponds association towards co-occurrence, equals to or less than 0 corresponds mutual exclusivity; p < 0,05 is significant association.
Figure 1. Mutations in HLA class I-related genes in MSI colorectal cancers from the DFCI cohort.Citation21 MSI colorectal cancers included in the DFCI cohort are numbered from 1 to 91, each mutation in the corresponding patient is shown by different colors that are indicated below the Fig.; darker colors display pathogenic mutations; whereas, lighter colors indicate mutations with unclear significance. Lack of mutations in the respective tumors and genes is displayed by gray color. Detailed information about the mutations can be found in Supplementary .
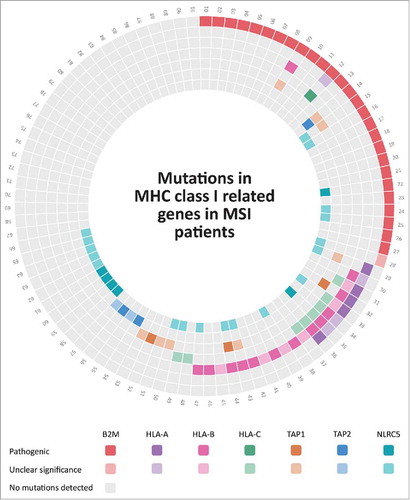
Figure 2. Mutations of HLA class I-related genes as possible immune evasion mechanisms in MSI colorectal cancer. The analyzed HLA class I-related genes (B2M, HLA-A, HLA-B, HLA-C, TAP1, TAP2 and NLRC5) play a major role in HLA class I-mediated antigen processing and presentation. Analysis of the DFCI cohort revealed at least one mutation in 72% of the analyzed MSI colorectal cancers, with 54% of the tumors presenting with pathogenic mutations. The percentages of pathogenic mutations and overall mutations [in brackets] are shown in the respective frames.
![Figure 2. Mutations of HLA class I-related genes as possible immune evasion mechanisms in MSI colorectal cancer. The analyzed HLA class I-related genes (B2M, HLA-A, HLA-B, HLA-C, TAP1, TAP2 and NLRC5) play a major role in HLA class I-mediated antigen processing and presentation. Analysis of the DFCI cohort revealed at least one mutation in 72% of the analyzed MSI colorectal cancers, with 54% of the tumors presenting with pathogenic mutations. The percentages of pathogenic mutations and overall mutations [in brackets] are shown in the respective frames.](/cms/asset/cc94e9cf-0781-4d2b-b548-6f6d93a39d91/koni_a_1445453_f0002_oc.jpg)
To evaluate further a possible functional role of the mutations affecting genes encoding components of HLA class I antigens, analysis of mutual exclusivity was performed. Mutual exclusivity was significant for B2M and HLA-B genes and co-occurence was observed for HLA-A and HLA-B; HLA-A and HLA-C; and HLA-B and HLA-C genes.
In addition to mutations predicted to directly alter HLA class I antigens, mutations interfering with the cellular antigen processing and presentation machinery were detected in the genes coding for TAP1 (12.1%) and TAP2 (5.5%). Moreover, we observed a surprisingly high number of mutations affecting the gene coding for the recently described HLA class I transactivator NLRC5 in 24 (26.4%) out of 91 MSI colorectal cancer. Although truncating mutations of the NLRC5 gene were only present in 6 (6.6%) of the tumors, this frequency was still higher than the frequency of truncating mutations in the TAP1 and TAP2 genes (3.3% each) previously described as mutant in MSI colorectal cancer.Citation24
Comparison of MSI and non-MSI colorectal cancer
In order to examine whether the high frequency of mutations affecting genes involved in cellular antigen processing and presentation detected in MSI colorectal cancer was specifically associated with the MSI phenotype, we compared mutation frequencies between MSI and non-MSI colorectal cancers. For all examined candidate genes, a significantly elevated mutation frequency was observed in MSI compared to non-MSI colorectal cancers ().
Table 2. Comparison of mutation frequencies between MSI and non-MSI colorectal cancers.
Table 3. List of primers used.
Consequences of NLRC5 mutations in MSI colorectal cancer samples
To further examine a potential functional role of NLRC5 mutations in MSI cancer, we examined the consequence of NLRC5 mutations in our own collection of MSI colorectal cancer specimens (n = 95).
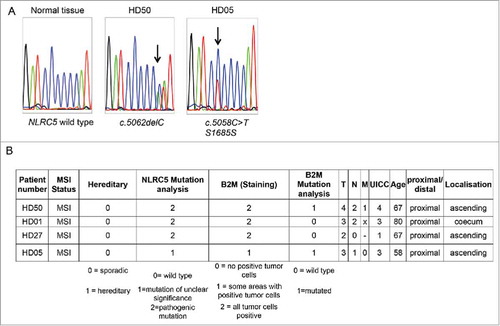
We focused on potential mutational hot spots in the gene by screening for coding microsatellite sequences contained in the gene using www.seltarbase.org.Citation25 The detected cMS were evaluated for known somatic mutations using the COSMIC database, revealing a C6 coding microsatellite located in position 5057 to 5062 as a potential mutation hot spot.
Four (4.2%) out of 95 tumors displayed mutations affecting the C6 cMS located in the NLRC5 gene (). Three identified mutations were one-basepair deletions (c.5062del) leading to a truncated NLRC5 protein; one mutation was a silent mutation within the C6 repeat (c.5058C>T, S1685S). Although in our own collection of tumors, which could be evaluated for B2M protein expression by immunohistochemistry, the tumors harboring NLRC5 frameshift mutations were B2M-positive, statistically significant evidence for mutual exclusivity was not reached in the DFCI cohort (log odds ratio -0.450, p = 0.283).
Figure 3. NLRC5 mutations in MSI tumor samples. A. Representative sequencing results spanning the C6 coding microsatellite in the NLRC5 coding region. B. Information about NLRC5-mutated tumors.
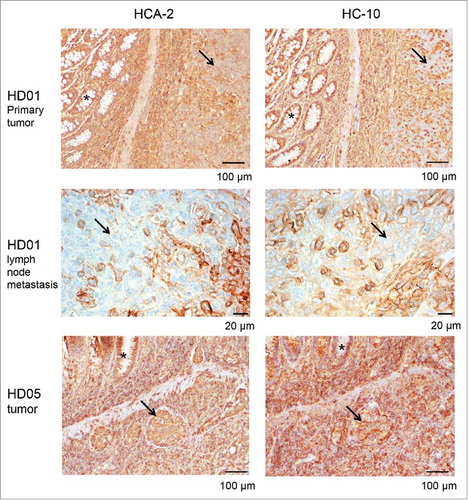
In order to evaluate the consequences of NLRC5 mutations on HLA class I antigen expression in MSI colorectal cancer, we performed staining of the respective tumors, including metastases if available, for HLA class I heavy chains using the mouse monoclonal antibodies HC-10 (predominantly recognizing HLA-B and HLA-C) and HCA-2 (predominantly recognizing HLA-A). Representative staining results are displayed in . Consistently, primary tumors and lymph node metastases harboring NLRC5 frameshift mutations displayed low levels of HLA class I antigen expression on the membrane (), suggesting a potential functional significance of NLRC5 for HLA class I antigen expression in MSI colorectal cancer. In contrast, no reduction of HLA class I antigen expression was observed in the tumor harboring the c.5058C>T point mutation ().
Figure 4. HLA class I heavy chain expression in NLRC5-mutant MSI colorectal cancers. HD01 tumor cells (arrows) harboring the NLRC5c.5062del frameshift mutation (upper panel) show reduced expression of HLA class I antigens (HC10, left panel; HC-A2, right panel) when compared to normal epithelial crypts (asterisks) and surrounding stroma. Reduced HLA class I antigen expression was also detectable in the corresponding lymph node metastases (middle panel, left and right). In contrast, tumor HD05 that harbored the c.5058C>T point mutation presented with strong HLA class I antigen expression in tumor cells (lower panel, arrows) as well as adjacent normal crypts (asterisks).
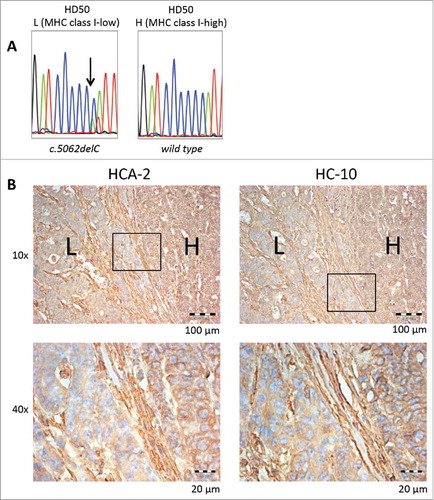
Interestingly, one of the tumor samples displayed regional heterogeneity of HLA class I antigen expression, as visualized by HCA-2 staining (). In order to evaluate a potential contribution of NLRC5 mutations on this regional diversity, regions with low and high HLA class I antigen expression levels were microdissected and evaluated for NLRC5 mutations separately. Remarkably, the NLRC5c.5062del frameshift mutation was only detectable in the region with low levels of HLA class I antigen expression, but absent in the region with high HLA class I antigen expression levels ().
Figure 5. Regional NLRC5 mutation and corresponding HLA class I antigen expression. NLRC5 sequencing results of microdissected HLA class I low (“L”) and high (“H”) regions are shown in A, and IHC staining of HD50 tumor samples with HCA-2 and HC-10 antibody in different magnifications is shown in B. Regions with high HLA class I antigen expression (H) showed the wild type C6 repeat of NLRC5, whereas the NLRC5c.5062del frameshift mutation was restricted to the area with reduced HLA class I antigen expression.
Discussion
The enhanced immunogenicity of mismatch repair-deficient tumors has been shown in a variety of studies, and the direct link between the MSI phenotype and the high load of mutational neoantigens has been demonstrated (reviewed by ref.3). The initially high immunogenicity of developing MSI tumors most likely is the reason for immune evasion phenomena that are frequently detectable in manifest MSI colorectal cancers,Citation10 most commonly mediated by mutations of the B2M gene. EchterdiekCitation15 and Janikovits et al.Citation16 reported that B2M mutations are detected especially in the tumors where there is a strong immunoselection pressure due to highly activated immune microenvironment, underlining the concept of immuno-editing in MSI cancers.
Although several distinct mechanisms have been reported that can lead to an impairment of HLA class I-mediated antigen presentation in MSI cancers, in the majority of MSI cancers no alterations had been found that may explain immune evasion.
In the present study we therefore focused on a more comprehensive overview of potential HLA class I-related immune evasion mechanisms in MSI colorectal cancers. Screening the public DFCI database for somatic mutations in MSI colorectal cancer we detected B2M mutations in approximately 30% of the specimens. This frequency is in line with previous reports.Citation13,Citation26
Interestingly, mutations of the genes HLA-A, -B, and -C encoding HLA class I heavy chains were observed in 23 out of 91 MSI cancers. Most of the mutations affecting these genes were truncating mutations, suggesting that they lead to non-functional protein products. Mutations of HLA-A, -B, and -C were positively correlated (coefficients A vs B, B vs C, A vs C), and they occurred less frequently in tumors harboring B2M mutations, the negative correlation being significant for HLA-B and B2M mutations (Log odds ratio: -2,359, p = 0,005). These results strongly suggest that the observed mutations affecting HLA class I antigen components are in fact mediating immune evasion and reflecting immuno-selection. As B2M mutations lead to a complete breakdown of the HLA class I antigen expression on the surface of affected cells, additional mutations in HLA class I heavy chain-encoding genes would not provide the cells with evolutionary growth advantages mediated by lack of HLA class I antigens, which are anyway lost as a consequence of B2M inactivation. The significantly higher mutation frequencies in genes involved in HLA class I-mediated antigen presentation in MSI compared to non-MSI colorectal cancers provide further support of the hypothesis that MSI cancers undergo a process of immune selection. This concept is also in line with the observation that different MSI tumor types show distinct immune evasion patterns; for example, JAK1 mutations, which are comparatively rare in MSI colorectal cancer, are frequently observed in MSI endometrial cancer, likely reflecting distinct immune selection conditions.Citation27-Citation29
The observation of several different mutations affecting genes involved in HLA class I-mediated antigen presentation also has clinical implications. It underlines that even mutations with a minor overall contribution can have significant effects on the potential responsiveness towards immune therapy. Clinical decision making will in the future have to account for the diverse mutational patterns in order to realize personalized, individual therapeutic concepts in the clinical setting.
These results also indirectly prove that MSI tumor development takes place in an environment, in which T cells, particularly CD8-positive T cells that can recognize HLA class I antigens on the tumor cell surface, are capable of controlling the outgrowth of HLA class I antigen-positive MSI cancer cells or even directly kill them. On the basis of this observation, potential tolerance mediated by recurrent stimulation with MSI-induced neoantigens, as discussed to occur in Lynch syndrome mutation carriers,Citation3 appears extremely unlikely. Therefore, Lynch syndrome seems to be in stark contrast to mouse models of colorectal cancer,Citation30 in which recurrent stimulation with neoplasia-specific antigens leads to the induction of humoral immune responses and tolerance. One potential explanation is that in the respective mouse model, tumor cells express only one dominant antigen (SV40 large T), whereas emerging MSI cancers can generate a broad variety of mutation-induced neoantigens, which potentially prevents immune tolerance. Further studies, particularly in mouse models of Lynch syndrome, are required to address this issue in more depth.
Besides mutations affecting HLA class I antigen-encoding genes, mutations of genes coding for proteins involved in cellular antigen processing were detected. In addition to mutations affecting transporters of antigen presentation TAP1 and TAP2 that had previously been described in MSI colorectal cancer,Citation4 mutations of the NOD-like receptor family CARD domain containing 5 (NLRC5) gene were detected in a subset of MSI colorectal cancers of the TCGA/DFCI databases. NLRC5 codes for an essential transactivator of HLA class I-related genes.Citation31 Nlrc5-deficient mice displayed severe loss of MHC class I and APM gene expression.Citation32-Citation36 It has been shown that NLRC5 directly transactivates B2M, HLA A/B/C/D/E/F/G and TAP1.Citation37 Therefore, any mutations in NLRC5 would plausibly result in defects of MHC class I presentation. Correspondingly, NLRC5 alterations including loss of function mutations, promoter hypermethylation and copy number loss are frequently observed in solid cancers.Citation38
So far, to the best of our knowledge, NLRC5 mutations had not been described in MSI cancers. We therefore evaluated potential functional consequences of NLRC5 mutations in archival tissue specimens of MSI colorectal cancer focusing on a C6 microsatellite in the coding region of the NLRC5 gene.
Most important evidence for a functional role of NLRC5 mutations in immune evasion of MSI colorectal cancer comes from three observations: (1) all tumors presenting with truncating NLRC5 mutations did not harbor B2M mutations and had retained B2M expression. Although the number of NLRC5-mutant tumors was too low to draw any definitive conclusions, the observation is compatible with the hypothesis that NLRC5 mutations only provide affected cells with a growth advantage if HLA class I antigen-mediated recognition of tumor cells by CD8-positive T cells was still functional before the NLRC5 mutation occurred. (2) NLRC5-mutant tumors all showed reduced HLA class I antigen expression, suggesting that NLRC5 inactivation leads to lower HLA class I-related gene expression, as described previously both in vitroCitation31 and in vivo.Citation32-Citation36 (3) This hypothesis is strongly supported by our observation that partially reduced HLA class I antigen-expression co-localized with NLRC5 mutations.
Although we provide evidence that NLRC5 mutations can contribute to immune evasion of MSI cancers, the frequency of these mutations, compared to mutations of the B2M gene, is low. This may reflect that NLRC5 mutations in contrast to B2M mutations only lead to a partial loss of HLA class I-mediated antigen presentation, which is in line with the observation that we still can detect residual levels of HLA class I antigen expression in NLRC5-mutant MSI cancers. It has also been reported that unlike MHC class I deficiency, Nlrc5 knockout mice display a mosaic deficiency in MHC class I expression in different cells and tissues.Citation39 However, NLRC5 mutations were more frequent than the previously described mutations affecting APM component-encoding genes such as TAP1 and TAP2.Citation24
In summary, our results provide strong and additional support to the model that MSI cancers develop through a stringent process of immune selection. This is reflected by the fact that alterations affecting the HLA class I antigen processing and presentation pathway are detectable in more than two out of three MSI colorectal cancer lesions, with a high degree of mutual exclusivity. NLRC5 mutations likely represent a novel mechanism of immune evasion in B2M-wild type MSI colorectal cancers. Monitoring the broad variety of immune evasion phenomena typical of MSI cancers will be of particular importance to more precisely predict success or failure of immune checkpoint blockade in MSI cancer patients.
Materials and methods
Database search
The DFCI databaseCitation21 was used to determine mutation patterns of commonly mutated colon cancer genes APC and KRAS in MSI cancer samples (www.cbioportal.org, status: January 31st, 2017).Citation22,Citation23
Namely, mutations of the genes encoding the HLA heavy chains HLA-A, HLA-B and HLA-C, the HLA class I light chain B2M, the transporters of antigen presentation TAP1 and TAP2, and the HLA class I transactivator NLRC5 were analyzed by filtering MSI cancers in the DFCI database. Analysis of mutual exclusivity and co-occurrence of the selected HLA class I-related genes was performed by cBioportal version 1.7.0.
Patients and tumor samples
The study examines two distinct collections of MSI colorectal cancers, first, samples from the publicly available DFCI databaseCitation21 (labeled from 01 to 91), and a collection of formalin-fixed, paraffin-embedded tumor specimens obtained from the Department of Applied Tumor Biology, Institute of Pathology, University Hospital Heidelberg (labeled “HD” followed by a number). Tumors were collected in frame of the German HNPCC Consortium, the study was approved by the institutional Ethics Committee, and informed or written consent was obtained from all patients.
DNA isolation from FFPE tissues
Genomic tumor DNA was isolated from FFPE tissue blocks after manual microdissection to enrich the tumor cell content, or to separate tumor regions with low and high HLA class I antigen expression. DNA was isolated by QIAamp DNA FFPE tissue kit (56404; Qiagen) according to manufacturer´s protocol. Following DNA isolation, to measure the concentration and quality of DNA Nanodrop spectrophotometer(Nanodrop Technologies) was used.
Sequencing
Following DNA isolation, PCR amplification was performed using oligonucleotide primers specific for the amplification of B2M and a repetitive microsatellite region located in the NLRC5 gene. PCR samples were run on agarose gel to confirm the products and purified by QIAquick PCR purification kit (Qiagen, Hilden, Germany). Sanger sequencing was performed for B2M and NLRC5 coding regions as described previouslyCitation13 using the oligonucleotide primers listed in . The products were visualized using an ABI prism 3130xl Genetic Analyzer (Applied Biosytems, Darmstadt, Germany), and sequence analysis was performed using the Sequencing Analysis Software v.6 (Applied Biosystems). Part of the B2M mutation data have been published previously.Citation15
Immunohistochemistry
2 μm FFPE tissue sections of tumor resection specimens were deparaffinized and rehydrated by xylene and sequential decreasing percentages of ethanol. Immunohistochemical staining was performed as described previously.Citation12 Following epitope retrieval using 10 mM of citric acid monohydrate solution, pH 6.0, in a microwave oven three times for 5 minutes at 560 W, endogenous peroxidase was quenched by 2% hydrogen peroxide solution. After blocking nonspecific binding by incubation of the slides with serum, tissue sections were incubated with the mouse monoclonal antibodies HCA-2 (AM33034PU-N; Acris) and HC-10 (AM33035PU-N; Acris) that recognize a variety of human HLA-A and HLA-B proteins,Citation40 both at a dilution of 1:150 at 4°C overnight. Next, secondary antibody biotin coupled horse anti-mouse IgG (1:100 dilution, BA2000; Vector) was applied on the slides and Vectastain Elite ABC kit (Vector) was used to amplify the signal. To develop the slides, liquid DAB and substrate chromogen system (K3468; Dako) was utilized. Hematoxylin/Eosin staining was used to counterstain the slides. The stained samples were visualized using an Olympus BX43 microscope and the cell^D software (version 2.6, Olympus, Duesseldorf, Germany).
Abbreviations used
| APM | = | antigen presentation machinery |
| B2M | = | beta2-microglobulin |
| cMS | = | coding microsatellite |
| FFPE | = | formalin-fixed paraffin-embedded tissue |
| FSP | = | frameshift peptide |
| MSI | = | microsatellite instability |
| NLRC5 | = | NOD-like receptor family CARD domain containing 5 |
| TAP | = | transporter associated with antigen processing |
Disclosure of interest
The authors declare no conflict of interest.
supp_data.zip
Download Zip (58.7 KB)Acknowledgments
The expert technical assistance of Beate Kuchenbuch, Petra Höfler, and Lena Ehret is gratefully acknowledged. We thank Sascha Wahlbrink for the graphical representation of data.
Additional information
Funding
References
- Kloor M, Staffa L, Ahadova A, Von Knebel Doeberitz M. Clinical significance of microsatellite instability in colorectal cancer. Langenbeck's Archives of Surgery. 2014;399(1):23–31. doi:10.1007/s00423-013-1112-3. PMID:24048684
- Jasperson KW, Tuohy TM, Neklason DW, Burt RW. Hereditary and Familial Colon Cancer. Gastroenterology. 2010;138(6):2044–58. doi:10.1053/j.gastro.2010.01.054. PMID:20420945
- Kloor M, Von Knebel Doeberitz M. The immune biology of microsatellite-unstable cancer. Trends Cancer. 2016;2(3):121–33. doi:10.1016/j.trecan.2016.02.004. PMID:28741532
- Buckowitz A, Knaebel H-P, Benner A, Bläker H, Gebert J, Kienle P, von Knebel Doeberitz M, Kloor M. Microsatellite instability in colorectal cancer is associated with local lymphocyte infiltration and low frequency of distant metastases. Br J Cancer. 2005;92(9):1746–53. doi:10.1038/sj.bjc.6602534. PMID:15856045
- Dolcetti R, Viel A, Doglioni C, Russo A, Guidoboni M, Capozzi E, Vecchiato N, Macrì E, Fornasarig M, Boiocchi M. High Prevalence of Activated Intraepithelial Cytotoxic T Lymphocytes and Increased Neoplastic Cell Apoptosis in Colorectal Carcinomas with Microsatellite Instability. Am J Pathol. 1999;154(6):1805–13. doi:10.1016/S0002-9440(10)65436-3. PMID:10362805
- Lee K, Tosti E, Edelmann W. Mouse models of DNA mismatch repair in cancer research. DNA Repair. 2016;38:140–6. doi:10.1016/j.dnarep.2015.11.015. PMID:26708047
- Lee V, Murphy A, Le DT, Diaz LA. Mismatch Repair Deficiency and Response to Immune Checkpoint Blockade. The Oncologist. 2016;21(10):1200–11. doi:10.1634/theoncologist.2016-0046. PMID:27412392
- Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372(26):2509–20. doi:10.1056/NEJMoa1500596. PMID:26028255
- Teixidó C, González-Cao M, Karachaliou N, Rosell R. Predictive factors for immunotherapy in melanoma. Ann Trans Med. 2015;3(15):208. doi:10.3978/j.issn.2305-5839.2015.05.07 PMID:26488004
- Kloor M, Michel S, von Knebel Doeberitz M. Immune evasion of microsatellite unstable colorectal cancers. Int J Cancer. 2010;127(5):1001–10. doi:10.1002/ijc.25283. PMID:20198617
- Bicknell DC, Kaklamanis L, Hampson R, Bodmer WF, Karran P. Selection for beta 2-microglobulin mutation in mismatch repair-defective colorectal carcinomas. Curr Biol. 1996;6(12):1695–7. doi:10.1016/S0960-9822(02)70795-1. PMID:8994836
- Kloor M, von Knebel Doeberitz M, Gebert JF. Molecular testing for microsatellite instability and its value in tumor characterization. Expert review of molecular diagnostics. 2005;5(4):599–611. doi:10.1586/14737159.5.4.599. PMID:16013977
- Kloor M, Michel S, Buckowitz B, Rüschoff J, Büttner R, Holinski-Feder E, Dippold W, Wagner R, Tariverdian M, Benner A, et al. Beta2-microglobulin mutations in microsatellite unstable colorectal tumors. Int J Cancer. 2007;121(2):454–8. doi:10.1002/ijc.22691. PMID:17373663
- Benitez R, Godelaine D, Lopez-Nevot MA, Brasseur F, Jiménez P, Marchand M, Oliva MR, van Baren N, Cabrera T, Andry G, et al. Mutations of the beta2-microglobulin gene result in a lack of HLA class I molecules on melanoma cells of two patients immunized with MAGE peptides. Tissue Antigens. 1998;52(6):520–9. doi:10.1111/j.1399-0039.1998.tb03082.x. PMID:9894850
- Echterdiek F, Janikovits J, Staffa L, Müller M, Lahrmann B, Frühschütz M, Hartog B, Nelius N, Benner A, Tariverdian M, et al. Low density of FOXP3-positive T cells in normal colonic mucosa is related to the presence of beta2-microglobulin mutations in Lynch syndrome-associated colorectal cancer. OncoImmunology. 2016;5(2):e1075692. doi:10.1080/2162402X.2015.1075692. PMID:27057447
- Janikovits J, Müller M, Krzykalla J, Körner S, Echterdiek F, Lahrmann B, Grabe N, Schneider M, Benner A, Doeberitz M von K, et al. High numbers of PDCD1 (PD-1)-positive T cells and B2M mutations in microsatellite-unstable colorectal cancer. OncoImmunology. 2018;7(2):e1390640. doi:10.1080/2162402X.2017.1390640. PMID:29308317
- Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, Lu S, Kemberling H, Wilt C, Luber BS, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357(6349):409–13. doi:10.1126/science.aan6733. PMID:28596308
- Shin DS, Zaretsky JM, Escuin-Ordinas H, Garcia-Diaz A, Hu-Lieskovan S, Kalbasi A, Grasso CS, Hugo W, Sandoval S, Torrejon DY, et al. Primary resistance to PD-1 blockade mediated by JAK1/2 mutations. Cancer Discovery. 2017;7(2):188–201. doi:10.1158/2159-8290.CD-16-1223. PMID:27903500
- Rosenberg SA, Yang JC, Robbins PF, Wunderlich JR, Sherry RM, Schwartzentruber DJ, Topalian SL, Nicholas P, Filie A, Chang R, et al. Cell transfer therapy for cancer: lessons from sequential treatments of a patient with metastatic melanoma. J Immunother. 2003;26(5):385–93. doi:10.1097/00002371-200309000-00001. PMID:12973027
- Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, Torrejon DY, Abril-Rodriguez G, Sandoval S, Barthly L, et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. New England Journal of Medicine. 2016;375(9):819–29. doi:10.1056/NEJMoa1604958. PMID:27433843
- Giannakis M, Mu X, Shukla S, Qian Z, Cohen O, Nishihara R, Bahl S, Cao Y, Amin-Mansour A, Yamauchi M, et al. Genomic Correlates of Immune-Cell Infiltrates in Colorectal Carcinoma. Cell reports. 2016;15(4):857–65. doi:10.1016/j.celrep.2016.03.075.
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio Cancer Genomics Portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discovery. 2012;2(5):401–4. doi:10.1158/2159-8290.CD-12-0095. PMID:22588877
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Science Signaling. 2013;6(269):pl1–pl1. doi:10.1126/scisignal.2004088. PMID:23550210
- Kloor M. Immunoselective Pressure and Human Leukocyte Antigen Class I Antigen Machinery Defects in Microsatellite Unstable Colorectal Cancers. Cancer Res. 2005;65(14):6418–24. doi:10.1158/0008-5472.CAN-05-0044. PMID:16024646
- Woerner SM, Yuan YP, Benner A, Korff S, Von Knebel Doeberitz M, Bork P. SelTarbase, a database of human mononucleotide-microsatellite mutations and their potential impact to tumorigenesis and immunology. Nucleic Acids Res. 2009;38(SUPPL.1):682–9. doi:10.1093/nar/gkp839
- Koelzer VH, Baker K, Kassahn D, Baumhoer D, Zlobec I. Prognostic impact of β-2-microglobulin expression in colorectal cancers stratified by mismatch repair status. J Clin Pathol. 2012;65(11):996–1002. doi:10.1136/jclinpath-2012-200742. PMID:22859396
- Sveen A, Johannessen B, Tengs T, Danielsen SA, Eilertsen IA, Lind GE, Berg KCG, Leithe E, Meza-Zepeda LA, Domingo E, et al. Multilevel genomics of colorectal cancers with microsatellite instability—clinical impact of JAK1 mutations and consensus molecular subtype 1. Genome Medicine. 2017;9(1):46. doi:10.1186/s13073-017-0434-0. PMID:28539123
- Ren Y, Zhang Y, Liu RZ, Fenstermacher DA, Wright KL, Teer JK, Wu J. JAK1 truncating mutations in gynecologic cancer define new role of cancer-associated protein tyrosine kinase aberrations. Sci Rep. 2013;3:3042. doi:10.1038/srep03042. PMID:24154688
- Albacker LA, Wu J, Smith P, Warmuth M, Stephens PJ, Zhu P, Yu L, Chmielecki J. Loss of function JAK1 mutations occur at high frequency in cancers with microsatellite instability and are suggestive of immune evasion Reis RM, editor. PLOS ONE. 2017;12(11):e0176181. doi:10.1371/journal.pone.0176181. PMID:29121062
- Czéh M, Loddenkemper C, Shalapour S, Schön C, Robine S, Goldscheid E, Stein H, Schüler T, Willimsky G, Blankenstein T. The immune response to sporadic colorectal cancer in a novel mouse model. Oncogene. 2010;29(50):6591–602. doi:10.1038/onc.2010.388. PMID:20818425
- Meissner TB, Li A, Biswas A, Lee K-H, Liu Y-J, Bayir E, Iliopoulos D, van den Elsen PJ, Kobayashi KS. NLR family member NLRC5 is a transcriptional regulator of MHC class I genes. Proc Natl Acad Sci. 2010;107(31):13794–9. doi:10.1073/pnas.1008684107.
- Biswas A, Meissner TB, Kawai T, Kobayashi KS. Cutting Edge: Impaired MHC Class I Expression in Mice Deficient for Nlrc5/Class I Transactivator. J Immunol. 2012;189(2):516–20. doi:10.4049/jimmunol.1200064. PMID:22711889
- Robbins GR, Truax AD, Davis BK, Zhang L, Brickey WJ, Ting JP-Y. Regulation of Class I Major Histocompatibility Complex (MHC) by Nucleotide-binding Domain, Leucine-rich Repeat-containing (NLR) Proteins. J Biol Chem. 2012;287(29):24294–303. doi:10.1074/jbc.M112.364604. PMID:22645137
- Staehli F, Ludigs K, Heinz LX, Seguin-Estevez Q, Ferrero I, Braun M, Schroder K, Rebsamen M, Tardivel A, Mattmann C, et al. NLRC5 Deficiency Selectively Impairs MHC Class I- Dependent Lymphocyte Killing by Cytotoxic T Cells. J Immunol. 2012;188(8):3820–8. doi:10.4049/jimmunol.1102671. PMID:22412192
- Tong Y, Cui J, Li Q, Zou J, Wang HY, Wang R-F. Enhanced TLR-induced NF-κB signaling and type I interferon responses in NLRC5 deficient mice. Cell Research. 2012;22(5):822–35. doi:10.1038/cr.2012.53. PMID:22473004
- Yao Y, Wang Y, Chen F, Huang Y, Zhu S, Leng Q, Wang H, Shi Y, Qian Y. NLRC5 regulates MHC class I antigen presentation in host defense against intracellular pathogens. Cell Research. 2012;22(5):836–47. doi:10.1038/cr.2012.56. PMID:22491475
- Chelbi ST, Guarda G. NLRC5, a promising new entry in tumor immunology. Journal for ImmunoTherapy of Cancer. 2016;4(1):39. doi:10.1186/s40425-016-0143-z. PMID:27437103
- Yoshihama S, Roszik J, Downs I, Meissner TB, Vijayan S, Chapuy B, Sidiq T, Shipp MA, Lizee GA, Kobayashi KS. NLRC5/MHC class I transactivator is a target for immune evasion in cancer. Proc Natl Acad Sci. 2016;113(21):5999–6004. doi:10.1073/pnas.1602069113.
- Chelbi ST, Dang AT, Guarda G. Emerging Major Histocompatibility Complex Class I-Related Functions of NLRC5. Adv Immunol. 2017;133:89–119. doi:10.1016/bs.ai.2016.11.003
- de Kruijf EM, van Nes JGH, Sajet A, Tummers QRJG, Putter H, Osanto S, Speetjens FM, Smit VTHBM, Liefers GJ, van de Velde CJH, et al. The Predictive Value of HLA Class I Tumor Cell Expression and Presence of Intratumoral Tregs for Chemotherapy in Patients with Early Breast Cancer. Clin Cancer Res. 2010;16(4):1272–80. doi:10.1158/1078-0432.CCR-09-1844. PMID:20145162
