ABSTRACT
The immune system plays an essential role in eradicating cancer in concert with various treatment modalities. In the absence of autologous tumor material, no standardized method exists to assess T cell responses against the many antigens that may serve as cancer rejection antigens. Thus, development of methods to screen for therapy-induced anti-tumor responses is a high priority that could help tailor therapy. Here we tested whether a tumor-derived antigen source called DRibbles®, which contain a pool of defective ribosomal products (DRiPs), long-lived and short-lived proteins (SLiPs) and danger-associated molecular patterns (DAMPs), can be used to identify tumor-associated antigen (TAA)-specific responses in patients before or after immunotherapy treatment. Protein content, gene expression and non-synonymous – single nucleotide variants (ns-SNVs) present in UbiLT3 DRibbles were compared with prostate adenocarcinomas and the prostate GVAX vaccine cell lines (PC3/LNCaP). UbiLT3 DRibbles were found to share proteins, as well as match tumor sequences for ns-SNVs with prostate adenocarcinomas and with the cell lines PC3 and LNCaP. UbiLT3 DRibbles were used to monitor anti-tumor responses in patients vaccinated with allogeneic prostate GVAX. UbiLT3-DRibble-reactive CD8+ T-cell responses were detected in post-vaccine PBMC of 6/12 patients (range 0.85–22% of CD8+ cells) after 1 week in vitro stimulation (p = 0.007 vs. pre-vaccine). In conclusion, a cancer-derived autophagosome-enriched preparation, packaging over 100 proteins over-expressed in prostate cancer into microvesicles containing DAMPs, could be used to identify CD8+ T cells in peripheral blood from patients after prostate GVAX vaccination and may represent a general method to monitor anti-cancer T cell responses following immunotherapy.
Introduction
Tumor cells express self and altered-self proteins (neo-antigens) and present these to the immune system by means of MHC-peptide complexes. The majority of the peptides that are presented on tumor human leukocyte antigen (HLA) molecules are likely derived from short-lived proteins (SLiPs) and defective ribosomal products (DRiPs) bound to HLA and transported to the cell surface.Citation1-Citation3 While these peptides may be readily presented by the tumor cells, the immune system has been poorly educated to recognize these, since these rapidly degraded products are less likely available for cross-presentation by professional antigen presenting cells (APCs) such as dendritic cells (DCs). Hence, the peptide repertoire that DCs use to prime the adaptive immune system might not be representative of the dominant epitopes tumor cells actually present, resulting in poor tumor-cell recognition. Moreover, in the presence of tumor-induced suppression, function of both DCs and T cells can be hampered, further compromising elimination of tumor cells by the immune system.Citation4,Citation5 Many immunotherapy regimens aim to convert the tolerant tumor microenvironment into an immune stimulatory one, thereby promoting tumor cell recognition and killing.Citation6,Citation7
Primary and metastatic prostate cancers have been characterized by sequencing, copy-number alteration and expression profiling.Citation8,Citation9 Recently, 333 primary prostate cancers were characterized and found to have an overall mutation burden of 0.94 mutations/Mb (median, range 0.04–28 per megabase), a lower mutation burden than many other epithelial tumors.Citation8,Citation10 150 metastatic prostate cancers were found to have an overall mutation rate of 4.4 mutations/Mb, though some had nearly 50 mutations/MbCitation9. We hypothesize that immune responses that develop in patients with objective clinical responses upon immunotherapy treatment are directed against a wide variety of antigenic determinants overexpressed by their cancer. In the case of prostate cancer, we reported that a patient, who experienced a complete response following treatment with ipilimumab, developed strong humoral immune responses against a number of proteins that had not been identified previously as possible tumor antigens. This included one protein, HIBCH, strongly over-expressed by the patient's tumor and found to be mutated in 5 percent of prostate cancers.Citation11 In another study, patients with prostate cancer that experienced a clinical response following treatment with GM-CSF and ipilimumab, exhibited some shared antibody responses, and a CD4T cell response was detected against one of these shared proteins, PAK6.Citation12 Neither HIBCH nor PAK6 had previously been considered to be tumor-associated antigens. While the concept that mutations or non-synonymous single nucleotide variants (ns-SNVs) in random proteins can serve as tumor-rejection antigens has been described, and is driving significant enthusiasm for novel immunotherapy strategies,Citation13-Citation17 this is in part a consequence of knowing the neo-epitopes present in a specific tumor and looking for an immune response against those antigens. While clearly an important step, this approach excludes the possibility of detecting immune responses against SLiPs/DRiPs and a spectrum of proteins known to be over-expressed by cancers, including many that were “prioritized” by a National Cancer Institute panel based on parameters that included known immunogenicity and in some cases, association of immune responses with therapeutic effects.Citation18 While protein arrays are allowing an evaluation of humoral immunity against more than 15,000 proteins,Citation12,Citation19 our ability to monitor T cell immunity has not kept pace. In 2017, the generally applied immune monitoring strategies focus on identifying responses against selected well-known antigens, the specific antigen(s) used as a vaccine or, when tumor sequence data is available, evaluate for response to possible neo-epitopes.Citation20 These approaches will miss immune responses against other over-expressed targets, whether or not they are mutated, will generally miss epitope spreading, and might underestimate the magnitude of the induced anti-cancer immune response.
Our lab developed autophagosome-enriched vaccines, called DRibbles®, which are generated from tumor cells or cell lines through inhibition of proteasomal degradation and lysosomal fusion. This results in accumulation of proteins including SLiPs and DRiPs in stable double-membrane autophagosomal vesicles that are isolated through cell disruption and differential centrifugation.Citation21-Citation24 A human allogeneic autophagosome vaccine, DPV-001, comprising DRibbles derived from an adenocarcinoma (UbiLT6) and a mixed histology adenocarcinoma/squamous cell cancer (UbiLT3) cell line, has recently been tested in a randomized multi-center Phase II study for adjuvant treatment of definitively treated stage IIIA/B NSCLC patients and a pilot study for men with advanced prostate cancer.Citation24 Gene expression analysis of the two cell lines, UbiLT3 and UbiLT6, revealed that they highly express genes for a wide range of antigens common to adenocarcinomas and squamous cancers (unpublished data).
While allogeneic DRibbles are not designed to encompass patient-specific neo-epitopes, whole exome-sequencing (WES) analysis identified sequences in the UbiLT3 and UbiLT6 cell lines that match the tumor sequence for a wide variety of tumor somatic mutations. Additionally, the potential of ns-SNVs to generate altered peptide ligands, may help break tolerance against non-altered proteins that are over-expressed by a patient's cancer.Citation25 In some cases, the vaccine, or monitoring agent (in this case UbiLT3 DRibbles) may match some ns-SNV somatic mutations. In addition, in vitro analyses showed that DRibbles derived from the UbiLT3 cell line contain agonist activity for multiple Toll-like receptors (TLRs (TLR-2,-3,-4,-7,-9) (TLH unpublished data), which can promote the activation of DCs and monocytes.Citation26 In this study, we analyzed transcript levels for antigens over-expressed in prostate cancers compared to the patients' normal tissue (The Cancer Genome Atlas (TCGA) PRAD Provisional database – 498 samples) for which protein was confirmed to be present in the UbiLT3 DRibble vaccine. We anticipated, also based on our in vivo mouse studies, that APCs would be required for the processing and cross-presentation of the DRibble-derived antigens,Citation22,Citation23,Citation27 allowing this approach to be used for all patients and not being restricted to HLA-A2+ patients, as is often the case for peptide-based monitoring approaches. This is, to our knowledge, the first report in which human tumor cell-line derived autophagosomes are used to monitor responses against non-viral antigens in human patient samples. Our data show that in some but not all prostate GVAX treated patients, IFN-γ producing CD8+ T cells could be identified in post vaccine PBMC after one week in vitro-stimulation with UbiLT3 DRibbles.
Results
Identification of overlapping gene expression, ns-SNVs and protein between UbiLT3 DRibbles and Prostate cancers
In order to determine whether the well-characterized UbiLT3 DRibble product could be used to monitor vaccine-induced anti-tumor T-cell responses in clinical samples from prostate cancer patients, we identified overlapping gene expression and protein products between prostate adenocarcinomas, UbiLT3 DRibbles, and the GVAX vaccine cell lines (PC3 and LNCaP). As is common for patients with metastatic prostate cancer, no material was available for gene expression analysis. Instead, publically available gene expression profiles, and ns-SNVs in prostate cancers compared to patients' normal tissue, were obtained from TCGA. Mass spectrometry identified 5286 proteins in UbiLT3 DRibbles. Gene expression data (RNA-Seq) was available in TCGA prostate adenocarcinoma (PRAD) provisional database for 5233 of the proteins. Proteins confirmed to be present in UbiLT3 DRibbles by mass spectrometry were queried (http://www.cbioportal.org/public-portal/) against gene expression data (z score ≥±2) for PRAD cases (n = 498) to identify the degree of overlap between genes overexpressed in tumor specimens compared to the patients' normal tissue and proteins in DRibbles used to monitor T-cell responses (average = 235.2 genes) (). Every patient's tumor over-expressed genes for which the protein is in UbiLT3 DRibbles, with 329 of 498 tumors over-expressing more than 100 genes coding for proteins confirmed to be present in UbiLT3 DRibbles. PRAD tumors overexpressed (z-score ≥ 2) an average of 734.5 genes, with a range from 57–2975. UbiLT3 DRibbles contained an average of 235.2 of the overexpressed genes per patient sample, range 5–1327. In samples categorized as having evidence of metastasis in the associated clinical data, the average number of overexpressed genes contained in UbiLT3 DRibbles increased to 330 (data not shown). The percent of overexpressed genes present in UbiLT3 ranged from 4% to 61%. There were at least 81 proteins identified in UbiLT3 DRibbles where the gene is over-expressed in ≥10% of the TCGA PRAD samples (z-score ≥ 2). Publically available gene expression data for the LNCaP and PC3 cell lines (TCGA, CCLE microarray) showed overexpression (z-score ≥ 2) of 5 and 24 of those genes, respectively (Supplemental Table S1).
Figure 1. Tumor-antigen overlap between the UbiLT3 DRibbles and Prostate cancer specimens in the TCGA database. A. Over-expressed genes (z-score ≥2) from RNA-Seq expression data on 498 prostate adenocarcinoma tumor specimens, compared to the patients' normal tissue (TCGA), analyzed for overlap with proteins identified in UbiLT3 DRibbles by mass spectrometry. B. Mutation data from WES of 307 prostate adenocarcinoma specimens (TCGA) for ns-SNVs in tumor specimens with respect to the patients' normal tissue was compared to the sequence of UbiLT3 for UbiLT3 sequences matching the tumor sequence. C. PC3 cell line sequences matching sequence overlap between UbiLT3 and PRAD ns-SNVs in panel B. D. LNCaP cell line sequences matching sequence overlap between UbiLT3 and PRAD ns-SNVs in panel B.
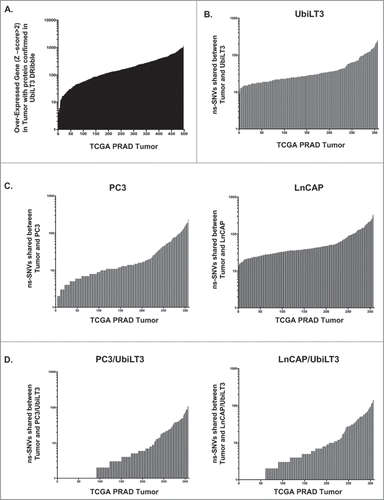
Since epitopes derived from mutations might be more immunogenic due to the creation of neo-epitopes, whole exome-sequencing data of the UbiLT3 cell line was compared to ns-SNVs identified in available prostate adenocarcinoma samples and PC3/LNCaP WES databases. Whole exome sequencing of the UbiLT3 cell line identified ns-SNVs in UbiLT3 with respect to the hg19 reference genome (9532 ns-SNVs; 1342 ns-SNVs filtered with (hg19) dbsnp (common) variants). VCF files for 307 specimens in the TCGA PRAD provisional database of exome-sequenced tumors were analyzed to determine unique ns-SNVs in tumor specimens with respect to the patients' normal tissue. Unique ns-SNVs were compared to the UbiLT3 sequence to identify potential mutations matching the UbiLT3 sequence using CLC Genomics Workbench 7.5 (Average = 44.38) (). Ns-SNVs were identified in all specimens where UbiLT3 matched the unique tumor sequence, with a range from 9 to 256. VCF files for PC3 and LNCaP cell lines were compared to the TCGA PRAD provisional database () for shared ns-SNVs (PC3 Average = 28; Range 1–234 and LNCaP Average = 59.6; Range 14–340 shared ns-SNVs). UbiLT3 and PC3 or UbiLT3 and LNCaP sequences were compared to PRAD specimens to identify corresponding ns-SNVs (). PRAD specimens had an average of 11.23 ns-SNVs found in PC3 and UbiLT3. Shared ns-SNVs ranged from 0 to 108. PC3 and UbiLT3 shared 3067 ns-SNVs (254 ns-SNVs filtered with (hg19) dbsnp (common) variants). PRAD specimens had an average of 15.34 ns-SNVs found in both LNCaP and UbiLT3. Shared ns-SNVs ranged from 0 to 148. LNCaP and UbiLT3 shared 4306 ns-SNVs (419 ns-SNVs filtered with (hg19) dbsnp (common) variants). UbiLT3 DRibbles contain 847 proteins covering 1310 of 6407 ns-SNVs present in PC3. 468 proteins cover 633 out of the 3067 ns-SNVs shared between PC3 and UbiLT3. 1745 UbiLT3 DRibble proteins cover 2881 of 11938 ns-SNVs present in LNCaP. 651 proteins cover 893 of 3067 ns-SNVs shared between LNCaP and UbiLT3. The mutation status of proteins identified by mass spectrometry and antigenicity/processing of mutations and ns-SNVs has yet to be determined.
Shared TAA in UbiLT3 DRibbles can be recognized by CD8+ T cells post prostate GVAX vaccination
The protein overlap and the presence of relevant ns-SNVs in DRibbles generated from the UbiLT3 cell line and the genes over-expressed by prostate cancers, prompted us to test whether UbiLT3 DRibbles could be used to detect tumor-antigen-specific T cells in blood samples from patients with prostate cancer. As a control, DRibbles were prepared from human primary kidney cells using the same protocol.
To test whether UbiLT3 DRibbles could be used to monitor the generation of a cancer-vaccine induced immune response, we isolated PBMC from patients with metastatic CRPC, who were vaccinated with prostate GVAX. From the 17 patients included in this trial, post GVAX PBMC samples (week 12) were available from 12/17 patients. No objective clinical responses were observed in any of the patients treated on this trial. These patients were very sick and did not receive any checkpoint blockade in addition to the prostate GVAX vaccine to overcome tumor-induced immune suppression.Citation28,Citation29 Two patients in cohort C and one patient in cohort B displayed an increase in prostate-specific antigen doubling time (PSA-DT) post-treatment, suggestive of reduced tumor growth. Additional clinical assessment and immune monitoring performed for this trial will be described elsewhere (Curti et al., and Puri et al. manuscripts in preparation).
As an initial screen, pre- and post treatment PBMC from four patients were cultured ex vivo in the presence of 25μg/ml DRibbles derived from normal kidney cells (NK) or UbiLT3 or vehicle control for 40 hours, after which supernatants were harvested and IFN-γ secretion was assessed (). No IFN-γ was detected in supernatants from pre-vaccine PBMC (data not shown), but IFN-γ secretion was detected in 3 out of 4 post-vaccine PBMC samples upon incubation with UbiLT3 Dribbles, but not vehicle, or NK DRibbles. In this experiment, the 3 patients in whom IFN-γ secretion was detected upon culture with UbiLT3 DRibbles also displayed increased or stabilized PSA-DT upon therapy (PSA-DT of 1.0, 1.3 and 3.4, for patients 9, 10 and 12, respectively).
Figure 2. UbiLT3 DRibble-specific CD8+ T cell responses in post GVAX PBMC. 2A. IFN-γ responses determined by CBA in supernatants of post-GVAX PBMC samples from four patients harvested after 40 hours ex vivo stimulation with vehicle, NK DRibbles (NK) or UbiLT3 DRibbles (UbiLT3). PSA-DT increases or decreases are depicted. 2B. Shown are the percentages of IFN-γ producing CD8+ T cells in 12 patients after 1 week in vitro stimulation of pre- (white bars) and post- (black bars) PBMC. Primary stimuli are as depicted on the x-axis and the secondary stimulus was 20 μg/ml UbiLT3 DRibbles. Duplicate wells were measured for each condition. The different treatment arms (A,B,C) and the ratio in PSA-DT are indicated in the graphs. N.a. = not available. 2C. Combined CD8+ IFN-γ responses for UbiLT3- (left) and NK DRibble prime/boost (right) for all patients (n = 12) and healthy donors (HD) (n = 6) tested. A 2-tailed Wilcoxin signed rank test was performed to determine a significant difference in response between pre-and post PBMC (p < 0.007).
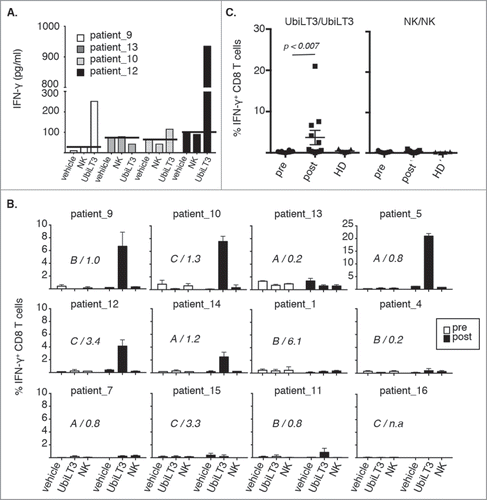
Next, we tested whether stimulation with UbiLT3 DRibbles induced IFN-γ producing CD8+ T cells. In order to increase the chance of detecting robust CD8+ T cell responses, a one-week in vitro stimulation protocol was applied. Of the twelve evaluable patients, IFN-γ producing CD8+ T cells responsive to antigens present within UbiLT3 DRibbles could be identified in in vitro-stimulated post-vaccine samples from 6/12 patients with a mean of 7.1% IFN-γ+ CD8+ T cells (range 0.85-22%) ( and ). IFN-γ producing CD8+ T cells were never observed in pre-vaccine PBMC or in post-vaccine PBMC stimulated with vehicle or NK DRibbles as primary stimulus followed by re-stimulation with UbiLT3 DRibbles () or NK DRibble prime/boost (). UbiLT3-responding CD8+ T cells were not detected in healthy donor (HD) PBMC, suggesting that these CD8 responses were vaccine induced and therefore likely to recognize antigens present in the UbiLT3 DRibbles (). CD8+ T-cell responses to UbiLT3 DRibbles in the patients tested were independent of the treatment arm, since the patients with a CD8 response to UbiLT3 Dribbles were present in all three arms (). There was no correlation between PSA-DT increase or decrease upon treatment and the ability to generate UbiLT3-recognizing IFN-γ producing CD8+ T cells, nor was there a prevalence for a specific HLA-type amongst the responding patients or an overlap in HLA-type with UbiLT3 (data not shown). Increased IFN-γ production in the UbiLT3 prime/boost condition was also apparent when looking at the IFN-γ mean fluorescence intensity levels of the post GVAX CD8± T cells compared to the pre GVAX and healthy control samples (Supplementary Fig. S2A). Moreover, antigen-independent CD3 stimulation, by means of plate-bound cross-linking using the anti-CD3 antibody OKT3 after UbiLT3 priming (UbiLT3 prime/OKT3 boost), showed that pre GVAX, post GVAX and healthy donor CD8± T cells were all equally equipped to produce IFN-γ, while no response was observed when cells were primed with NK DRibbles and boosted with UbiLT3 DRibbles (Supplementary Fig. S2A). Dividing the post GVAX samples into those patients without a CD8± T cell response to UbiLT3 prime/boost and those with a response, displayed significantly enhanced IFN-γ levels in the UbiLT3 prime/boost setting, but not in the UbiLT3/OKT3 or NK/UbiLT3 control conditions (Supplementary Fig. S2B).
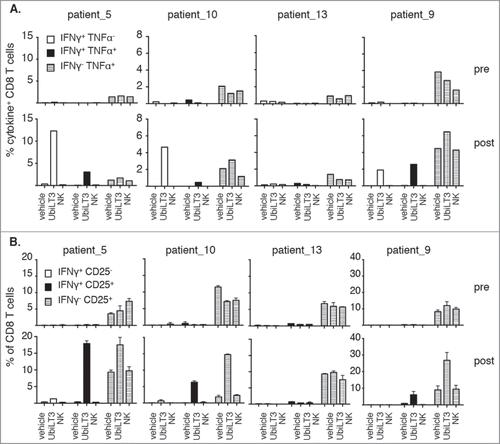
Of the twelve patients, we selected four patients (three UbiLT3-responding patients and one non-responding patient) to assess multi-functionality of the CD8± T cells by measuring IFN-γ and TNF-α co-expression. TNF-α producing cells were seen upon primary stimulation with vehicle or NK DRibbles and boost with UbiLT3 DRibbles, though frequencies increased under UbiLT3 DRibble prime/boost conditions in post-GVAX PBMC. In contrast, TNF-α+ IFN-γ+ cells as well as TNF-α− IFN-γ+ cells were specifically induced upon UbiLT3 DRibble prime/boost (). The patient in whom no IFN-γ+ CD8+ T cells had been detected after in vitro stimulation previously (patient-13) also exhibited no UbiLT3-induced TNF-α response. A similar pattern was observed looking at CD8+ T cell activation by means of CD25 expression (). In concurrence with the IFN-γ data, increased expression of CD25 on CD8+ T cells upon UbiLT3 DRibble stimulation was only observed in post GVAX PBMC and not in pre-vaccination samples.
Figure 3. UbiLT3-recognizing CD8T cells are poly-functional. 3A. Combined production of IFN-γ and TNF-α are depicted as the percentage of CD8+ T cells producing IFN-γ only (white bars), IFN-γ and TNF-α (black bars) or TNF-α only (striped bars). 3B. Induction of CD25 expression on CD8+ T cells, either or not in conjunction with IFN- γ release: IFN-γ+ CD25− (white bars), IFN-γ+ CD25+ (black bars) or IFN-γ− CD25+ (striped bars). 3A, B. The upper rows show data for pre-GVAX PBMC and the lower rows for post-GVAX PBMC. Duplicate wells were measured for each condition. Primary stimuli are as depicted on the x-axes and T cells were boosted using 20μg/ml UbiLT3 DRibbles.
Interferon-γ responses by CD4+ T cells were analyzed; however no UbiLT3-specific responses could be detected in pre- or post-vaccine PBMC samples after in vitro stimulation (Supplementary Fig. S3). In contrast, when DRibbles were generated from UbiLT3 cells transduced to stably express the CMV pp65 protein,Citation30 a CD4+ T cell response was observed both in pre-vaccine and post-vaccine PBMC in those patients who had apparently come in contact with this virus (Supplementary Fig. S3A), indicating that patient CD4+ T cells can recognize DRibble-derived antigens. Using these pp65 UbiLT3 DRibbles, we could also demonstrate that antigens derived from DRibbles are better processed and identified than the same antigen provided as protein or UbiLT3-pp65 cell lysate in an overnight stimulation protocol using PBMC from CMV-responsive donors (Supplementary Fig. S3B and data not shown).
No functional differences between pre- and post-vaccine CD11 c+ APC in presenting DRibble-derived antigens to CD8+ T cells
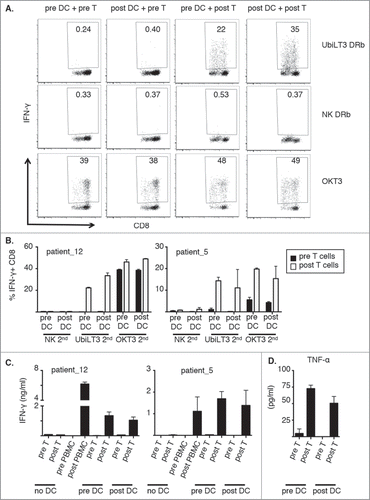
The prostate GVAX vaccine, through its secretion of the monocyte/DC-activating cytokine GM-CSF, could potentially have altered the functional abilities of the APC present within the post-vaccine PBMC, such as antigen (cross)-presentation and stimulation of T-cell proliferation. To assess qualitative differences between pre- and post-vaccine APC, pre/post crisscross experiments were performed with selected patient samples. These crisscross experiments showed that both pre- and post CD11 c+ cells were able to present UbiLT3 DRibble-derived antigens to CD8+ T cells but that only post GVAX-vaccine CD8+ T cells were able to respond by producing IFN-γ ( and ). In contrast, both pre- and post CD8+ T cells were able to produce IFN-γ in response to CD3 cross-linking, confirming that pre CD8+ T cells were functionally capable of secreting IFN-γ ( and ). The intracellular IFN-γ data were confirmed when looking at secreted IFN-γ in parallel cultures (). In addition, in these cultures TNF-α secretion was elevated upon prime/boost with UbiLT3 DRibbles (). TNF-α secretion was high in both pre- and post T cell conditions upon boost with OKT3 and no TNF-α was produced when NK DRibbles were used to boost the T cells (data not shown).
Figure 4. Both pre- and post-vaccine CD11 c+ cells can stimulate post vaccine CD8+ T cells to recognize UbiLT3 DRibble-derived tumor antigens. 4A. Dot plots from a pre/post crisscross experiment of one representative patient (out of 4 tested), looking at IFN-γ secretion by CD8+ T cells after the secondary stimulation with UbiLT3 DRibbles (top), NK DRibbles (middle) or plate-bound OKT3 (bottom) for the different combinations of pre- or post-DC with pre- or post T cells as indicated. 4B. Combined data of duplicate wells analyzed for two representative patients (out of 4 tested). The graph on the left corresponds with the patient data shown in 4A. 4C. Amounts of secreted IFN-γ in ng/ml. Data are shown for the 2 patients shown in B (out of 2 tested). 4D. TNF-α secretion data in pg/ml combined for the two patient samples depicted in 4B and C.
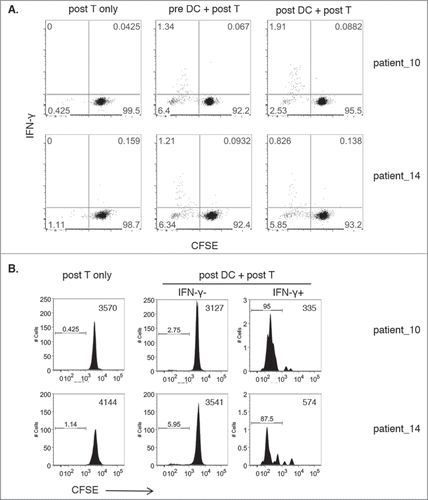
Similar crisscross results were obtained using two additional donors with lower frequencies of DRibble-responsive IFN-γ producing CD8+ T cells. In these experiments, isolated T cells were stained with carboxyfluorescein succinimidyl ester (CFSE) prior to co-culture with sorted, UbiLT3-stimulated, pre- or post CD11 c+ cells to determine whether IFN-γ producing T cells proliferated over the course of the one week stimulation. As shown in , IFN-γ+ CD8+ T cells, 1.34% (pre-DC) and 1.91% (post-DC) of post T cells for patient-10 and 1.21% (pre-DC) and 0.89% (post-DC) of post T cells for patient-14, showed low CFSE levels, indicating that the IFN-γ secreting cells are within the proliferated cell fraction, while the majority of the IFN-γ negative CD8+ T cells had not proliferated (). Indeed, gating specifically on IFN-γ positive cells in the post DC/post T cell conditions showed that 95% (patient-10) and 87.5% (patient-14) of all IFN-y positive cells had proliferated as shown in the histogram plots ().
Figure 5. Proliferation of DRibble-responsive CD8+ T cells. 5A. Dot plots show IFN-γ release (y-axis) versus CFSE levels gated on CD8+ T cells of post-GVAX isolated T cells co-cultured with sorted pre-GVAX or post-GVAX UbiLT3 DRibble-stimulated DC for two patients. Post-GVAX T cells only were taken along as negative controls. 5B. Histogram plots show CFSE levels of T cells only and gated IFN-γ− CD8+ and IFN-γ+ CD8+ T cells from co-cultures of post-GVAX UbiLT3 DRibble-stimulated DC and post-GVAX T cells.
Discussion
With increased recognition of the importance of the immune system on the disease course of cancers,Citation31-Citation36 approaches to study the mounting anti-cancer immunity in the absence of accessible autologous tumor tissue for read-out purposes are limited. It is crucial to find better ways to monitor the magnitude, and ideally the broadness, of therapy-generated T cell immune responses.Citation20 Here we show that an allogeneic, tumor-derived autophagosome preparation derived from a mixed histology adenocarcinoma/squamous cancer cell line, can be used as a monitoring tool to detect vaccine-induced CD8+ T cell responses in PBMC from patients with prostate cancer. This approach provides an alternative to MHC-multimer or peptide-based approaches to capture the breadth of the anti-cancer immune response, especially for those cases where screening of autologous tumors for over-expressed proteins or neo-epitopes is impossible or impractical due to absence or limited availability of material. In addition, our data confirm that multiple tumor histologies, particularly adenocarcinomas, display substantial overlap in gene expression (e.g. NSCLC, UbiLT3 cell line, and prostate cancer), and that these overlapping characteristics can be exploited for immune monitoring purposes.
Tumor mutations that provide neo-antigens the immune system can distinguish from self-antigens have been shown to be important in many clinical responses.Citation13,Citation15,Citation37 Here we suggest that over expressed cancer proteins, as well as over-lapping ns-SNVs between tumor cell lines and a cancer type can also be exploited for immune monitoring purposes. In this study UbiLT3 DRibbles were used to monitor responses to GVAX in mCRPC patients. No gene expression data for these patients was available, but by comparing the proteins in the UbiLT3 DRibbles to gene expression data for 498 PRAD patients (available through the TCGA), we found that the percent of overexpressed genes for which the protein is present in the UbiLT3 DRibbles ranged from 4% to 61%. A screening library of DRibbles from a wider range of tumor cell lines with unique gene expression, SLiPS, DRiPs and neo-epitopes may further increase the ability to monitor vaccine-specific responses in patients with various types of cancer. Our group recently showed that isolated ubiquitinated proteins, representing SLiPs targeted for proteasomal degradation, can also serve as a promising source of tumor antigen and can be recognized by tumor-antigen specific T cells.Citation38 The broad range of potential epitopes represented in a library of DRibble tumor cell lines increases the chances of detecting neo-antigen responses in addition to tumor antigens from SLiPs, DRiPs, overexpressed proteins and cancer-testis antigens. GuhaThakurta et al. showed that humoral responses against non-targeted tumor antigens could be detected in prostate cancer patients treated with Sipuleucel-T and correlated with improved overall survival.Citation19 Potentially, our monitoring approach could pick up T cell responses generated against non-targeted over-expressed proteins shared between a patient's prostate cancer and UbiLT3 DRibbles. Moreover, in the case of breast and prostate cancer there is evidence of immune responses against a number of shared non-mutated proteins expressed by these cancers, encouraging the development and testing of multivalent vaccines against non-mutated targets.Citation12,Citation39 Of note, we have observed that the amount of immune-activating danger signals contained within DRibble-preparations varies among different cell lines. UbiLT3 DRibbles, which contain danger signals activating multiple TLRs (TLR-2,-3,-4,-7,-9), were found to activate human monocytes and DC.Citation26 Of note, for this study DRibbles were prepared from the LNCaP prostate cancer cell line as well. However, we were unable to detect LNCaP responding T cells using the described IVS culture method. This could be explained due to low activation induction of monocytes/DCs using the LNCaP DRibbles compared to the UbiLT3 DRibbles. Also, in a clinical trial where mCRPC patients received GVAX in combination with ipilimumabCitation28 no T cell responses in IFN-γ ELIspot were observed after 10-day IVS with LNCaP cells, whereas responses were seen using PC3 cells as stimulators (unpublished data: personal communication with Prof. TD. de Gruijl). This suggests that the LNCaP cell line might not be very immunogenic. Therefore, our current study employed the more immune-activating, and well-characterized UbiLT3 DRibbles instead. The presence of multiple TLR ligands in UbiLT3 DRibbles may also explain the superior response against DRibble-derived pp65 antigen compared to loading of PBMC with pp65 protein (Fig. S3B). The fact that the response against the short peptide was much higher than the response against pp65 DRibbles in one of the two patients tested, might just reflect a total pp65 peptide concentration difference since DRibbles will also contain other antigens or the lack of need for antigen-uptake and processing of short peptide sequences. In a separate study, we have optimized the protocol for viral antigen recognition using DRibbles.Citation30
To date, little data are available on the magnitude of tumor-specific CD8+ T cell responses in patients with prostate cancer, when looking at intracellular cytokines and monitoring for more than one antigen. However, there are reports of vaccine-induced, antigen-specific CD8+ T cell responses detected by IFN-γ ELIspot against specific peptide pools.Citation40 In the majority of the clinical trials, the focus is on serum PSA responses, humoral responses as well as T cell activation or CD4-responses.Citation19,Citation28,Citation29,Citation41-Citation44 No autologous tumors were available from the patients from this trial, making it impossible to test whether the CD8+ T cells recognizing antigens in the UbiLT3 DRibble vaccine could also recognize and kill autologous tumor cells. The lack of primary tumor also prevented the search for patient-specific neo-epitopes and CD8+ T cells recognizing such epitopes.Citation13,Citation37,Citation45 In ongoing research, we are looking for ways to evaluate the tumor-killing abilities of DRibble-responsive T cells in other cancer types where autologous tumor cells are available. Also, we aim to perform comparable monitoring studies in a cohort of patients in whom objective clinical responses were observed after therapy to determine whether the presence or magnitude of DRibble-responsive CD8+ T cells correlate with improved clinical outcome. Had the patients in our trial received checkpoint inhibitors in conjunction with prostate GVAX, more clinical responses might have been obtained. It could thus be that the T cells, detected (and expanded) in our in vitro assay, were present in vivo, but could not effectively eliminate cancer cells due to the suppressive tumor microenvironment.
In conclusion, our data show that an autophagosome-based vaccine preparation, UbiLT3 DRibbles, shares tumor antigens with prostate cancers and can be used to detect vaccine-induced CD8+ T cell responses. We postulate that tumor-cell derived DRibble preparations, due to their content of SLiPs and DRiPsCitation22,Citation23 as well as danger signals,Citation27 could be used as a practical immune monitoring tool for multiple types of cancers to assess the magnitude of therapy-induced T cell responses and may be useful as a vaccine for patients with prostate cancer.
Patients and Methods / Materials and Methods
Clinical trial
A total of seventeen patients with mCRPC were enrolled to receive the prostate GVAX vaccine at our institute. This was a single center, US Department of Defense-funded clinical trial open at Providence Cancer Center between 2005 and 2007 (NCT00122005). An apheresis product was obtained from all patients before treatment (pre sample) and from 12/17 patients at week 12 (post sample). The 5 patients that went off-study before the week-12 apheresis were not included in the current study. Patients were randomized into three cohorts; patients received up to 12 prostate GVAX vaccines (two week intervals) with either no additional chemotherapy (cohort A), 350 mg/m2 cyclophosphamide on days 1–3 (cohort B) or 350 mg/m2 cyclophosphamide and 20 mg/m2 fludarabine on days 1–3 (cohort C). Men receiving nonmyeloablative chemotherapy received hematopoietic reconstitution with an adoptive transfer of an autologous apheresis product 2 days following completion of nonmyeloablative chemotherapy (See supplementary Fig. S1 for trial flow chart).
Collection of patient- and healthy donor blood samples
Leukapheresis products or blood draws were also obtained from anonymous healthy volunteers. Written informed consent was obtained from all patients and healthy donors in accordance with the Declaration of Helsinki. PBMCs were isolated using a density gradient separation method and were cryopreserved and stored in a monitored LN2 storage system under standardized operating procedures. Approval was obtained from the institutional review board (IRB) of Providence Portland Medical Center for all studies.
Protein expression and sequencing analysis
The UbiLT3 cell line used was developed at the Earle A. Chiles Research Institute and the UbiLT3 DRibble vaccine was performed by UbiVac. The Cell line identity testing and confirmation that it was absent from specified pathogens, was performed by BioReliance. Proteins in the UbiLT3 DRibble vaccine were TMT labeled and identified by mass spectrometry (OHSU Proteomics Core) on a Thermo Thermo Scientific Orbitrap Fusion, 2D-RPRP with 18 fractions, 120 min/fraction. The mass spectrometry data was processed with SEQUEST/PAWS using SPROT_HUMAN database. 5286 proteins were detected with a minimum of 2 unique peptides/protein. Proteins detected in UbiLT3 were screened against 498 prostate adenocarcinoma patient samples (TCGA PRAD, Provisional) and the Cancer Cell Line Encyclopedia in The Cancer Genome Atlas (TCGA) database using the cBio Portal (http://www.cbioportal.org/public-portal/) for genes with expression up-regulated (z score > 2) in tumor compared to the patients' normal tissue.
UbiLT3 exomic DNA was sequenced at 30x read depth (Otogenetics – Illumina Iibrary prep/QC; Agilent human V4/V5, 51 Mb capture/QC; HiSeq2000 NGS). Ns-SNVs were determined with respect to the hg19 reference genome. Publically available VCF files for 307 Prostate adenocarcinoma patients from the TCGA database (https://tcga-data.nci.nih.gov/tcga/ – from the PRAD study using the BI Mutation calling pipeline) were used to identify ns-SNVs present only in the tumor sample compared to the patient's normal tissue. Ns-SNVs unique to the tumor were compared to UbiLT3 ns-SNVs to identify shared ns-SNVs using the CLC Genomics Workbench 7.5.
The prostate GVAX vaccine consisted of PC3 and LNCaP cell lines. PC3 and LNCaP exome sequence data were obtained from the Cancer Cell Line Encyclopedia database (broadinstitute.org/ccle/home). Ns-SNVs for PC3 and LNCaP were determined with respect to hg19 and compared to unique ns-SNVs in the 307 prostate adenocarcinoma tumors described above. VCF files for PC3 (6407 ns-SNVs (reference hg19); 748 ns-SNVs filtered with (hg19) dbsnp (common) variants) and LNCaP (11938 ns-SNVs (reference hg19); 3802 ns-SNVs filtered with (hg19) dbsnp (common) variants) cell lines were compared to the TCGA PRAD provisional database for shared ns-SNVs. Shared ns-SNVs between prostate tumors and PC3 or LNCaP were compared to UbiLT3 to identify ns-SNVs shared between the tumors and PC3/UbiLT3 or LNCaP/UbiLT3.
In vitro stimulation of PBMC with DRibbles
Autophagosome-enriched DRibbles were prepared from the UbiLT3 cell line, primary kidney cells or UbiLT3 cells transduced to stably express the CMV pp65 protein or GFP control protein (plasmids p6 A-O'PB-IPWS-pp65 and pmax-GFP provided by Dr. Hong-Ming- Hu), similarly to the preparation of the clinical-grade DPV-001 vaccine, with minor alterations to what was described previously for the preparation of mouse DRibbles.Citation22,Citation23 Briefly, tumor cells were treated with 100 nM bortezomib (Velcade) (Millenium) and 10 mM NH4Cl (Hospira, 0409-6043-01) for 24 hours in a 5% CO2 incubator at 37°C. At 24 hours post treatment, cells were harvested and disrupted using a Bioruptor (Diagenode). Cells were centrifuged at 625xg (4°C) to pellet and discard the cellular debris. The supernatant was centrifuged at 2000xg (4°C) to pellet larger protein aggregates and remaining cellular debris. The supernatant was centrifuged at 12,000xg (4°C) for 20 minutes. The final DRibble pellet was resuspended in 6% hetastarch, protein content measured by BCA (Pierce, 23277) and DRibbles brought to 1 mg/mL.
Pre- and post-vaccine PBMC were thawed in warm HBSS supplemented with 1% human serum albumin (HSA; Baxter, 2G0201), were rested for 15 minutes at RT and were stimulated for 18–20 h with 75μg/ml UbiLT3 or control normal kidney (NK) DRibbles or a corresponding volume (75μl/ml) of 6% hetastarch (vehicle) (Hospira, 00409-7248-03) in sterile polypropylene 4 ml tubes, in a maximum volume of 1 ml (4 million cells/ml) in X-Vivo 15 media (Lonza, 04–418Q). Cells were cultured at 37°C and 5% CO2. After 18–20 h, cells were washed and taken up in media consisting of 50% X-Vivo 15 and 50% RPMI 1640 supplemented with 1.25% HSA, 2 mM L-glutamine (Lonza, BW17-605E), 1 mM sodium-pyruvate (Lonza, BW13-115E) and 50 μg/ml gentamicin-sulfate (Lonza, 17–518Z) (RPMI/HSA). Cells were transferred to 96-well round-bottom plates at a concentration of 1 million cells/ 200μl per well in duplicate. On day 4, 5 cU/ml IL-2 (Prometheus, NDC 65483-116-07) was added. On day 7, cells from each well were divided into 3 wells, and re-stimulated with plate-bound anti-CD3 (OKT3; Miltenyi Biotec, 170-076-124) as positive control (200μl of 10 μg/ml per well coated for for 4 hours at 37°C or for 18 hrs at 4°C), 20μg/ml NK DRibbles and 20μg/ml UbiLT3 DRibbles. After 5 hours, 5μg/ml Brefeldin-A (Sigma-Aldrich, B7651) was added and intracellular cytokine analysis was performed 12 hours later. For the analysis of secreted cytokines in the culture supernatant, cells were treated similarly as above, unless indicated otherwise in the text, without the addition of Brefeldin-A. Supernatants were harvested 18–20 hours after the start of the re-stimulation and stored at -80°C until further use.
Criss-cross IVS experiment
Pre- and post-PBMC were stimulated for 20 h with vehicle or UbiLT3 DRibbles as described above, after which CD11 c+ cells were isolated by flow cytometric sorting (CD11 c PerCP-eFluor710, eBiosciences: 46-0116-42; clone 3.9) from the DRibble-stimulated samples using a custom-build BD FACS Aria II (BD Biosciences) and T cells were isolated by means of untouched isolation from the vehicle cultured pre- and post PBMC using the pan T cell isolation kit II (Miltenyi Biotec: 130-096-535). Next, 50,000 CD11 c+ cells in 100μl RPMI/HSA and 200,000 T cells in 100μl X-Vivo15 were co-cultured for 7 days, combining pre- or post CD11 c+ cells with pre- or post T cells. Unsorted vehicle- and UbiLT3 DRibble stimulated PBMC samples were taken along as assay controls. After 7 days, cultures were re-stimulated with UbiLT3 DRibbles, NK DRibbles or OKT3 as indicated above and intracellular IFN-γ production and overall cytokine secretion were determined 18–20 hours later.
Magnetic-bead isolated pre- and post T cells were taken up in HBSS supplemented with 5% FBS at a concentration of 2 million cells/ml and were incubated with 1μM CFSE (Invitrogen, C34554) in a 37°C water bath for 7 minutes. Cells were washed three times with HBSS/FBS before resuspending in X-vivo15 media and the addition of 200,000 CFSE-labeled T cells to 50,000 UbiLT3 stimulated pre- or post CD11 c+ cells. Proliferation was assessed at the time of intracellular cytokine staining on day 8 after antigen re-stimulation. For intracellular cytokine staining protocol see below.
Flow cytometry analysis of surface markers and intracellular cytokines
For intracellular cytokine analysis, re-stimulated cells were harvested and stained for surface markers and viability using antibodies directed against CD3 (V500; BD Pharmingen: 560770; clone SP34-2), CD8 (BV421; BD Horizon: 562428; clone RPA-T8), CD4 (PE-Cy7; BD Pharmingen: 557852; clone SK3/Leu3a), CD14 (PE-TR; Invitrogen: MHCD1417; clone TuK4), CD25 (APC; Miltenyi Biotec: 130-098-213; clone 4E3) and yellow fixable live-dead dye (Invitrogen). After surface staining, cells were permeabilized and fixed using the BD fix/perm kit and incubated with anti-IFN-γ (PE; BD Pharmingen: 554701; clone B27) labeled antibodies for 30 minutes at 4°C. In some experiments cells were stained for intracellular TNF-α (APC; BD Pharmingen: 340534; clone 6401.1111) and not surface CD25. Cells were washed and analyzed on the Aria II flow cytometer. When feasible, 30,000 events were taken up for each tube. Analyses were performed using FlowJo software version 9.7.6 (Treestar).
Supernatant cytokine analysis by cytometric bead array
A Th1/Th2 cytometric bead array (CBA) was used for the detection of secreted IFN-γ, TNF-α, IL-2, IL-5, IL-10 and IL-4 (BD Biosciences: 550749). A BD HTS plate-reader attached to an LSR II BD flow cytometer running FACS Diva software was used for automated acquiring of the samples, acquiring 2000 bead events per well. BD Cytometric Bread Array 1.4 was used to analyze the data.
Statistical analysis
Prism6 software was used to generate graphs and perform statistical analysis. For the comparison of IFN-γ producing CD8+ T cells between pre- and post PBMC, a Wilcoxon matched-pairs signed rank test (two-sided) was used, based on nonparametric distribution. Conditions were considered statistically significant when p < 0.05.
Abbreviations
| APC | = | antigen presenting cell |
| CFSE | = | carboxyfluorescein succinimidyl ester |
| DAMPs | = | danger-associated molecular patterns |
| DC | = | dendritic cell |
| DRiPs | = | defective ribosomal products |
| HLA | = | human leukocyte antigen |
| PBMC | = | peripheral blood mononuclear cells |
| MHC | = | major histocompatibility complex |
| PRAD | = | prostate adeno |
| ns-snv | = | non-synonymous single-nucleotide variant |
| PSA-DT | = | prostate-specific antigen doubling time |
| SLiPs | = | short lived proteins |
| TAA | = | tumor-associated antigen |
| TCGA | = | The Cancer Genome Atlas |
| WES | = | whole exome sequencing |
Disclosures
Dr. BA. Fox and Dr. HM. Hu have ownership interests in UbiVac Inc. Dr. TL. Hilton is employed by UbiVac Inc.
Suppl_mat.zip
Download Zip (876.7 KB)Acknowledgments
The authors would like to thank the patients who participated in the clinical trial from which material was used for this study, and their families. We would also like to thank the nurses, staff, the Immune Monitoring Laboratory at the EACRI for the isolation of PBMC and accurate storage of healthy donor and patient samples and the Proteomics Shared Resource facility at Oregon Health and Science University in Portland, Oregon for performing the mass spectrometric analysis of the UbiLT3 DRibbles.
Additional information
Funding
References
- Yewdell JW. DRiPs solidify: progress in understanding endogenous MHC class I antigen processing. Trends Immunol. 2011;32:548–58. doi:10.1016/j.it.2011.08.001. PMID:21962745.
- Dolan BP, Bennink JR, Yewdell JW. Translating DRiPs: progress in understanding viral and cellular sources of MHC class I peptide ligands. Cell Mol Life Sci. 2011;68:1481–9. doi:10.1007/s00018-011-0656-z. PMID:21416150.
- Yewdell JW, Nicchitta CV. The DRiP hypothesis decennial: support, controversy, refinement and extension. Trends Immunol. 2006;27:368–73. doi:10.1016/j.it.2006.06.008. PMID:16815756.
- Motz GT, Coukos G. Deciphering and reversing tumor immune suppression. Immunity. 2013;39:61–73. doi:10.1016/j.immuni.2013.07.005. PMID:23890064.
- Baghdadi M, Yoneda A, Yamashina T, Nagao H, Komohara Y, Nagai S, Akiba H, Foretz M, Yoshiyama H, Kinoshita I, et al. TIM-4 glycoprotein-mediated degradation of dying tumor cells by autophagy leads to reduced antigen presentation and increased immune tolerance. Immunity. 2013;39:1070–81. doi:10.1016/j.immuni.2013.09.014. PMID:24315994.
- Gallois A, Bhardwaj N. Dendritic cell-targeted approaches to modulate immune dysfunction in the tumor microenvironment. Front Immunol. 2013;4:436. doi:10.3389/fimmu.2013.00436. PMID:24339825.
- Kershaw MH, Devaud C, John LB, Westwood JA, Darcy PK. Enhancing immunotherapy using chemotherapy and radiation to modify the tumor microenvironment. Oncoimmunology. 2013;2:e25962. doi:10.4161/onci.25962. PMID:24327938.
- Network TCGAR, Abeshouse A, Ahn J, Akbani R, Ally A, Amin S, Andry CD, Annala M, Aprikian A, Armenia J, et al. The molecular taxonomy of primary prostate cancer. Cell. 2015;163:1011–25. doi:10.1016/j.cell.2015.10.025. PMID:26544944.
- Robinson D, Van Allen EM, Wu Y-M, Schultz N, Lonigro RJ, Mosquera J-M, Montgomery B, Taplin M-E, Pritchard CC, Attard G, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–28. doi:10.1016/j.cell.2015.05.001. PMID:26000489.
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SAJR, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Børresen-Dale A-L, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21. doi:10.1038/nature12477. PMID:23945592.
- Graff JN, Puri S, Bifulco CB, Fox BA, Beer TM. Sustained complete response to CTLA-4 blockade in a patient with metastatic, castration-resistant prostate cancer. Cancer Immunol Res. 2014;2:399–403. doi:10.1158/2326-6066.CIR-13-0193. PMID:24795352.
- Kwek SS, Dao V, Roy R, Hou Y, Alajajian D, Simko JP, Small EJ, Fong L. Diversity of antigen-specific responses induced in vivo with CTLA-4 blockade in prostate cancer patients. J Immunol. 2012;189:3759–66. doi:10.4049/jimmunol.1201529. PMID:22956585.
- Gubin MM, Zhang X, Schuster H, Caron E, Ward JP, Noguchi T, Ivanova Y, Hundal J, Arthur CD, Krebber W-J, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014;515:577–81. doi:10.1038/nature13988. PMID:25428507.
- Lu Y-C, Robbins PF. Article in press. Seminars Immunol. 2015;1–6.
- Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348:69–74. doi:10.1126/science.aaa4971. PMID:25838375.
- Fritsch EF, Rajasagi M, Ott PA, Brusic V, Hacohen N, Wu CJ. HLA-binding properties of tumor neoepitopes in humans. Cancer Immunol Res. 2014;2:522–9. doi:10.1158/2326-6066.CIR-13-0227. PMID:24894089.
- Yadav M, Jhunjhunwala S, Phung QT, Lupardus P, Tanguay J, Bumbaca S, Franci C, Cheung TK, Fritsche J, Weinschenk T, et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature. 2014;515:572–6. doi:10.1038/nature14001. PMID:25428506.
- Cheever MA, Allison JP, Ferris AS, Finn OJ, Hastings BM, Hecht TT, Mellman I, Prindiville SA, Viner JL, Weiner LM, et al. The prioritization of cancer antigens: a national cancer institute pilot project for the acceleration of translational research. Clin Cancer Res. 2009;15:5323–37. doi:10.1158/1078-0432.CCR-09-0737. PMID:19723653.
- GuhaThakurta D, Sheikh NA, Fan LQ, Kandadi H, Meagher TC, Hall SJ, Kantoff PW, Higano CS, Small EJ, Gardner TA, et al. Humoral immune response against nontargeted tumor antigens after treatment with sipuleucel-T and its association with improved clinical outcome. Clin Cancer Res. 2015;21:3619–30. doi:10.1158/1078-0432.CCR-14-2334. PMID:25649018.
- Yuan J, Hegde PS, Clynes R, Foukas PG, Harari A, Kleen TO, Kvistborg P, Maccalli C, Maecker HT, Page DB, et al. Novel technologies and emerging biomarkers for personalized cancer immunotherapy. J ImmunoTherapy Cancer. 2016;4:3. doi:10.1186/s40425-016-0107-3.
- Li Y, Wang L-X, Pang P, Cui Z, Aung S, Haley D, Fox BA, Urba WJ, Hu H-M. Tumor-derived autophagosome vaccine: mechanism of cross-presentation and therapeutic efficacy. Clin Cancer Res. 2011;17:7047–57. doi:10.1158/1078-0432.CCR-11-0951. PMID:22068657.
- Li Y, Wang L-X, Pang P, Twitty C, Fox BA, Aung S, Urba WJ, Hu H-M. Cross-presentation of tumor associated antigens through tumor-derived autophagosomes. Autophagy. 2009;5:576–7. doi:10.4161/auto.5.4.8366. PMID:19333005.
- Twitty CG, Jensen SM, Hu H-M, Fox BA. Tumor-derived autophagosome vaccine: induction of cross-protective immune responses against short-lived proteins through a p62-dependent mechanism. Clin Cancer Res. 2011;17:6467–81. doi:10.1158/1078-0432.CCR-11-0812. PMID:21810919.
- Page DB, Hulett TW, Hilton TL, Hu H-M, Urba WJ, Fox BA. Glimpse into the future: harnessing autophagy to promote anti-tumor immunity with the DRibbles vaccine. J ImmunoTherapy Cancer. 2016;4:25. doi:10.1186/s40425-016-0130-4.
- Buhrman JD, Jordan KR, U'ren L, Sprague J, Kemmler CB, Slansky JE. Augmenting antitumor T-cell responses to mimotope vaccination by boosting with native tumor antigens. Cancer Res. 2013;73:74–85. doi:10.1158/0008-5472.CAN-12-1005. PMID:23161490.
- Xing Y, Cao R, Hu H-M. TLR and NLRP3 inflammasome-dependent innate immune responses to tumor-derived autophagosomes (DRibbles). Cell Death Dis. 2016;7:e2322. doi:10.1038/cddis.2016.206. PMID:27490927.
- Yi Y, Zhou Z, Shu S, Fang Y, Twitty C, Hilton TL, Aung S, Urba WJ, Fox BA, Hu H-M, et al. Autophagy-assisted antigen cross-presentation: autophagosome as the argo of shared tumor-specific antigens and DAMPs. Oncoimmunology. 2012;1:976–8. doi:10.4161/onci.20059. PMID:23162777.
- van den Eertwegh AJM, Versluis J, van den Berg HP, Santegoets SJAM, van Moorselaar RJA, van der Sluis TM, Gall HE, Harding TC, Jooss K, Lowy I, et al. Combined immunotherapy with granulocyte-macrophage colony-stimulating factor-transduced allogeneic prostate cancer cells and ipilimumab in patients with metastatic castration-resistant prostate cancer: a phase 1 dose-escalation trial. Lancet Oncol. 2012;13:509–17. doi:10.1016/S1470-2045(12)70007-4. PMID:22326922.
- Santegoets SJAM, Stam AGM, Lougheed SM, Gall H, Scholten PET, Reijm M, Jooss K, Sacks N, Hege K, Lowy I, et al. T cell profiling reveals high CD4+CTLA-4 + T cell frequency as dominant predictor for survival after prostate GVAX/ipilimumab treatment. Cancer Immunol Immunother. 2013;62:245–56. doi:10.1007/s00262-012-1330-5. PMID:22878899.
- Ye W, Xing Y, Paustian C, van de Ven R, Moudgil T, Hilton TL, Fox BA, Urba WJ, Zhao W, Hu H-M. Cross-presentation of viral antigens in dribbles leads to efficient activation of virus-specific human memory T cells. J Translational Med. 2014;12:100. doi:10.1186/1479-5876-12-100.
- Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–23. doi:10.1056/NEJMoa1003466. PMID:20525992.
- Ott PA, Hodi FS, Robert C. CTLA-4 and PD-1/PD-L1 blockade: new immunotherapeutic modalities with durable clinical benefit in melanoma patients. Clin Cancer Res. 2013;19:5300–9. doi:10.1158/1078-0432.CCR-13-0143. PMID:24089443.
- Couzin-Frankel J. Breakthrough of the year 2013. Cancer immunotherapy. Science. 2013;342:1432–3. doi:10.1126/science.342.6165.1432. PMID:24357284.
- Wolchok JD, Kluger H, Callahan MK, Postow MA, Rizvi NA, Lesokhin AM, Segal NH, Ariyan CE, Gordon R-A, Reed K, et al. Nivolumab plus ipilimumab in advanced melanoma. N Engl J Med. 2013;369:122–33. doi:10.1056/NEJMoa1302369. PMID:23724867.
- Gajewski TF, Schreiber H, Fu Y-X. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14:1014–22. doi:10.1038/ni.2703. PMID:24048123.
- Angell H, Galon J. From the immune contexture to the immunoscore: the role of prognostic and predictive immune markers in cancer. Curr Opin Immunol. 2013;25:261–7. doi:10.1016/j.coi.2013.03.004. PMID:23579076.
- Kvistborg P, Philips D, Kelderman S, Hageman L, Ottensmeier C, Joseph-Pietras D, Welters MJP, van der Burg S, Kapiteijn E, Michielin O, et al. Anti-CTLA-4 therapy broadens the melanoma-reactive CD8+ T cell response. Sci Transl Med. 2014;6:254ra128. doi:10.1126/scitranslmed.3008918. PMID:25232180.
- Yu G, Moudgil T, Cui Z, Mou Y, Wang L, Fox BA, Hu H-M. Ubiquitinated proteins isolated from tumor cells are efficient substrates for antigen cross-presentation. J Immunother. 2017;40:155–63. doi:10.1097/CJI.0000000000000165. PMID:28368960.
- Marquez JP, Stanton SE, Disis ML. The antigenic repertoire of premalignant and high-risk lesions. Cancer Prev Res. 2015;8:266–70. doi:10.1158/1940-6207.CAPR-14-0314.
- McNeel DG, Dunphy EJ, Davies JG, Frye TP, Johnson LE, Staab MJ, Horvath DL, Straus J, Alberti D, Marnocha R, et al. Safety and immunological efficacy of a DNA vaccine encoding prostatic acid phosphatase in patients with stage D0 prostate cancer. J Clin Oncol. 2009;27:4047–54. doi:10.1200/JCO.2008.19.9968. PMID:19636017.
- Sanda MG, Smith DC, Charles LG, Hwang C, Pienta KJ, Schlom J, Milenic D, Panicali D, Montie JE. Recombinant vaccinia-PSA (PROSTVAC) can induce a prostate-specific immune response in androgen-modulated human prostate cancer. Urology. 1999;53:260–6. doi:10.1016/S0090-4295(98)00539-1. PMID:9933036.
- Chen H, Liakou CI, Kamat A, Pettaway C, Ward JF, Tang DN, Sun J, Jungbluth AA, Troncoso P, Logothetis C, et al. Anti-CTLA-4 therapy results in higher CD4+ICOShi T cell frequency and IFN-gamma levels in both nonmalignant and malignant prostate tissues. Proc Natl Acad Sci USA. 2009;106:2729–34. doi:10.1073/pnas.0813175106. PMID:19202079.
- Liakou CI, Kamat A, Tang DN, Chen H, Sun J, Troncoso P, Logothetis C, Sharma P. CTLA-4 blockade increases IFNgamma-producing CD4+ICOShi cells to shift the ratio of effector to regulatory T cells in cancer patients. Proc Natl Acad Sci USA. 2008;105:14987–92. doi:10.1073/pnas.0806075105. PMID:18818309.
- Gerritsen WR. The evolving role of immunotherapy in prostate cancer. Ann Oncol. 2012;23 Suppl 8:viii22–7. doi:10.1093/annonc/mds259. PMID:22918924.
- van Buuren MM, Calis JJ, Schumacher TN. High sensitivity of cancer exome-based CD8 T cell neo-antigen identification. Oncoimmunology. 2014;3:e28836. doi:10.4161/onci.28836. PMID:25083320.
