ABSTRACT
CTCL follows different courses depending on the clinical stage at the time of diagnosis. Patients with early stage Mycosis Fungoides (MF) variant of CTCL may experience an indolent course over decades, whereas patients with advanced MF and Sézary Syndrome (SS) disease at diagnosis, often succumb within 5 years. Even within early stage CTCL/MF, a minority of patients will progress to more advanced stages. We recently generated RNA sequencing data on 284 CTCL-relevant genes for 157 patients and identified differentially expressed genes across stages I-IV. In this study, we aim to validate robust molecular markers linked to disease progression and survival. We performed multiple hypothesis testing-corrected analysis of variance (ANOVA) on the expression of individual genes across all CTCL samples and early stage (≤IIA) CTCL/MF patients. We used in silico immune cell-type deconvolution from gene expression data to estimate immune cell populations. Based on the analysis of all CTCL samples, we identified TOX, FYB, and CD52 as predictors of disease progression and poor survival. Among early stage (≤IIA) CTCL/MF patients, these 3 genes, along with CCR4, were valuable to predict disease progression. We validated these 4 genes in 3 independent, external Sézary Syndrome patient cohorts with RNA-Sequencing data. In silico immune cell-type deconvolution revealed that neutrophil infiltration in early stage MF conveyed a higher risk for disease progression. Also, NK cell infiltration in late stage MF/SS correlated with improved survival. TOX, FYB, CCR4 and CD52 are robust disease progression and decreased survival biomarkers in CTCL.
Introduction
Cutaneous T-Cell Lymphomas (CTCL) display a characteristic malignant clonal T lymphocyte skin infiltrate.Citation1 They remain uncommon (about 10 to 11 cases per million person-years in the United States and Canada).Citation2-Citation7 Most patients present with skin-limited CTCL that follows a smoldering course and does not progress to affect the blood or visceral organs, but approximately 15 to 20% of patients will progress to a potentially lethal disease with multi-organ involvement and survival of less than 5 years.Citation2,Citation8 Cutaneous lymphomas have a wide range of clinical presentations, including mycosis fungoides (MF) and the hematogenous/leukemic form, Sézary Syndrome (SS).Citation9 In addition to polymorphic clinical variants, even classic CTCL/MF can mimic benign, more common inflammatory conditions such as eczema, contact or drug induced spongiotic dermatitis, lichen planus and psoriasis, contributing to as much as 6-year delay in diagnosis from the initial presentation.Citation10
Predicting disease progression and overall survival in CTCL are challenging endeavors, but could help stratify early stage CTCL patients as those with indolent course and those at a higher risk for progression towards advanced stages.Citation11 Clinical disease stage at the time of diagnosis directly influences prognosis,Citation12 as expected. Search for molecular biomarkers has yielded clues into disease etiology and progression. Gene expression profiling studies have identified an association between adverse prognosis and high expression of TOX, GTSF1, NOTCH1, CCR4, ITK, FYB, SYC1, LCK or miR155, miR21, and let-7i differentially-expressed genes (DEGs).Citation9,Citation13-Citation20 Recently, a miRNA classifier, based on a linear combination of 3 miRNAs (miR-106b-5p, miR-148a-3p, and miR-338-3p), successfully separated early stage CTCL patients into two groups (high risk and low risk) with different outcomes based on disease progression and survival.Citation21 From gene expression data, computational methods to enumerate individual cell types, a process called RNA deconvolution, can also reveal prognostic biomarkers.Citation22 One such popular deconvolution algorithm, based on support vector regression and thus less sensitive to outliers, is CIBERSORT.Citation23
Recently we obtained targeted RNA-sequencing-based gene expression data from 157 patients with either CTCL or benign skin dermatoses (including skin tags, eczema, psoriasis among others),Citation24 mostly from formalin-fixed paraffin-embedded (FFPE) clinical samples. Our primary analysis validated previously identified DEGsCitation9,Citation24 and confirmed their ability to distinguish benign from CTCL samples.Citation24 We also performed subgroup analysis to identify additional DEGs and explored biases arising from processing FFPE samples on the TruSeq targeted RNA sequencing platform.Citation25
In this study, we use our TruSeq gene expression data from our previously characterized CTCL patient cohort to identify robust disease progression and survival markers, followed by external validation in publicly-available sequencing-based datasets generated from Sézary Syndrome patients.Citation26-Citation28 We also perform RNA deconvolution to identify immune cell types with possible clinical implications.
Results
High expression of TOX, FYB, and CD52 in CTCL samples correlates with greater risk of disease progression and poor survival
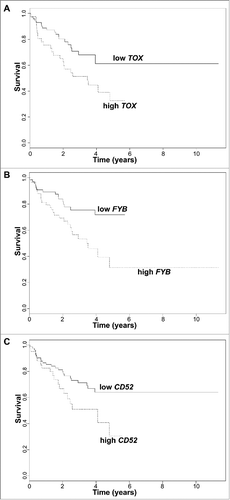
To robustly identify genes for which differential expression is strongly associated with disease progression, we performed one-way ANOVA across 134 Formalin Fixed and Paraffin Embedder (FFPE) samples of lesional CTCL skin from 110 patients with available clinical data from our cohortCitation24 (Supplementary Table 1 for clinical characteristics of CTCL patients). After multiple hypothesis correction using Benjamini-Hochberg method,Citation29 high expression of TOX (corrected p = 0.005), FYB (p = 0.005) and CD52 (p = 0.015) were associated with CTCL disease progression (). Survival analyses for these 3 genes confirmed their role in CTCL prognostication, as significantly-decreased disease-specific survival was observed for high expression of TOX (p = 0.020; log-rank test), FYB (p = 0.004), and CD52 (p = 0.032) (). Hazard ratios adjusted for age (along with 95% confidence intervals) were 2.03 (1.10–3.74) for TOX, 2.44 (1.29–4.60) for FYB, and 1.94 (1.05–3.59) for CD52.
Figure 1. Gene expression presented in Transcripts per million (TPM) according to disease progression status for TOX (A), FYB (B), and CD52 (C) in all CTCL samples. Quartile boxplots are shown.
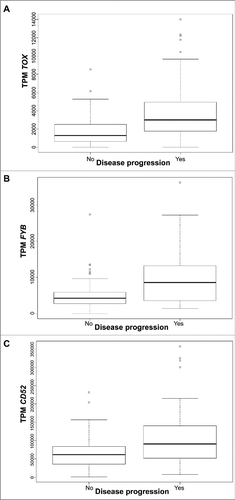
Figure 2. Survival analysis plots of all CTCL samples according to gene expression for TOX (A), FYB (B), and CD52 (C). Samples are dichotomized based on gene expression levels in TPM after finding an optimal cut-off; samples with TPM below cut-off labelled as low (solid line) and samples with TPM above cut-off labelled as high (dotted line). Survival plots correspond to Cox proportional hazards regression models.
High expression of TOX, FYB, CD52, and CCR4 in early stage CTCL samples is associated with disease progression
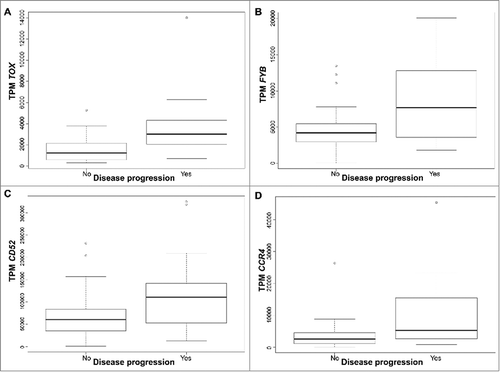
Identification of molecular biomarkers involved in discrimination between early stage (≤IIA) CTCL/MF patients, who progress and the majority, who do not is a challenging task.Citation11 We used one-way ANOVA analysis across a subset (64 FFPE samples of lesional CTCL/MF skin from 57 patients) of CTCL samples that corresponded to early stage disease (≤IIA),Citation24,Citation25 to identify genes that could be associated with disease progression to advanced (≥IIB) stages. Four genes were found to be significant at the p < 0.01 level: the three genes previously highlighted TOX (p < 0.001), FYB (p < 0.001), and CD52 (p = 0.008), plus an additional gene, CCR4 (p = 0.003) (). High expression of these four genes heralds disease progression in early stage CTCL/MF.
Figure 3. Gene expression presented in Transcripts per million (TPM) according to disease progression status for TOX (A), FYB (B), CD52 (C), and CCR4 (D), in early stage (≤IIA) CTCL samples. Quartile boxplots are shown.
While stage IA patients have normal life expectancy, those with stage IB experience a mild decrease in survival.Citation30 Analyzing a subset of these patients, specifically those with stage IB disease who either progressed to stage IIB (8 patients) or remained in stage IB (15 patients), higher expression of the four aforementioned biomarkers was observed in samples from patients who progressed from stage IB to IIB compared to those who did not (TOX mean TPM 3122 vs. 1578; CCR4 mean TPM 7570 vs. 5074; FYB mean TPM 9574 vs. 4503; CD52 mean TPM 103,543 vs. 73,074).
Higher RNA-sequencing expression of TOX, FYB, CD52 and CCR4 in Sézary patients compared to controls from external cohorts
To validate our expression based-biomarkers, we searched PubMed and NCBI Gene Expression Omnibus (GEO), NBI Short Read Archive (SRA) and EMBL-EBI European Genome-Phenome Archive (EGA) for RNA sequencing-based datasets pertaining to CTCL/MF/SS. We obtained RNA-Seq data from three studies focusing on Sézary Syndrome patients,Citation26-Citation28,Citation31 encompassing 14 affected patients and 3 healthy controls for Ungewickell et al. and Choi et al. combined, and 34 affected patients and 5 healthy controls for Wang et al. All samples analyzed in these three studies were from peripheral blood. Using non-parametric resampling to enable scale-free comparisons between different study designs and to decrease batch effect,Citation32 gene expression levels were compared for TOX, FYB, CD52 and CCR4 between affected (i.e., CTCL) and non-affected individuals. All four genes were significantly enriched amongst Sézary syndrome patients in Ungewickell et al. and Choi et al.: TOX (p < 10−5), FYB (p = 0.004), CD52 (p < 10−5), and CCR4 (p < 10−5) (). For Wang et al., more samples had TPM = 0 for the genes of interest, affecting results given the non-parametric approach. The ratio of mean TPM Sézary syndrome to mean TPM controls was at least >3 for all four genes (); TOX (p < 10−5), FYB (p = 0.0002) and CD52 (p < 10−5) were enriched in Sézary syndrome patients. Overall, these results indicate that our prognostication biomarkers can be applied to other cohorts as well.
Table 1A. External validation of CTCL progression-associated genes in two pooled independent cohorts of Sézary patients with RNA-Sequencing gene expression data (Ungewickell et al., Choi et al.).
Table 1B. External validation of CTCL progression-associated genes in the Wang et al. independent cohort of Sézary patients with RNA-Sequencing gene expression data.
In silico immune cell-type enumeration using RNA deconvolution
Leukocyte infiltration of malignant tumors, often described as the tumor micro-environment, has clinical prognostic implications.Citation33 Measurement of cell heterogeneity using conventional techniques such as immunohistochemistry can suffer from artefacts or prove impractical.Citation34 Gene expression signatures from pure cell lines can be used to enumerate cell composition of admixed, heterogeneous samples using RNA deconvolution.Citation34 RNA deconvolution can be used by itself as a novel biomarker.Citation22 Malignant inflammation from the tumor micro-environment influences CTCL tumorigenesis.Citation35 We first used CIBERSORT,Citation23 a robust algorithm, based on support vector regression, to determine the immune composition of our 134 CTCL FFPE samples obtained from 110 patients and then tested whether there was an association with disease progression in early stage (i.e. stage ≤IIA) CTCL/MF and survival advantage in late stage (≥IIB) CTCL (MF and SS).
In early stage CTCL, tumors from patients who progressed show a higher degree of neutrophilic infiltration (ANOVA; p = 0.032; ). Previous reports have indicated a link between high expression of IL-17A and IL-17F, neutrophil infiltration of CTCL tumors and increased risk of CTCL progression.Citation36,Citation37 Unfortunately, IL-17 genes were not included in our TruSeq panel. We observed that high levels of IL-17A and IL-17F, but also IL-17C, IL-17D and IL-17E mRNA, were found amongst Sézary patients (p < 10−5) along with a lower expression of IL-17B (p < 10−5) () for the two pooled cohorts of Ungewickell et al. and Choi et al. For the Wang et al. Sézary cohort, more samples had IL-17 genes with TPM = 0. The ratio of mean TPM Sézary to mean TPM controls was at least >1.5 for IL-17B, IL-17D and IL-17F expression (); IL-17A (p < 10−5), IL-17B (p < 10−5) and IL-17F (p < 10−5) were enriched in Sézary patients.
Figure 4. (A) Neutrophil immune cell fraction obtained in silico using CIBERSORT according to disease progression status in early stage (≤IIA) CTCL samples. Quartile boxplots are shown. (B) Survival analysis plots of late stage (IV) CTCL samples according to NK T lymphocyte fraction. Samples are dichotomized according to NK T lymphocyte immune cell fraction obtained in silico using CIBERSORT; samples with fraction below 0.05 labelled as low (solid line) and samples with fraction above 0.05 labelled as high (dotted line). Survival plot corresponds to Cox proportional hazards regression models.
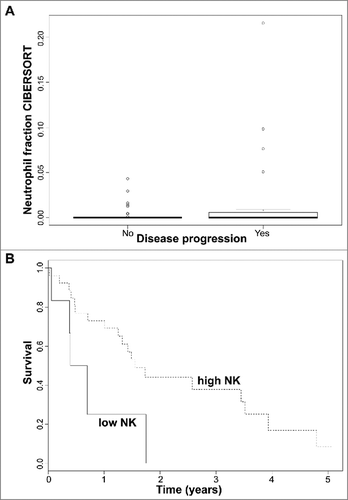
Table 2A. IL-17A through IL-17F expression in two pooled independent cohorts of Sézary patients with RNA-Sequencing gene expression data (Ungewickell et al., Choi et al.).
Table 2B. IL-17A through IL-17F expression in the Wang et al. independent cohort of Sézary patients with RNA-Sequencing gene expression data.
In late stage CTCL (stage IV), patient samples lacking NK T lymphocyte infiltration were associated with a faster demise (50% survival about 0.5 years if NK T cell <5% vs. more or less 1.5 years if NK T cell > 5%, as obtained by CIBERSORT) (Log-rank test; p = 0.032; ). This is consistent with reactive NK T lymphocytes as cytotoxic effectors to control CTCL progression, an infiltration linked with a better prognosis.Citation38
For immune cell type enumeration, we did not observe any significant association between 2 other cell types (CD8+ T lymphocytes or regulatory T lymphocytes) and disease progression/survival.
Discussion
We previously reported TOX, FYB, GTSF1, and CCR4 differentially-expressed genes (DEGs) when comparing CTCL and benign dermatoses, conditions that mimics this cancer.Citation24 In this study, we confirmed their importance to clinical prognostication by showing that TOX and FYB are associated with early stage disease progression. Also, in all CTCL samples across stage I-IV disease these genes were associated with disease progression and poor disease-related survival. In other cohorts, high expression of TOX, a protein implicated in T-cell development through chromatin regulation, is known to lead to CTCL tumor progression and poor overall prognosis,Citation39 where as a minimal level of TOX mRNA expression can be observed in normal skin or benign dermatoses.Citation16,Citation40 When over-expressed, FYB, a T cell adapter protein involved in T cell activation, has also been associated with advanced disease in other cohorts.Citation9,Citation30 In our cohort, CCR4 was found to be significantly associated with disease progression in early stage CTCL only. CCR4, a chemokine involved in T cell homing to the skin, is expressed in approximately 40% of CTCL patients,Citation41 and correlates well with skin infiltration by these cells.Citation42 High expression of CCR4 mRNA has been linked to disease progression in other cohorts.Citation9,Citation43 Importantly, CCR4 is an actionable target.Citation42 Mogamulizumab, a humanized anti-CCR4 monoclonal antibody, showed overall response rates (complete and partial responses) of 28.6% in MF patients, and 47.1% in Sézary patients, during a phase I/II clinical trial.Citation44 It can significantly reduce levels of CCR4-expressing malignant T cells.Citation45
We also found that high expression of CD52 leads to disease progression in early stage CTCL/MF, and to disease progression and decreased survival in all CTCL patients, a novel finding. CD52 is a glycosylphosphatidylinositol-anchored negatively-charged glycoprotein with potential roles in T cell migration and co-stimulation of the immune response.Citation46,Citation47 CD52 is highly expressed in both SS and MF compared to controls, in the latter more specifically in lesional skin.Citation48 Alemtuzumab, a humanized anti-CD52 monoclonal antibody, has demonstrated efficacy in MF and SS patients with an overall response rate of 55%,Citation49 but other studies in advanced stage CTCL, treatment-refractory patients showed a moderate response rate (38%), but short time to disease progression following treatment.Citation50 Of note, in chronic lymphocytic leukemia, response rates to alemtuzumab were correlated with the level of CD52 expression.Citation51
Importantly, we have successfully validated our four biomarkers with RNA-Seq data in Sézary patients from other cohorts,Citation26-Citation28,Citation31 as RNA-sequencing-based gene expression studies from skin CTCL clinical samples are sparse. Despite the fact that different tissues were analyzed in our study (skin tissue biopsy samples) and in the external validation cohort studies (peripheral blood samples), biomarkers were reproducibly enriched in skin CTCL samples and in blood Sézary samples, strongly supporting their involvement in tumorigenesis in both conditions.
RNA deconvolution from gene expression data also helped us identify clinically-relevant biomarkers.Citation22 Our in silico immune cell-type enumeration using RNA deconvolution with the CIBERSORT algorithmCitation23 yielded two cellular biomarkers related to CTCL tumorigenesis. First, advanced stage CTCL samples with the least (<5%) NK T lymphocyte infiltration showed decreased survival times. NK T lymphocytes play a role in a cytotoxic response to skin malignant infiltration, resulting in better prognosis.Citation38 An in vitro cellular model from primary tumors demonstrated that activation of NK T cells, such as with an anti-KIR3DL2 monoclonal antibody, leads to an anti-tumoral NK cytotoxic activity.Citation52 Second, neutrophilic infiltration of early stage CTCL lesions is linked to a progression to more advanced disease stages (>IIB) in our cohort of patients. Reports from independent cohorts are consistent with this observation in CTCL.Citation36,Citation37 Increased neutrophil infiltration, often calculated as a high neutrophil to lymphocyte ratio, usually heralds disease progression and decreased survival in other cancers, including melanoma.Citation53
Neutrophils are often recruited by cytokines produced by Th17 lymphocytes. We have found that IL-17A and IL-17F had increased RNA-Seq expression in Sézary patients compared to normal patients in one external cohort.Citation27 IL-17A and IL-17F were both expressed preferentially in lesional skin in CTCL patients,Citation37 but only IL-17F has been linked to disease progression.Citation37 A recent in vitro study showed that increased angiogenesis might be the result of IL-17F release.Citation54 IL-17 expression in CTCL is likely driven by external triggers (S. aureus bacterial toxins, UV radiation, dermatophytes, etc.) and other factors in the tumor microenvironment.Citation55 Based on recent clinical experience, it appears that blocking IL-17 signaling does not slow CTCL progression and must be further addressed with caution. Through clinical reportsCitation56 and personal clinical experience, where CTCL patients misdiagnosed with erythrodermic psoriasis received up to 1 year of secukinumab treatment that blocks IL-17 signaling, no disease improvement was observed and this medication failed to halt CTCL progression.
In conclusion, in this study, we confirmed the value, and cost-effectiveness, of performing TruSeq targeted RNA sequencing on clinically-obtained biopsy samples to uncover gene expression-based markers of survival and disease progression for cutaneous cancers. The biomarkers we identified robustly in our CTCL patients were validated in external cohorts, linked to CTCL tumorigenesis, and (for CCR4 and CD52) can be targeted by monoclonal antibodies. In the future, when precision medicine becomes more routine in the clinical setting, these molecular biomarkers may play an important role in stratifying patients at higher risk of disease progression that would benefit from early systemic therapy, while avoiding unnecessary side effects from these treatment modalities in patients at low risk of disease progression and mortality.
Methods
Patients and samples
Patients originated from the University of Texas MD Anderson Cancer Center (MDACC) under IRB-approved research protocols PA12-0267, PA12-0497 and Lab97-256, and from McGill University/McGill University Health Centre (MUHC) under IRB-approved protocols A09-M106-13A and 13-201-GEN, as previously described.Citation24 This study followed recommendations of the Research Ethics Board of the McGill University/McGill University Health Centre, with written informed consent from all subjects in accordance with the Declaration of Helsinki, and followed recommendations of the MD Anderson Cancer Center (MDACC) Research Ethics Board, which exempted us from obtaining written informed consent from patients, who earlier signed a hospital consent allowing their stored biopsy samples to be used for research. The diagnosis and clinical staging were established according to the diagnostic criteria of CTCL.Citation57 Follow-up times according to disease progression status are shown in Supplementary Fig. 1.
Data acquisition
Transcripts Per Million (TPM) from previously processed TruSeq data was re-analyzed; this data has been deposited in NCBI SRA under accession number SRP114956.Citation24,Citation25 The following de-identified clinical data variables from the CTCL patients cohort were considered: disease progression as a dichotomic variable, and survival/vital status as a dichotomic variable. Only time from sample biopsy to either death or last follow-up were considered. If exact date was not available, then the mid-point was considered (i.e. June 2011 would become June 15 2011). Samples were grouped as early (stage ≤IIA) vs. intermediate (stages IIB and III) vs. advanced (stage IV) CTCL, as previously described.Citation24,Citation25
Identification of disease progression and survival markers
One-way Analysis of Variance (ANOVA) test was performed for every gene, comparing disease progression status (defined as progression to a higher clinical stage), on all CTCL samples. P-values were then corrected for multiple hypothesis testing using Benjamini-Hochberg methodCitation29 and genes with corrected P-value <0.05 were considered. Disease-specific survival analyses were performed using Cox proportional hazards regression in R (packages “survival”, “KMsurv” and “OIsurv”); variables included vital status, time to death or last follow-up from sample biopsy date, age (adjustment parameter) and dichotomized gene expression based on an arbitrary TPM cut-off for each gene. For disease-specific survival analysis, p-values were computed using log-rank test.
We specifically looked at early stage MF samples, to confirm whether the same genes were found to be associated with disease progression status and whether other genes would be significant. We performed one-way ANOVA as described above. The same 3 genes, plus only one additional gene, were found using p-value <0.01 as a cut off point.
Cell-type enumeration using RNA deconvolution
CIBERSORT was used to perform RNA deconvolution using the standard LM22 leukocyte signature matrix, obtained from 22 immune pure cell linesCitation23 and 100 permutations. One-way ANOVA for disease progression and survival analyses were performed for early stage MF and late stage MF/SS, respectively.
External validation of disease progression and survival markers
We obtained publicly-available RNA-Seq data for Sézary Syndrome patients.Citation26-Citation28,Citation31 To prevent batch effectCitation32 from comparing different studies with different designs and annotations, we have converted TPM to a normalized rank, more specifically for each external case by taking the number of genes with a higher TPM value divided by the total number of reported genes in that sample. This gives rise to a non-parametric rank fraction value. We next computed p-values comparing the means of non-parametric rank fraction values between Sézary patients and control patients using a non-parametric resampling method (Bootstrapping with sample = 100 and number of iterations = 100,000). We inspected distributions from resampling means, for each gene and by patient type (disease vs. control), as a quality control procedure, histograms of which all followed normal distributions. Data from Ungewickell et al. and Choi et al. were pooled together given their small sample sizes, whereas data from Wang et al. had a sufficient number of Sézary syndrome and control samples and, hence, was analyzed separately. Two of the three studies did not provide clinical data on patient characteristics.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Suppl_mat.zip
Download Zip (16.3 KB)Acknowledgments
We thank Mr. Philippe Thibault for his assistance with the bioinformatical analysis. We thank Ms. Maria Cardenas for her assistance with data access management. We thank Dr. Paul Khavari and Ms. Aparna Bhudari for kindly sharing previously-published RNA-sequencing data.
Additional information
Funding
References
- Lamberg SI, Bunn PA, Jr. Cutaneous T-cell lymphomas. Summary of the Mycosis Fungoides Cooperative Group-National Cancer Institute Workshop. Arch Dermatol. 1979;115(9):1103–5. Epub 1979/09/01. doi:10.1001/archderm.1979.04010090053026. PMID:39515.
- Han T, Abdel-Motal UM, Chang DK, Sui J, Muvaffak A, Campbell J, Zhu Q, Kupper TS, Marasco WA. Human anti-CCR4 minibody gene transfer for the treatment of cutaneous T-cell lymphoma. PloS one. 2012;7(9):e44455. doi:10.1371/journal.pone.0044455. PMID:22973452.
- Saunes M, Nilsen TI, Johannesen TB. Incidence of primary cutaneous T-cell lymphoma in Norway. Br J Dermatol. 2009;160(2):376–9. Epub 2008/09/24. doi:10.1111/j.1365-2133.2008.08852.x. PMID:18808419.
- Ghazawi FM, Netchiporouk E, Rahme E, Tsang M, Moreau L, Glassman S, Provost N, Gilbert M, Jean SE, Pehr K, et al. Comprehensive analysis of cutaneous T-cell lymphoma (CTCL) incidence and mortality in Canada reveals changing trends and geographic clustering for this malignancy. Cancer. 2017;123(18):3550–67. doi:10.1002/cncr.30758. PMID:28493286.
- Litvinov IV, Tetzlaff MT, Rahme E, Habel Y, Risser DR, Gangar P, Jennings MA, Pehr K, Prieto VG, Sasseville D, et al. Identification of geographic clustering and regions spared by cutaneous T-cell lymphoma in Texas using 2 distinct cancer registries. Cancer. 2015;121(12):1993–2003. doi:10.1002/cncr.29301. PMID:25728286.
- Litvinov IV, Tetzlaff MT, Rahme E, Jennings MA, Risser DR, Gangar P, Netchiporouk E, Moreau L, Prieto VG, Sasseville D, et al. Demographic patterns of cutaneous T-cell lymphoma incidence in Texas based on two different cancer registries. Cancer Med. 2015;4(9):1440–7. doi:10.1002/cam4.472. PMID:26136403.
- Ghazawi FM, Netchiporouk E, Rahme E, Tsang M, Moreau L, Glassman S, Provost N, Gilbert M, Jean SE, Roshdy O, et al. Distribution and clustering of cutaneous T-cell lymphoma (CTCL) cases in Canada during 1992 to 2010. J Cutan Med Surg. 2017:1203475417745825. doi:10.1177/1203475417745825. PMID:29241349.
- Talpur R, Singh L, Daulat S, Liu P, Seyfer S, Trynosky T, Wei W, Duvic M. Long-term outcomes of 1,263 patients with mycosis fungoides and Sezary syndrome from 1982 to 2009. Clin Cancer Res. 2012;18(18):5051–60. doi:10.1158/1078-0432.CCR-12-0604. PMID:22850569.
- Litvinov IV, Netchiporouk E, Cordeiro B, Dore MA, Moreau L, Pehr K, Gilbert M, Zhou Y, Sasseville D, Kupper TS. The use of transcriptional profiling to improve personalized diagnosis and management of cutaneous T-cell lymphoma (CTCL). Clin Cancer Res. 2015;21(12):2820–9. doi:10.1158/1078-0432.CCR-14-3322. PMID:25779945.
- Kirsch IR, Watanabe R, O'Malley JT, Williamson DW, Scott LL, Elco CP, Teague JE, Gehad A, Lowry EL, LeBoeuf NR, et al. TCR sequencing facilitates diagnosis and identifies mature T cells as the cell of origin in CTCL. Sci Transl Med. 2015;7(308):308ra158. doi:10.1126/scitranslmed.aaa9122. PMID:26446955.
- Weed J, Girardi M. The difficult–and often delayed–diagnosis of CTCL. Sci Transl Med. 2015;7(308):308fs41. doi:10.1126/scitranslmed.aad2518. PMID:26446952.
- Talpur R, Sui D, Gangar P, Dabaja BS, Duvic M. Retrospective analysis of prognostic factors in 187 cases of transformed mycosis fungoides. Clinical Lymphoma, Myeloma Leuk. 2016;16(1):49–56. doi:10.1016/j.clml.2015.11.010. PMID:26702474.
- Sandoval J, Diaz-Lagares A, Salgado R, Servitje O, Climent F, Ortiz-Romero PL, Pérez-Ferriols A, Garcia-Muret MP, Estrach T, Garcia M, et al. MicroRNA expression profiling and DNA methylation signature for deregulated microRNA in cutaneous T-cell lymphoma. J Invest Dermatol. 2015;135(4):1128–37. doi:10.1038/jid.2014.487. PMID:25405321.
- Ralfkiaer U, Hagedorn PH, Bangsgaard N, Lovendorf MB, Ahler CB, Svensson L, Kopp KL, Vennegaard MT, Lauenborg B, Zibert JR, et al. Diagnostic microRNA profiling in cutaneous T-cell lymphoma (CTCL). Blood. 2011;118(22):5891–900. doi:10.1182/blood-2011-06-358382. PMID:21865341.
- Kamstrup MR, Gjerdrum LM, Biskup E, Lauenborg BT, Ralfkiaer E, Woetmann A, Ødum N, Gniadecki R. Notch1 as a potential therapeutic target in cutaneous T-cell lymphoma. Blood. 2010;116(14):2504–12. Epub 2010/06/12. doi:10.1182/blood-2009-12-260216. PMID:20538790.
- Litvinov IV, Cordeiro B, Huang Y, Zargham H, Pehr K, Dore MA, Gilbert M, Zhou Y, Kupper TS, Sasseville D. Ectopic expression of cancer-testis antigens in cutaneous T-cell lymphoma patients. Clin Cancer Res. 2014;20(14):3799–808. Epub 2014/05/23. doi:10.1158/1078-0432.CCR-14-0307. PMID:24850846.
- Litvinov IV, Netchiporouk E, Cordeiro B, Zargham H, Pehr K, Gilbert M, Zhou Y, Moreau L, Woetmann A, Ødum N, et al. Ectopic expression of embryonic stem cell and other developmental genes in cutaneous T-cell lymphoma. Oncoimmunology. 2014;3(11):e970025. doi:10.4161/21624011.2014.970025. PMID:25941598.
- Litvinov IV, Tetzlaff MT, Thibault P, Gangar P, Moreau L, Watters AK, Netchiporouk E, Pehr K, Prieto VG, Rahme E, et al. Gene expression analysis in Cutaneous T-Cell Lymphomas (CTCL) highlights disease heterogeneity and potential diagnostic and prognostic indicators. Oncoimmunology. 2017;6(5):e1306618. doi:10.1080/2162402X.2017.1306618.
- Litvinov IV, Zhou Y, Kupper TS, Sasseville D. Loss of BCL7A expression correlates with poor disease prognosis in patients with early-stage cutaneous T-cell lymphoma. Leuk Lymphoma. 2012;54(3):653–4. Epub 2012/08/04. doi:10.3109/10428194.2012.717695. PMID:22856870.
- Litvinov IV, Zhou Y, Pehr K, Kupper TS, Sasseville D. Gene expression analysis in CTCL patients validates the clinical importance of novel oncogenes and tumor suppressor genes. J Invest Dermatol. 2013;133:S62–S. PMID:WOS:000317698900361.
- Lindahl LM, Besenbacher S, Rittig AH, Celis P, Willerslev-Olsen A, Gjerdrum LMR, Krejsgaard T, Johansen C, Litman T, Woetmann A, et al. Prognostic miRNA classifier in early-stage mycosis fungoides: development and validation in a Danish nationwide study. Blood. 2018;131(7):759–70. Epub 2017/12/07. doi:10.1182/blood-2017-06-788950. PMID:29208599.
- Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, Nair VS, Xu Y, Khuong A, Hoang CD, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21(8):938–45. Epub 2015/07/21. doi:10.1038/nm.3909. PMID:26193342.
- Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nature methods. 2015;12(5):453–7. Epub 2015/03/31. doi:10.1038/nmeth.3337. PMID:25822800.
- Litvinov IV, Tetzlaff MT, Thibault P, Gangar P, Moreau L, Watters AK, Netchiporouk E, Pehr K, Prieto VG, Rahme E, et al. Gene expression analysis in Cutaneous T-Cell Lymphomas (CTCL) highlights disease heterogeneity and potential diagnostic and prognostic indicators. Oncoimmunology. 2017;6(5):e1306618. doi:10.1080/2162402X.2017.1306618. PMID:28638728.
- Lefrançois P, Tetzlaff MT, Moreau L, Watters AK, Netchiporouk E, Provost N, Gilbert M, Ni X, Sasseville D, Duvic M, et al. TruSeq-Based gene expression analysis of formalin-fixed paraffin-embedded (FFPE) cutaneous T-cell lymphoma samples: subgroup analysis results and elucidation of biases from FFPE sample processing on the TruSeq platform. Front Med. 2017;4:153. doi:10.3389/fmed.2017.00153.
- Ungewickell A, Bhaduri A, Rios E, Reuter J, Lee CS, Mah A, Zehnder A, Ohgami R, Kulkarni S, Armstrong R, et al. Genomic analysis of mycosis fungoides and Sezary syndrome identifies recurrent alterations in TNFR2. Nat Genet. 2015;47(9):1056–60. doi:10.1038/ng.3370. PMID:26258847.
- Choi J, Goh G, Walradt T, Hong BS, Bunick CG, Chen K, Bjornson RD, Maman Y, Wang T, Tordoff J, et al. Genomic landscape of cutaneous T cell lymphoma. Nat Genet. 2015;47(9):1011–9. Epub 2015/07/21. doi:10.1038/ng.3356. PMID:26192916.
- Wang L, Ni X, Covington KR, Yang BY, Shiu J, Zhang X, Xi L, Meng Q, Langridge T, Drummond J, et al. Genomic profiling of Sezary syndrome identifies alterations of key T cell signaling and differentiation genes. Nat Genet. 2015;47(12):1426–34. doi:10.1038/ng.3444. PMID:26551670.
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodol). 1995;57(1):289–300.
- Shin J, Monti S, Aires DJ, Duvic M, Golub T, Jones DA, Kupper TS. Lesional gene expression profiling in cutaneous T-cell lymphoma reveals natural clusters associated with disease outcome. Blood. 2007;110(8):3015–27. Epub 2007/07/20. blood-2006-12-061507 [pii]. doi:10.1182/blood-2006-12-061507. PMID:17638852.
- Lee CS, Ungewickell A, Bhaduri A, Qu K, Webster DE, Armstrong R, Weng WK, Aros CJ, Mah A, Chen RO, et al. Transcriptome sequencing in Sezary syndrome identifies Sezary cell and mycosis fungoides-associated lncRNAs and novel transcripts. Blood. 2012;120(16):3288–97. Epub 2012/09/01. doi:10.1182/blood-2012-04-423061. PMID:22936659.
- Goh WWB, Wang W, Wong L. Why batch effects matter in Omics data, and how to avoid them. Trends Biotechnol. 2017;35(6):498–507. Epub 2017/03/30. doi:10.1016/j.tibtech.2017.02.012. PMID:28351613.
- Whiteside TL. The tumor microenvironment and its role in promoting tumor growth. Oncogene. 2008;27(45):5904–12. Epub 2008/10/07. doi:10.1038/onc.2008.271. PMID:18836471.
- Shen-Orr SS, Gaujoux R. Computational deconvolution: extracting cell type-specific information from heterogeneous samples. Curr Opin Immunol. 2013;25(5):571–8. Epub 2013/10/24. doi:10.1016/j.coi.2013.09.015. PMID:24148234.
- Krejsgaard T, Lindahl LM, Mongan NP, Wasik MA, Litvinov IV, Iversen L, Langhoff E, Woetmann A, Odum N. Malignant inflammation in cutaneous T-cell lymphoma-a hostile takeover. Semin Immunopathol. 2017;39(3):269–82. doi:10.1007/s00281-016-0594-9. PMID:27717961.
- Ciree A, Michel L, Camilleri-Broet S, Jean Louis F, Oster M, Flageul B, Senet P, Fossiez F, Fridman WH, Bachelez H, et al. Expression and activity of IL-17 in cutaneous T-cell lymphomas (mycosis fungoides and Sezary syndrome). Int J Cancer. 2004;112(1):113–20. doi:10.1002/ijc.20373. PMID:15305382.
- Krejsgaard T, Litvinov IV, Wang Y, Xia L, Willerslev-Olsen A, Koralov SB, Kopp KL, Bonefeld CM, Wasik MA, Geisler C, et al. Elucidating the role of interleukin-17F in cutaneous T-cell lymphoma. Blood. 2013;122(6):943–50. doi:10.1182/blood-2013-01-480889. PMID:23801634.
- Bouaziz JD, Ortonne N, Giustiniani J, Schiavon V, Huet D, Bagot M, Bensussan A. Circulating natural killer lymphocytes are potential cytotoxic effectors against autologous malignant cells in sezary syndrome patients. J Invest Dermatol. 2005;125(6):1273–8. Epub 2005/12/16. doi:10.1111/j.0022-202X.2005.23914.x. PMID:16354199.
- Huang Y, Litvinov IV, Wang Y, Su MW, Tu P, Jiang X, Kupper TS, Dutz JP, Sasseville D, Zhou Y. Thymocyte selection-associated high mobility group box gene (TOX) is aberrantly over-expressed in mycosis fungoides and correlates with poor prognosis. Oncotarget. 2014;5(12):4418–25. doi:10.18632/oncotarget.2031. PMID:24947046.
- Huang Y, Su MW, Jiang X, Zhou Y. Evidence of an oncogenic role of aberrant TOX activation in cutaneous T-cell lymphoma. Blood. 2015;125(9):1435–43. doi:10.1182/blood-2014-05-571778. PMID:25548321.
- Hristov AC, Vonderheid EC, Borowitz MJ. Simplified flow cytometric assessment in mycosis fungoides and Sezary syndrome. Am J Clin Pathol. 2011;136(6):944–53. Epub 2011/11/19. doi:10.1309/AJCP09OTJOYAVZZK. PMID:22095381.
- Ishida T, Iida S, Akatsuka Y, Ishii T, Miyazaki M, Komatsu H, Inagaki H, Okada N, Fujita T, Shitara K, et al. The CC chemokine receptor 4 as a novel specific molecular target for immunotherapy in adult T-Cell leukemia/lymphoma. Clin Cancer Res. 2004;10(22):7529–39. Epub 2004/12/01. doi:10.1158/1078-0432.CCR-04-0983. PMID:15569983.
- Ferenczi K, Fuhlbrigge RC, Pinkus J, Pinkus GS, Kupper TS. Increased CCR4 expression in cutaneous T cell lymphoma. J Invest Dermatol. 2002;119(6):1405–10. Epub 2002/12/18. doi:10.1046/j.1523-1747.2002.19610.x. PMID:12485447.
- Duvic M, Pinter-Brown LC, Foss FM, Sokol L, Jorgensen JL, Challagundla P, Dwyer KM, Zhang X, Kurman MR, Ballerini R, et al. Phase 1/2 study of mogamulizumab, a defucosylated anti-CCR4 antibody, in previously treated patients with cutaneous T-cell lymphoma. Blood. 2015;125(12):1883–9. Epub 2015/01/22. doi:10.1182/blood-2014-09-600924. PMID:25605368.
- Ni X, Jorgensen JL, Goswami M, Challagundla P, Decker WK, Kim YH, Duvic MA. Reduction of regulatory T cells by Mogamulizumab, a defucosylated anti-CC chemokine receptor 4 antibody, in patients with aggressive/refractory mycosis fungoides and Sezary syndrome. Clin Cancer Res. 2015;21(2):274–85. Epub 2014/11/08. doi:10.1158/1078-0432.CCR-14-0830. PMID:25376389.
- Xia MQ, Hale G, Lifely MR, Ferguson MA, Campbell D, Packman L, Waldmann H. Structure of the CAMPATH-1 antigen, a glycosylphosphatidylinositol-anchored glycoprotein which is an exceptionally good target for complement lysis. Biochem J. 1993;293(Pt 3):633–40. Epub 1993/08/01. PMID:7688956.
- Watanabe T, Masuyama J, Sohma Y, Inazawa H, Horie K, Kojima K, Uemura Y, Aoki Y, Kaga S, Minota S, et al. CD52 is a novel costimulatory molecule for induction of CD4+ regulatory T cells. Clin Immunol. 2006;120(3):247–59. Epub 2006/06/27. doi:10.1016/j.clim.2006.05.006. PMID:16797237.
- Hahtola S, Tuomela S, Elo L, Hakkinen T, Karenko L, Nedoszytko B, Heikkilä H, Saarialho-Kere U, Roszkiewicz J, Aittokallio T, et al. Th1 response and cytotoxicity genes are down-regulated in cutaneous T-cell lymphoma. Clin Cancer Res. 2006;12(16):4812–21. Epub 2006/08/18. doi:10.1158/1078-0432.CCR-06-0532. PMID:16914566.
- Lundin J, Hagberg H, Repp R, Cavallin-Stahl E, Freden S, Juliusson G, Rosenblad E, Tjønnfjord G, Wiklund T, Osterborg A. Phase 2 study of alemtuzumab (anti-CD52 monoclonal antibody) in patients with advanced mycosis fungoides/Sezary syndrome. Blood. 2003;101(11):4267–72. Epub 2003/01/25. doi:10.1182/blood-2002-09-2802. PMID:12543862.
- Kennedy GA, Seymour JF, Wolf M, Januszewicz H, Davison J, McCormack C, Ryan G, Prince HM. Treatment of patients with advanced mycosis fungoides and Sezary syndrome with alemtuzumab. Eur J Haematol. 2003;71(4):250–6. Epub 2003/09/03. doi:10.1034/j.1600-0609.2003.00143.x. PMID:12950233.
- Ginaldi L, De Martinis M, Matutes E, Farahat N, Morilla R, Dyer MJ, Catovsky D. Levels of expression of CD52 in normal and leukemic B and T cells: correlation with in vivo therapeutic responses to Campath-1H. Leuk Res. 1998;22(2):185–91. Epub 1998/05/21. doi:10.1016/S0145-2126(97)00158-6. PMID:9593475.
- Sicard H, Bonnafous C, Morel A, Bagot M, Bensussan A, Marie-Cardine A. A novel targeted immunotherapy for CTCL is on its way: anti-KIR3DL2 mAb IPH4102 is potent and safe in non-clinical studies. Oncoimmunology. 2015;4(9):e1022306. Epub 2015/09/26. doi:10.1080/2162402X.2015.1022306. PMID:26405593.
- Lino-Silva LS, Salcedo-Hernandez RA, Garcia-Perez L, Meneses-Garcia A, Zepeda-Najar C. Basal neutrophil-to-lymphocyte ratio is associated with overall survival in melanoma. Melanoma Res. 2017;27(2):140–4. Epub 2017/01/27. doi:10.1097/CMR.0000000000000333. PMID:28125448.
- Lauenborg B, Litvinov IV, Zhou Y, Willerslev-Olsen A, Bonefeld CM, Nastasi C, Fredholm S, Lindahl LM, Sasseville D, Geisler C, et al. Malignant T cells activate endothelial cells via IL-17F. Blood Cancer J. 2017;7(7):e586. Epub 2017/07/22. doi:10.1038/bcj.2017.64. PMID:28731459.
- Willerslev-Olsen A, Krejsgaard T, Lindahl LM, Litvinov IV, Fredholm S, Petersen DL, Nastasi C, Gniadecki R, Mongan NP, Sasseville D, et al. Staphylococcal enterotoxin A (SEA) stimulates STAT3 activation and IL-17 expression in cutaneous T-cell lymphoma. Blood. 2016;127(10):1287–96. doi:10.1182/blood-2015-08-662353. PMID:26738536.
- De A, Raychaudhury T, Rajagopalan M, Sarda A, Sharma N. A case of cutaneous T-cell lymphoma, masquerading as psoriasis, was given etanercept and secukinumab: emphasizing the need for biopsy confirmation before starting biologics. Indian J Dermatol. 2017;62(5):533–5. Epub 2017/10/06. doi:10.4103/ijd.IJD_311_17. PMID:28979022.
- Olsen E, Vonderheid E, Pimpinelli N, Willemze R, Kim Y, Knobler R, Zackheim H, Duvic M, Estrach T, Lamberg S, et al. Revisions to the staging and classification of mycosis fungoides and Sezary syndrome: a proposal of the International Society for Cutaneous Lymphomas (ISCL) and the cutaneous lymphoma task force of the European Organization of Research and Treatment of Cancer (EORTC). Blood. 2007;110(6):1713–22. doi:10.1182/blood-2007-03-055749. PMID:17540844.
