ABSTRACT
NKp46 (CD335) is a surface receptor shared by both human and mouse natural killer (NK) cells and innate lymphoid cells (ILCs) that transduces activating signals necessary to eliminate virus-infected cells and tumors. Here, we describe a spontaneous point mutation of cysteine to arginine (C14R) in the signal peptide of the NKp46 protein in congenic Ly5.1 mice and the newly generated NCRB6C14R strain. Ly5.1C14R NK cells expressed similar levels of Ncr1 mRNA as C57BL/6, but showed impaired surface NKp46 and reduced ability to control melanoma tumors in vivo. Expression of the mutant NKp46C14R in 293T cells showed that NKp46 protein trafficking to the cell surface was compromised. Although Ly5.1C14R mice had normal number of NK cells, they showed an increased number of early maturation stage NK cells. CD49a+ILC1s were also increased but these cells lacked the expression of TRAIL. ILC3s that expressed NKp46 were not detectable and were not apparent when examined by T-bet expression. Thus, the C14R mutation reveals that NKp46 is important for NK cell and ILC differentiation, maturation and function.
Significance
Innate lymphoid cells (ILCs) play important roles in immune protection. Various subsets of ILCs express the activating receptor NKp46 which is capable of recognizing pathogen derived and tumor ligands and is necessary for immune protection. Here, we describe a spontaneous point mutation in the signal peptide of the NKp46 protein in congenic Ly5.1 mice which are widely used for tracking cells in vivo. This Ncr1 C14R mutation impairs NKp46 surface expression resulting in destabilization of Ncr1 and accumulation of NKp46 in the endoplasmic reticulum. Loss of stable NKp46 expression impaired the maturation of NKp46+ ILCs and altered the expression of TRAIL and T-bet in ILC1 and ILC3, respectively.
Introduction
Natural killer (NK) cells are cytolytic and cytokine-producing cells that contribute to eradicate pathogen-infected cells and cancers thereby mediating frontline defense and immunosurveillance.Citation1,Citation2 The capacity to mediate these functions depends on the balance between inhibitory and activating signals. Activating receptors include the family of natural cytotoxicity receptors (NCRs) NKp46, NKp30 and NKp44.Citation3,Citation4 NKp46 is expressed by NK cells and group 1 and 3 innate lymphoid cells (ILCs).Citation5-Citation7 NKp46 is encoded by Ncr1 and associates with the ITAM-containing CD3ζ or FcRγ polypeptides. Several endogenous ligands of NKp46 have been described including the complement factor P,Citation8 the intracellular filamentous cytoskeletal protein vimentin expressed on the surface of Mycobacterium tuberculosis–infected monocytes,Citation9 and several viral proteins such as hemagglutinin and hemagglutinin neuraminidases of the influenza,Citation10,Citation11 Sendai,Citation12 Newcastle disease,Citation13 ectromelia and vaccinia viruses.Citation14 NKp46 was also shown to recognize PfEMP1 of Plasmodium falciparum,Citation14 an unknown ligand from Fusobacterium nucleatum,Citation15 adhesins from Candida glabrata.Citation16 This wide expression supports that the triggering of NKp46 is essential for effective immune responses.
Previous studies have shown that mutations of the Ncr1 gene can cause a reduction in the cell surface expression of the NKp46 protein. Various strains of mice have been generated that either lack Ncr1 or exhibit mutations that have been chemically induced.Citation12,Citation17,Citation18 These lines (Ncrgfp/gfp, Ncr1Noe/Noe and Ncr1iCre/iCre) exhibit different phenotypes, possibly as a consequence of their difference in NKp46 protein expression. For example, Ncr1Noe/Noe and Ncr1iCre/iCre strains harbor modest gene changes but preserve the protein while Ncr1gfp/gfp mice,Citation17 and Ncr1gfp/gfpKlrk1−/- double knockout mice which have defects in both NKp46 and NKG2D expression,Citation19 lack exons 5–7 resulting in a complete disruption of the protein.
Narni-Mancinelli et al.Citation18 first described the effect of the point mutation W32R in a colony of C57BL/6J mice carrying N-ethyl-N-nitrosourea (ENU)–induced mutations (called Noé mice). These mice had normal numbers of NK cells but showed an impaired expression of NKp46 on the cell surface. Moreover, Noé mice displayed an increased expression of the zinc-finger protein Helios which is involved in the regulation of lymphocyte development. Noé NK cells were hyper responsive to various stimuli in vitro and Noé mice were more resistant to infection with mouse cytomegalovirus.Citation18 Glasner et al.Citation20 further characterized the effect of the Noé mutation clarifying that the NKp46W32R protein could reach the surface of NK cells, but in a slow and unstable manner. This resulted in an accumulation of NKp46 in the endoplasmic reticulum (ER) and limited transport to the cell surface.Citation20 Recently, studies of human NK cells carrying the same mutation NKp46W32R led to similar conclusions.Citation21
Here we describe the analysis of two independent colonies of congenic CD45.1+ (Ly5.1) mice that exhibited a spontaneous single point mutation (C14R, designated Ly5.1C14R mice) in the signal sequence of the Ncr1 gene. This mutation impaired NKp46 surface expression in ILC subsets. The C14R mutation did not alter the overall expression of Ncr1 mRNA in mutant NK cells but impaired the surface expression of NKp46 in ILCs and was associated with alteration in ILC maturation. These results were also confirmed by a newly generated NCRB6C14R strain.
Results
Ly5.1 congenic mouse strains exhibit a reduced surface expression of NKp46
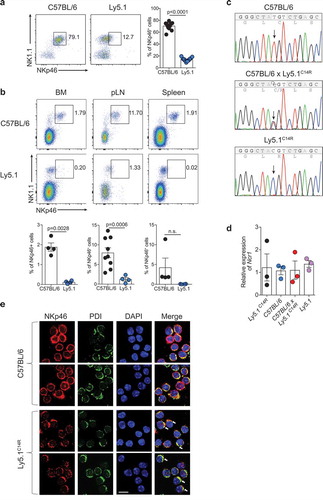
Ly5.1 (B6.SJL-Ptprca Pepcb/BoyJ) congenic mice expressing CD45.1 on the surface of all lymphocytes are classically used in combination with CD45.2-expressing C57BL/6 mice to establish experimental systems in which donor and host immune cells can be faithfully tracked both at steady-state and during an immune challenge. However, during analyses of these types of systems, it was noticed that Ly5.1+ mice consistently exhibited very poor expression of the receptor NKp46, a key phenotypic marker of NK cellsCitation22 and some subsets of ILCs, in multiple tissues including peripheral blood (), bone marrow, peripheral lymph nodes and spleen (). This observation was made in two mouse colonies completely independent of each other but which were originally derived from the Jackson Laboratory (Jax) imported in 2008 (University Hospital Erlangen) and 2010 (Walter and Eliza Hall Institute, WEHI) and maintained as closed colonies. This was distinct from newly imported mice (2012, University Hospital Erlangen; 2016, WEHI), also from Jax, which exhibited NKp46 expression equivalent to the C57BL/6 lines and F1 mice derived from the originally imported Ly5.1 line (C57BL/6 × Ly5.1WEHI) (SI Appendix, Figure S1A). These findings suggested that this alteration was a recessive trait limited to specific colonies that may have carried the mutation. While this mutation was tightly linked to the Ptprca locus on chromosome 1, Ncr1 is found on chromosome 7 indicating that the alteration was not within Ptprca itself.
Figure 1. Ly5.1 congenic mouse strain exhibits reduced surface expression of NKp46 that alters the localization of the NKp46 protein. (A) Dot plot showing the staining and frequency of NK1.1+NKp46+ cells in the peripheral blood lymphocytes of C57BL/6 and Ly5.1 mice. Data show representative plots gated on live lymphocytes CD3− CD19− (n = 12 mice/genotype). (B) Dot plots showing the expression and frequency quantification of NK1.1+NKp46+ cells in the bone marrow (BM), peripheral lymph node (pLN) and spleen of C57BL/6 and Ly5.1 mice. (A, B) Data show representative plots gated on live lymphocytes CD3−CD19− pooled from two to five independent experiments (n = 4–12 mice/genotype/tissue). (C) Sanger sequencing analysis of the Ncr1 gene in C57BL/6, C57BL/6 × Ly5.1C14R and Ly5.1C14R mice showing the position of the point mutation. (D) Relative levels of NKp46 transcripts in splenic NK cells of C57BL/6, Ly5.1C14R, WT Ly5.1 and C57BL/6 × Ly5.1C14R mice. Data show the mean ± SEM of 3–4 mice/genotype for one of three similar experiments. P values were calculated using an unpaired two-tailed Student’s t test. (E) NKp46 localization in primary NK cells. Representative images of NK cells isolated from C57BL/6 and mutant Ly5.1C14R mice stained with anti-NKp46 and anti-PDI primary antibodies, and AlexaFluor488-conjugated anti-goat and AlexaFluor546-conjugated anti-rabbit secondary antibodies (DAPI nuclear stain, blue; anti-NKp46, red; PDI, green). Images were obtained using confocal scanning microscopy. Arrows indicate ER localization. Scale bar, 10 μm.
A single amino acid change in the Ncr1 gene abrogates stable NKp46 surface expression
To understand the basis of this alteration, we used whole exome sequencing to examine C57BL/6 Ly5.1 (WEHI) and Ly5.1 (WEHI) from the mouse colonies in Melbourne, Australia. Assuming a recessive pattern of inheritance, 1042 SNPs were identified that were homozygous in the affected mice and heterozygous in the parental strain (SI Appendix, Dataset S1). Of these, 670 SNPs were classified as low impact mutations based on variant effector predictor analysis and as such were excluded from further analysis. Based on the observed phenotype, we initially concentrated on the candidate gene Ncr1. NKp46 is a 46 kDa type 1 transmembrane glycoprotein characterized by a short intracellular tail, a single transmembrane domain, and two extracellular Ig-like domains. Sequence analysis identified a single homozygous missense point mutation (T to C) in the signal peptide of Ncr1 in both strains of mice that exhibited low NKp46 protein expression. We confirmed by Sanger sequencing that the mutation was present in Ly5.1 WEHI (hereinafter referred to as Ly5.1C14R ) mice and C57BL/6 × Ly5.1C14R but not C57BL/6 mice and resulted in a single amino acid substitution of cysteine to arginine (C14R; ). In contrast, the RNA level of Ncr1 did not significantly differ in NK cells from C57BL/6, Ly5.1C14R, WT Ly5.1 and C57BL/6 × Ly5.1C14R mice (). It should be noted that subsequent to identification of this mutation in the established lines at each institution, new founders were imported to the WEHI (2016) to establish a new ‘unmutated’ colony. Breedings were kept entirely separate from the original colonies and progeny from the new breedings were analysed. Of these, 39 of 88 mice tested exhibited a reduction in NKp46 expression by flow cytometry.We further sequenced 29 mice from the new colony; 9 were homozygous wild-type for the Ncr1 gene and were used to reestablish the colony while the remainder were heterozygous. Thus, it appears that the mutation has inadvertently been retained undetected long term in parent colonies.
The C14R Ncr1 mutation significantly impairs surface NKp46 expression in 293T cells
As the discovery that a mutation in the leader sequence might affect NKp46 cell surface expression or trafficking was unexpected, we sought to demonstrate that ectopic expression of the NKp46C14R mutation itself was responsible for the alterations we observed in the mutated Ly5.1 mice. To this end, 293T cells were transfected with wild-type NKp46 or NKp46C14R cDNA, respectively, and the expression of NKp46 was tracked by flow cytometry. The surface expression of the NKp46C14R protein on 293T transfectants was significantly reduced compared with wild-type NKp46 while the total cellular NKp46 expression was at a comparable level (SI Appendix, Figure S1B and C), demonstrating that the impaired surface expression of NKp46 in Ly5.1 mice in fact is due to the C14R mutation in the NKp46 signal peptide. Analyses of freshly isolated naïve NK cells from wild-type or Ly5.1C14R mice by confocal laser scanning microscopy not only confirmed the reduced NKp46C14R surface localization, but also revealed a spotted intracellular distribution of the mutated NKp46C14R protein. Furthermore, the colocalization of NKp46 with protein disulfide isomerase (PDI), an enzyme in the endoplasmic reticulum, indicated an accumulation of NKp46C14R in that cell compartment ( and SI Appendix, S1 D). Thus the C14R mutation in the signal peptide of NKp46C14R resulted in disruption of NKp46 surface expression by failed trafficking within the cell.
Ly5.1C14R mice have normal numbers of total NK cells but altered distribution of ILC subsets
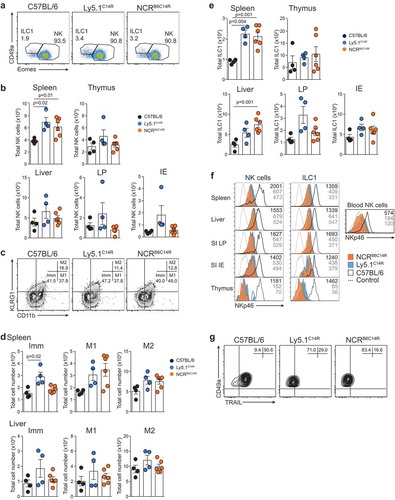
We then interrogate if the C14R mutation might impact the subsets of cells that normally express NKp46, i.e. NK cells, ILC1 and NCR+ ILC3s. Ly5.1C14R mice exhibited normal numbers of NK1.1+CD3− NK cells in the majority of the tissues but these numbers were significantly increased in the thymus compared with control mice (). We then examined the maturation status of NK cells by measuring the expression of KLRG1 and CD11b in C57BL/6 and Ly5.1C14R NK cells (). This revealed an increase in the immature (Imm) NK cell subsets in the spleen but not in the liver of Ly5.1C14R mice suggesting that steady-state signalling through NKp46 may act to modulate early NK cell maturation. Similar findings were observed when cells were analysed using CD27 and CD11b (SI Appendix, Figure S2A and B) and in Noé NK cells (SI Appendix, Figure S2C and D), a phenotype that was rescued by the expression of human NKp46.
Figure 2. Disruption of NK cell homeostasis and maturation of ILC1 and NK cells in Ly5.1C14R mice. (A) Dot plots showing the frequency of ILC1 and NK cells in the spleen. Data show representative plots gated on live NK1.1+ lymphocytes excluding T and B cells in C57BL/6, Ly5.1C14R and C57BL/6 × Ly5.1C14R mice. (B) Total number of NK cells in spleen, thymus, liver, small intestine lamina propria (LP) and within the intestinal intraepithelial compartment (IE). Data show the mean ± SEM pooled from three to six independent experiments (n = 6–15 mice/genotype); thymus are pooled from three to five independent experiments (n = 6–12 mice/genotype). P values were calculated using an unpaired two-tailed Student’s t test. (C) FACS plots showing the frequency of immature (Imm, KLRG1−CD11b−), mature 1 (M1, KLRG1−CD11b+) and mature 2 (M2, KLRG1+CD11b+) NK cells in splenic NK1.1+CD3−CD19− NK cells. (D) Total number of Imm, M1 and M2 NK cells in the spleen and liver of C57BL/6, Ly5.1C14R and C57BL/6 × Ly5.1C14R mice showing the mean ± SEM pooled from three to six independent experiments (n = 6–15 mice/genotype). P values were calculated using an unpaired two-tailed Student’s t test. (E) Total number of ILC1s in spleen, thymus, liver, small intestine lamina propria (LP) and within the intestinal intraepithelial compartment (IE). Data show the mean ± SEM pooled from three to six independent experiments (n = 6–15 mice/genotype); thymus data are pooled from three to five independent experiments (n = 6–12 mice/genotype). P values were calculated using an unpaired two-tailed Student’s t test. (F) Histograms showing the mean fluorescence intensity of NKp46 in various tissues for both NK cells and ILC1 for wild-type (black solid line), Ly5.1C14R (solid blue) and C57BL/6 × Ly5.1C14R (red solid line). CD3ε+ cells were used as a control for NKp46 expression (black dashed line). Data are representative of tissues analyzed in (A–E). (G) Expression of TRAIL on NK1.1+CD49a+CD3−CD19− hepatic ILC1 in C57BL/6, Ly5.1C14R and C57BL/6 × Ly5.1C14R (results shown in the upper panels) and in C57BL/6 controls compared to Ncr1gfp/gfp (results from experiment shown in the lower panels). Data show representative plots from three to five independent experiments and indicate the the frequency of expression (n = 6–12 mice/genotype).
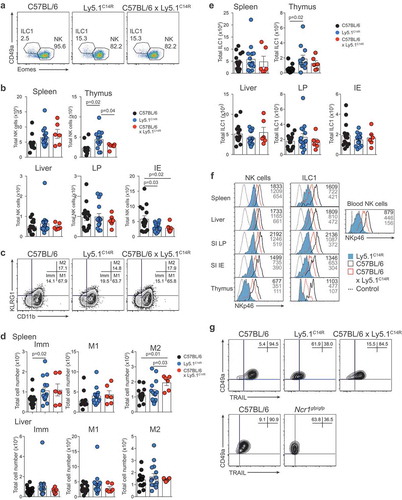
ILC1 were identified through the expression of the CD49a integrin and of the absence of expression of the transcriptional regulator Eomesodermin (Eomes) which is required for the development of NK cells.Citation23 This showed that similar to NK cells, ILC1 were also enriched in the thymus () although the level of expression of NKp46 in Ly5.1C14R mice was reduced by ~3-fold for both populations across all tissues when compared to that found in C57BL/6 mice and ~1.5-fold for C57BL/6 × Ly5.1C14R mice (). A similar reduction in the expression of NKp46 was seen in NK cells and ILC1s from tissues analysed in mixed bone marrow chimeras (SI Appendix, Figure S3). This effect was not seen in mice that carried only the six base pair mutation that encodes CD45.1 (CD45.1STEM) (SI Appendix, Figure S4) demonstrating that the NKp46 expression defect in Ly5.1C14R mice is intrinsic and not linked to Ly5.1 itself. In addition, hepatic ILC1 isolated from Ly5.1C14R mice lacked expression of the TNF-related apoptosis-inducing ligand, TRAIL (encoded by Tnfsf10) normally characteristic of this populationCitation24 (, upper panels). Such an effect was also seen in the Ncrgfp/gfp strain but not in Ly5.1STEM mice (, lower panels and SI Appendix, S4 B and C).
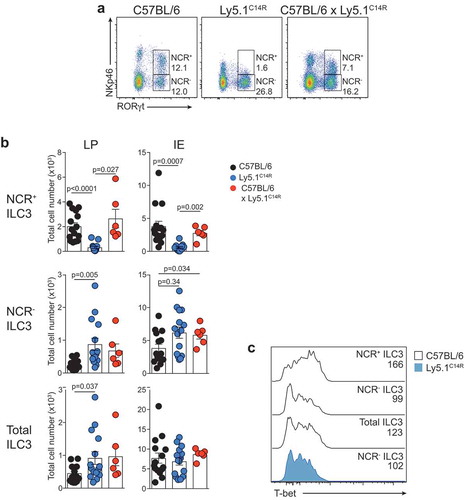
To extend these analyses, we investigated how loss of NKp46 influenced NCR+ and NCR− ILC3 subsets in the lamina propria (LP) and intraepithelial (IE) compartments of the small intestine (). As expected, NCR+ ILC3 were not detectable in the Ly5.1C14R mice () while ILC2 were present in normal numbers (data not shown). This was accompanied by an accumulation in both the frequency and number of NCR− ILC3. However, the overall number of Rorγt+ ILC3 was similar suggesting that loss of NKp46 did not adversely affect the development of total ILC3 at steady-state. Nevertheless, analyses of T-bet expression revealed that it was upregulated within the NCR+ subset in C57BL/6 mice as previously reported,Citation25,Citation26 while NCR− ILC3 from Ly5.1C14R mice did not show elevated levels of T-bet when compared with the NCR− population in C57BL/6 mice suggesting that NCR+ cells fail to develop ().
Figure 3. Ly5.1C14R mice have abnormal numbers of ILC3. (A) Dot plots showing the frequency of NCR+ and NCR− ILC3 in the LP of the small intestine of C57BL/6, Ly5.1C14R and C57BL/6 × Ly5.1C14R mice. Data show representative plots gated on live CD3−CD19− lymphocytes. (B) Enumeration of NCR+, NCR− and total ILC3 for LP and IE in the small intestine. Data showing the mean ± SEM pooled from three to six independent experiments (n = 6–15 mice/genotype). P values were calculated using an unpaired two-tailed Student’s t test. (C) Histograms show the mean fluorescence intensity of intracellular staining for T-bet in ILC3 subsets from the small intestine of C57BL/6 and Ly5.1C14R mice (n = 6 mice/genotype). P values were calculated using a Student’s t test.
Ly5.1C14R mice have an altered sensitivity to stimuli in vitro and fail to control tumors in vivo
To gain insight on the functional relevance of the C14R point mutation, the degranulation capacity of total NK cells from C57BL/6 and Ly5.1C14R mice was determined by staining for surface CD107a expression following exposure to various stimuli in vitro. As might be expected, NK cells from NKp46C14R mice were significantly less responsive to NKp46 stimulation. When NK cells were cross-linked through NK1.1, they showed a trend towards increased degranulation which was not statistically significant on the whole NK cell population (), but was statistically significant when restricted to the immature NK cell subset (SI Appendix, Figure S5), consistent with previous findings on Noé mice.Citation18 Regardless of this point, however, the in vitro anti-tumor NK cell cytolytic capacity was intact in NK cells from Ly5.1C14R mice ( and SI Appendix, Figure S5). Combined, these data suggest that NKp46C14R NK cells are functionally competent and can induce cell lysis through multiple pathways but exhibit defects in vitro when recognition requires engagement of NKp46.
Figure 4. Ly5.1C14R NK cells cells show altered sensitivity to stimuli in vitro but fail to control melanoma tumor development in vivo. (A) Degranulation capacity of total NK1.1+ NK cells determined by surface CD107a expression in C57BL/6 and Ly5.1C14R cells. Data show frequencies of CD107a+ NK cells ± SEM after coculture with various stimuli. Data shown from 2 independent experiments (n = 5 mice/genotype). (B) Cytolytic activity of C57BL/6 and Ly5.1C14R NK cells sensitized to B16F10 tumor cells. NK cells have been activated overnight with IL-2 (1000 U/ml). Data show the mean lysis ± SEM pooled from three independent experiments (n = 3 mice/genotype in each experiment). P values were calculated using a Student’s t test. (C) Representative whole mounts of the metastatic burden in the lungs of C57BL/6, Ly5.1C14R, Mcl-1fl/flNcr1iCre, C57BL/6 × Ly5.1C14R and Ly5.1 (Jax, 2017) mice 14 days after i.v injection of B16F10 melanoma cells. (D) Total tumor burden in the lungs of C57BL/6, Ly5.1C14R, Mcl-1fl/flNcr1iCre, C57BL/6 × Ly5.1C14R and Ly5.1 (Jax, 2017) mice shown in (C) 14 days after injection of B16F10 melanoma cells. Data show the mean ± SEM of tumor burden pooled from five independent experiments (n = 28–30 mice/genotype). Mcl-1fl/flNcr1iCre mice included in a single experiment (n = 5 mice/genotype) while C57BL/6 × Ly5.1C14R and Ly5.1 (Jax, 2016) mice were included in two experiments (n = 12 mice/genotype). P values were calculated using an unpaired two-tailed Student’s t test.
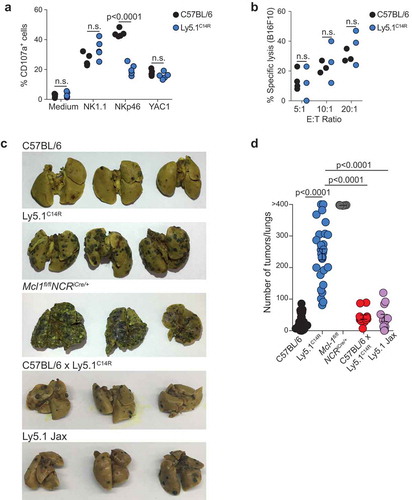
These findings suggested that despite the alteration of NKp46 in Ly5.1C14R, NK cells should be competent to protect against in vivo tumor challenge. To test this point, mice were inoculated with B16F10 melanoma cells which are known to be controlled by NK cells.Citation27,Citation28 While C57BL/6, C57BL/6 × Ly5.1C14R and Ly5.1 (Jax, 2017) mice were able to largely control tumor growth as measured by the number of primary lung tumors 14 days after tumor inoculation ( and ), mice that expressed the NKp46C14R single point mutation developed significantly more lung metastases, a condition that was accompanied by respiratory distress. These mice also presented metastases in other organs such as the kidney, liver and bone marrow (SI Appendix, Figure S5 D), a phenotype which was similar to that observed in Mcl-1fl/flNcr1iCre mice in which NCR+ cells are conditionally ablatedCitation29 ( and and SI Appendix, Figure S5D). Thus, although some function was preserved in the presence of the C14R mutation including the ability of NK cells to localize to the lung itself (SI Appendix, Figure S6), alteration of surface NKp46 in Ly5.1C14R mice strongly impaired the capacity of NK cells to control tumor development and escape in vivo.
C14R mutation disrupts cellular pathways associated with protein trafficking
To further dissect the molecular alterations that underpin the changes that occur when the NKp46 expression is altered, we analysed the transcriptome of naive C57BL/6 and Ly5.1C14R NK1.1+ NK cells by RNA-seq analyses (, SI Appendix, Dataset S2). This revealed that there was a significant enrichment in differentially regulated genes involved in intracellular trafficking compartments including the endoplasmic reticulum, endosome, ER to Golgi vesicle-mediated transport and Golgi apparatus in Ly5.1C14R cells (). In addition, alterations occurred in pathways associated with protein ubiquitination and transport together with enzymes associated with processing of proteins (eg. GTPase, peptidase activity, protein kinase). Although changes to NKp46 trafficking may have been expected, consistent with a similar alteration that occurs in Noé mice,Citation20,Citation21 broader changes in protein trafficking were not anticipated. Exome sequencing has uncovered that a broad gene set is altered in the Ly5.1C14R strain that warrants further investigation.
Figure 5. Altered molecular machinery in naïve Ly5.1C14R mutant NK cells affects antigen processing and protein trafficking pathways. (A, B) Enrichment clusters from genes upregulated and downregulated, respectively, in naïve Ly5.1C14R NK cells compared with C57BL/6 NK cells. (C) Gene ontology (GO) network analysis of significantly reduced gene expression levels in Ly5.1C14R NK cells (shown in B) via Metascape and visualized with Cytoskape (v3.1.2). (D) Heatmap of genes significantly up and downregulated in NK cells responding to B16F10 melanoma tumor cells seven days after challenge presented to show differences in gene expression patterns for C57BL/6 and Ly5.1C14R mice and the comparative gene expression found in naïve NK cells. (E) Gene ontology (GO) network analysis of significantly reduced gene expression levels in Ly5.1C14R NK cells isolated from day 7 lungs of mice challenged with B16F10 tumor cells. Nodes are coloured by p-value via Metascape and visualized with Cytoskape.
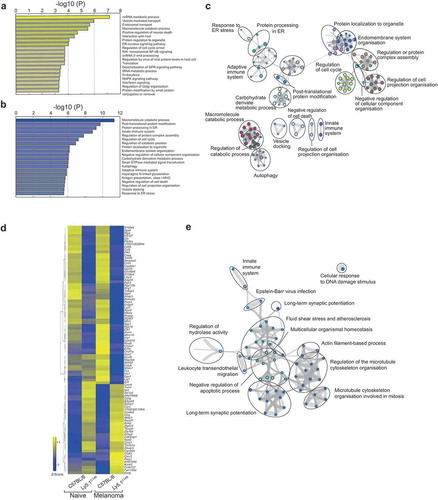
To determine how the NKp46C14R mutation might affect cells responding to a tumor challenge, we isolated total NK cells from the lungs of mice challenged with B16F10 tumor cells 7 days earlier and subjected them to RNAseq (SI Appendix, Dataset S3). Ninety-one genes were found to be differentially expressed between C57BL/6 and Ly5.1C14R in NK cells in response to melanoma (). Genes up regulated in C14R NK cells were associated with the mitochondrion and cell differentiation, while genes involved in protein processing in the endoplasmic reticulum, transcriptional regulators and cellular organization were downregulated (). However, genes such as Cxcr3 and Cxcr4, both of which are essential for infiltration and function of NK cells,Citation30–Citation32 and the transcriptional regulator Nab2 were upregulated in Ly5.1C14R. We observed that in Ly5.1C14R mice, the number of NK cells found in the lung were enriched ~2.1 fold indicating that the failure of tumor control was not driven by the inability to migrate to the lung.
Loss of NKp46 signaling is encoded by the point mutation C14R in Ncr1
It was clear from the analyses above that many more genes than would be anticipated are altered in Ly5.1C14R mice. Thus, we could not be completely certain that the effects identified in NKp46 expression in these mice are due solely to the C14R mutation detected in the Ncr1 gene. To determine whether this was indeed the case, we generated mice using the CRISPR/Cas9 technologyCitation33 that would carry only the C14R mutation on a C57BL/6 background. These mice are designated as the NCRB6C14R strain and were healthy, viable and bred normally to produce homozygous animals. Analyses of NKp46 surface expression in C57BL/6 and NCRB6C14R mice showed that the C14R mutation alone abrogated NKp46 binding phenocopying the Ly5.1C14R mice for this feature (). Similarly, the distribution of NK cells in NCRB6C14R mice were concordant with the pattern observed in Ly5.1C14R mice () while in both NCRB6C14R and Ly5.1C14R ILC1 were enriched in spleen and liver in four to five week old mice (). We also observed that NKp46+ ILC3 were reduced in NCRB6C14R similar to that observed in Ly5.1C14R mice. Notably across all organs examined, the surface expression of NKp46 was strongly reduced in NK cells and ILC1 and in blood NK cells (). This correlated with a strong reduction in TRAIL expression when C57BL/6 mice expressed the C14R mutation demonstrating that an alteration in NKp46 which affects the stability of the expression at the surface of the cell is sufficient to alter the induction of TRAIL in hepatic ILC1.
Figure 6. Loss of NKp46 surface expression depends on the C14R mutation in the Ncr1 gene. (A) Dot plots showing the frequency of ILC1 and NK cells in the spleen. Data show representative plots gated on live NK1.1+ lymphocytes excluding T and B cells. (B) Total number of NK cells in spleen, thymus, liver, small intestine lamina propria (LP) and within the intestinal intraepithelial compartment (IE). Data are pooled from two independent experiments and show the mean ± SEM (n = 4–6 mice/genotype). P values were calculated using an unpaired two-tailed Student’s t test. (C) FACS plots showing the frequency of immature (Imm, KLRG1−CD11b−), mature 1 (M1, KLRG1−CD11b+) and mature 2 (M2, KLRG1+CD11b+) NK cells in splenic NK1.1+CD3−CD19− NK cells. (D) Total number of Imm, M1 and M2 NK cells in the spleen and liver of C57BL/6, Ly5.1C14R and NCRB6C14R mice showing the mean ± SEM pooled from two experiments (n = 4–6 mice/genotype). P values were calculated using an unpaired two-tailed Student’s t test. (E) Total number of ILC1s in spleen, thymus, liver, small intestine lamina propria (LP) and within the intestinal intraepithelial compartment (IE). Data show the mean ± SEM pooled from two independent experiments (n = 4–6 mice/genotype). P values were calculated using an unpaired two-tailed Student’s t test. (F) Histograms showing the mean fluorescence intensity of NKp46 in various tissues for both NK cells and ILC1 for wild-type (black solid line), Ly5.1C14R (solid blue) and NCRB6C14R (solid orange). CD3ε+ cells were used as a control for NKp46 expression (black dashed line). Data are representative of tissues analyzed in (A-E). (G) Expression of TRAIL on NK1.1+CD49a+CD3−CD19− hepatic ILC1 in C57BL/6, Ly5.1C14R and NCRB6C14R. Data show representative plots from two independent experiments and indicate the the frequency of expression (n = 4–6 mice/genotype).
Discussion
Spontaneous mutations occur in eukaryotes at a rate of 0.1–100 per genome per sexual generation. In mice, nearly 5,000 spontaneous and induced mouse mutant alleles with clinically relevant phenotypes have been described in the Mouse Genome Informatics database and only about one third of these have characterized phenotypes.Citation34 In many cases, however, identification of these sequence alterations has served as a rich source of animal models for human genetic diseases.
Although the Ly5.1 (CD45.1) line should differ from its ‘wild-type’ counterpart by just five amino acids within the extracellular domain,Citation35 it is now clear that the B6.SJL-Ptprca Pepcb/BoyJ derived from Jax differs in many genes from the C57BL/6 line spanning ~40Mb and ~300 genes.Citation36 A single amino acid accounts for the difference between the CD45.1 and CD45.2 congenic markers that define these two strainsCitation36 and this difference has formed the basis for their use in extensive tracking experiments. These previously unappreciated alterations in the genome have most likely impacted experimental interpretations. We now add to this list the detection of a spontaneous mutation in the Ncr1 gene that has affected multiple mouse colonies in geographically distinct locations. This mutation may have arisen independently, but seems most likely to have arisen from an individual breeding founder from the Jax. As the C14R mutation of the Ncr1 gene remains undetected in heterozygous animals, this mutation has inadvertently been retained undetected long term in parent colonies. Using whole exome sequencing we identified a spontaneous single autosomal recessive mutation in the Ncr1 gene that significantly altered the expression of the NKp46 surface protein by interfering with its export to the cell surface. Subsequently, we generated C57BL/6 mice in which we introduced the C14R mutation in the Ncr1 gene negating the effect of other genes altered in the Ly5.1C14R strain. This directly confirmed that the change in NKp46 expression was solely driven by this mutation and will allow this specific mutation to be investigated in detail in future studies.
The C14R mutation in the Ncr1 gene of the mutant Ly5.1 lines occurred just proximal to the previously reported W32R mutation in the signal peptide which can typically affect the synthesis and/or secretion of a protein.Citation18,Citation21 We show that expression of Ncr1 mRNA was similar in NK cells of both wild-type, Ly5.1C14R mouse strains suggesting that the gene is effectively transcribed but that alterations occur after this stage. The Noé strain also showed poor cell surface expression of NKp46, and like the NKp46C14R NK cells, displayed no change in mRNA expression.Citation18 This was supported by analysis of transduced 293T cells where intracellular NKp46C14R and NKp46 expression were similar, but cell surface expression was strongly impaired in NKp46C14R. Genomic analyses revealed disruption in intracellular trafficking although a broader group of genes involved in transcriptional regulation and protein processing in the ER, endosome, Golgi apparatus and cellular organization were detected suggesting other potential defects in this mouse line. Collectively, however, our data suggest that in the mutants, NKp46 could be expressed on the cell surface but is unstable posing the possibility that an altered signaling pathway could be triggered by the constant internalization of NKp46. It also implies that the stability and/or level of NKp46 expression is important in signaling and prompts further dissection of the molecular control of NKp46 that appears to impact on ILC differentiation.
In the Ly5.1C14R mice, we observed that NK cell development in terms of cellularity was not impaired in the absence of NKp46. Neverthless, we show that the differentiation of NK cells and ILC1 were altered in both Ly5.1C14R and Noé mice. NK cell subsets were skewed towards an accumulation of less mature NK cells and NCR− ILC3s. The lack of NKp46 induced in Ncr1gfp/gfp mice has also recently been shown to be associated with an impaired NK cell maturation in response to mouse cytomegalovirus infection (MCMV).Citation37 This was associated with changes in CXCR3-driven migration of NK cells to the regional lymph nodes which can affect NK-dendritic cell crosstalk necessary for priming MCMV-specific CD4+ T cells.Citation38 In our study, we found that in contrast to this observation, Cxcr3 mRNA was upregulated, consistent with the higher expression of CXCR3 on the less mature CD27highM1 NK cells.Citation39,Citation40 Indeed, the number of NK cells that localized in the lung in response to a B16F10 tumor cells which are likely to express the ligand for NKp46 was not reduced in Ly5.1C14R mice (SI Appendix, Figure S4) and the cytolytic machinery was intact. Ncr1gfp/gfp mice appear to exhibit slightly reduced IFN-γ responses to NK1.1 stimulation in vitro and murine cytomegalovirus infection.Citation19,Citation37 Stimulation through the NKp46 receptor has been shown to drive IFN-γ production. This NK cell-driven IFN-γ production has recently been shown to induce the extracellular matrix protein fibronectin 1 that could modulate tumor formation.Citation41 In contrast, diminished NKp46 appears to regulate phosphorylation of Dok-1 to influence IFN-γ production and pathogen protection.Citation42 In our in vitro analyses, a reduction in IFN-γ was not observed in our mutant mice although signaling directly through NKp46 was strongly impaired. Thus, it appears that the complete loss of function of the Ncr1 allele in Ncr1gfp/gfp mice differs from the disruption induced by the introduction of a point mutation in the gene.
In addition to changes in NK cell differentiation, we observed that diminished NKp46 expression caused alterations in other ILCs. Although loss of NKp46 did not impair ILC1 numbers, we show that the apoptosis-inducing ligand TRAIL, which can be induced by TGF-βCitation43 and through viral binding to NKp46Citation44 was lacking implying that NKp46 may play a role in regulating TRAIL expression. NKp46 is also a key marker of one of the three subsets of ILC3.Citation18 In Ly5.1C14R mice, as expected NCR+ ILC3 were not detectable via NKp46 expression but we also did not see enrichment of expression of T-bet which is normally associated with induction of this subset,Citation25,Citation26 although ILC3 development itself was not affected implying that activation of NKp46 is important for the transcriptional program of these cells. This is supported by cell fate mapping experiments that suggest that instability of NKp46 expression in ILC3s found in the intestine reflects a major role of this receptor in tuning the very dynamic environmental signals encountered that drive ILC3 plasticity.Citation45,Citation46 Loss of Ncr1 has previously been shown not to affect ILC3 development with Ncr1gfp/gfpRag2−/- mice showing survival and clinical scores similar to control mice when challenged with the enteric infection Citrobacter rodentium.Citation47 In this setting, NKp46 expression did not appear to be essential for protection.
In summary, we have identified a commonly used mouse line, the Ly5.1C14R, that carries a point mutation in the Ncr1 gene together with a large number of previously unknown genes. The NCRB6C14R strain confirmed that the C14R Ncr1 mutation directly impaired the expression of surface NKp46 and alters differentiation of NK cells with increased accumulation of immature stages. In addition, increased numbers of ILC1 were found and these cells lacked TRAIL expression while NCR+ ILC3 were not detected. Thus, although reduced NKp46 expression did not disrupt either NK cell or ILC development, it exerted an important impact on normal differentiation, maturation and activation of these lineages.
Materials and methods
Mice
C57BL/6 and B6.SJL-Ptprca Pepcb/BoyJ (Ly5.1, originally obtained from The Jackson Laboratory), C57BL/6 × B6.SJL-PtprcaPepcb/BoyJ (C57BL/6 × Ly5.1 and B6.SJL-Ptprca Pepcb/BoyJAX (imported 2016 and sequence verified) mice were bred and maintained in-house at either the Walter and Eliza Hall Institute of Medical Research (WEHI, Australia) or the University Hospital Erlangen (Germany) under specific pathogen-free conditions. Mice used in Frankfurt were obtained from the Preclinical Experimental Animal Center (PETZ) of the Friedrich Alexander University (FAU) Erlangen-Nürnberg, Germany. Ncr1gfp/gfp, Noé mice and Mcl1fl/flNcr1iCre mice have been previously described.Citation17,Citation18,Citation29 CD45.1STEM miceCitation36 were bred at the Massachusetts General Hospital animal house (Boston). Male and female mice were used between the ages of six to eight weeks and were age matched unless otherwise stated. NCRB6C14R mice were used at four to five weeks of age. Animal experiments followed the National Health and Medical Research Council (NHMRC) Code of Practice for the Care and Use of Animals for Scientific Purposes guidelines and were approved by the Animal Ethics Committee of the respective institutions.
Generation of NCRB6C14R mice
A new C57BL/6 mouse strain that carried the C14R mutation in the Ncr1 gene (NCRB6C14R) was generated using CRISPR/Cas9 as previously described.Citation33 Briefly, one sgRNA of the sequence TAGGGCTATGTCTGAGCCAG (10ng/μl), an oligo donor of the sequence (gttgaatcaagagcagattggggggagacagcatgccattaaccctgttttctagGGCTACGACTGAGCCAGCGTATCAACACTGAAAAGGgtaagtccttccctcgaagtctcagggttgttcttatgggttca; 10ng/ul) and Cas9 mRNA (5ng/μl) were injected into the cytosol of C57BL/6J zygotes to generate NCRB6C14R point mutant mice.
Generation of mixed bone marrow chimeric mice
Mixed bone marrow chimeric mice were generated by reconstituting lethally irradiated (2 x 5.5Gy γ-irradiation, 137Cs source) with a mixture (1:1 ratio; total 10 × 106 bone marrow cells/mouse) of Ly5.1C14R (CD45.1+/+) and C57BL/6 × Ly5.1C14R (CD45.2+CD45.1+) bone marrow. Mice were analysed 6–8 wk after reconstitution.
Isolation of intestinal intraepithelial and lamina propria lymphocytes
Intestinal LPL were isolated from the intestine as previously described.Citation26,Citation48
Cell isolation and flow cytometric analyses
Single cell suspensions were generated by forcing organs through 70 μm sieves. Lymphocytes were isolated from liver using a 33% isotonic Percoll (Amersham Pharmacia Biotech) gradient centrifuged at 2000 rpm for 13 min at ambient temperature. Single cell suspensions were blocked with PBS containing 5 μg/ml anti-CD16/CD32 (2.4G2) and stained for 20–30 min on ice with fluorophore-conjugated antibodies, unless stated otherwise. For analysis of surface molecules, cells were stained with the following antibodies: anti-CD11b (M1/70) (BD Biosciences, or Miltenyi), anti-CD3ε (145-2C11) and anti-CD19 (1D3 and 6D5) (BioLegend), anti-KLRG1 (2F1), and anti-NK1.1 (PK136), anti-CD27 (LG.3A11), anti-CD45.1 (A20), anti-CD45.2 (104), anti-CD49a (HMα1), anti-B220 (RA3-6B2) (all from BD Biosciences), anti-NKp46 (29A1.4, eBioscience) and anti-CD49b (DX5, generated at The Walter and Eliza Hall Institute hybridoma facility). Live cells were identified by exclusion after staining with a fixable viability dye (BD Biosciences or BioLegend). Intracellular staining was performed using the Transcription Factor Staining Buffer Set (eBioscience) and monoclonal antibodies to EOMES (Dan11mag), GATA3 (TWAJ) (both from eBioscience) and RORγt (Q31-378, BD Biosciences). For IFN-γ detection, the monoclonal antibody XMG1.2 (BD Pharmingen) was used. Flow cytometry was performed using a LSRFortessa X-20 or Canto10C (BD Biosciences) and FlowJo analysis software (Treestar).
Murine melanoma cells and pulmonary metastasis
B16F10 murine melanoma cells (mycoplasma negative) were maintained in complete DMEM (Gibco) with 10% heat-inactivated FCS, 2 mM of L-Glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin. 2.5 × 105 B16F10 cells were injected into the tail vein of recipient mice. Fourteen days after injection, the lungs, liver, long bones and kidneys were harvested and fixed in Bouin’s solution. B16F10 metastases were counted using a dissecting microscope.Citation49
Exome sequencing
To identify the genetic mutation responsible for the reduced NKp46 expression in the Ly5.1C14R strain we performed exome sequencing on two mice from this strain and the C57BL/6 × Ly5.1C14R mice which expressed normal NKp46 protein levels (SI Appendix, Methods).
Sanger sequencing
To further confirm the Ncr1 mutation identified by exome sequencing we performed Sanger sequencing on C57BL/6, C57BL/6 × Ly5.1 and WEHI B6.SJL-Ptprca Pepcb/BoyJ mice (SI Appendix, Methods).
Quantitative PCR
RNA was isolated from sorted NK cells and NKp46 expression was determined as described in SI Appendix, Methods.
Cloning
NKp46 cDNA were generated from RNA isolated from purified NK cells of either C57BL/6 or Ly5.1C14R mice by RT-PCR and cloned into the plasmid RSV.5neo-FlagHis which provides C-terminal FLAG- and hexahistidine-tags.
Primers: NKp46 cDNA forward 5ʹ-GACTCCGCGGGCCACCATGCTGCCAACACTCACTG-3ʹ, NKp46 cDNA reverse 5ʹ-AGTCCTCGAG CAAGGCCCCAGGAGTTGC-3ʹ.
Transfection and flow cytometric analysis
293T cells were cultured in Dulbecco’s Modified Eagle’s Medium and were transfected with the appropriate RSV.5neo expression vectors containing NKp46 WT or mutant cDNA with Flag- and hexahistidine-tag using Applifect (AppliChem) (SI Appendix, Methods). Cells were fixed and permeabilized with BD Cytofix/Cytoperm (BD Biosciences) and subsequently stained with appropriate antibodies for 20 min at 4°C. Antibodies used: FLAG-tag mAb M2 (Sigma), anti-NKp46-PE mAb 29A1.4 (BioLegend), rat IgG2a-PE isotype control RTK2758 (BioLegend), mouse IgG1 isotype control N1G9, APC-conjugated F(ab)2-fragments of goat anti-mouse IgG (Jackson ImmunoResearch). Flow cytometry analysis was performed with a FACS Canto II (BD Biosciences, Heidelberg, Germany) and data analyzed using FlowJo (Tree Star, Ashland, OH).
Immunofluorescence staining
NK cells were isolated from the spleens of Ly5.1C14R and C57BL/6 mice and centrifuged on lysine coated slides, then fixed and stained with goat anti-mouse NKp46 antibody (R&D Systems, AF2225, 1:100) and rabbit anti-mouse PDI (Cell Signaling Technology, 3501S, 1:100) overnight, followed by AlexaFluor488-conjugated donkey anti-goat IgG (H + L) (ThermoFisher, A-11055, 1:250) and AlexFluor546-conjugated donkey anti-rabbit IgG (H + L) (ThermoFisher, A-10040, 1:200). Images were analyzed using LAS AF (Leica Application Suite – Advanced Fluorescence) software (SI Appendix, Methods).
NK cell stimulation
Plates were coated overnight at 4°C with antibodies against NK1.1 and NKp46 (10 μg/ml in PBS). 1.5 × 106 splenocytes were cultured in 200μl of complete RPMI1640 media containing 10% heat-inactivated FCS, 2mM of L-Glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin in the presence of Golgi Stop and soluble CD107a for 4h at 37°C. The YAC1 stimulation condition was obtained by adding 0.25 106 YAC1 cells to the well. Cells were subsequently stained and analyzed by flow cytometry with an LSR Fortessa and data analyzed using FlowJo (Tree Star, Ashland, OH).
Cytotoxicity assay
Europium release assay was used to assess the lytic capacity of murine NK cells as described in SI Appendix, Methods.
RNA-seq analysis
RNA-seq analyses was performed on NK cells isolated from the lungs of Ly5.1C14R and C57BL/6 mice seven days after challenge with B16F10 melanoma tumor cells. These strains were sourced from the sourced from the WEHI colonies. Samples were sequenced on an Illumina NextSeq 500 generating 75 bp paired end reads. Two biological replicates of each cell type were sequenced. RNA-seq Reads were aligned to the GRCm38/mm10 build of the Mus musculus genome using the Subread aligner.Citation50 Only uniquely mapped reads were retained. Genewise counts were obtained using featureCounts.Citation51 Reads overlapping exons in annotation build 38.1 of NCBI RefSeq database were included. Genes were excluded from downstream analysis if they failed to achieve a CPM (counts per million mapped reads) value of greater than 0.5 in at least two libraries. Counts were converted to log2 counts per million, quantile normalized and precision weighted with the voom function of the limma package.Citation52,Citation53 A linear model was fitted to each gene, and empirical Bayes moderated t-statistics were used to assess differences in expression.Citation54 A false discovery rate cut-off of 0.15 was applied for calling differentially expressed genes. Gene Ontology and gene enrichment analyses were performed using Metascape (http://metascape.org). Raw data files for the RNA sequencing have been deposited in the NCBI Gene Expression Omnibus under accession number GSE113030.
Highlights
A spontaneous mutation in the signal sequence of the Ncr1 gene arose in the CD45.1 mouse strain
Ncr1 C14R mutation impairs NKp46 surface expression in NK cells, ILC1 and ILC3
Destabilization of Ncr1 by the C14R mutation results in accumulation of NKp46 in the endoplasmic reticulum
Loss of stable NKp46 expression impairs the maturation of NKp46+ ILCs and impairs tumor control
Declaration of interest statement
E.V. is cofounder and employee of Innate Pharma.
Authors Contributions
M.E.F., S.W., A.G., T.Z., T.W., J.K., A.S., A.M., F.G., S.U., E.N-M., B.R., F.S., F.F., S.T., M.A.F., J.G., S.N.W., K.L., A.G.C. and F.F.A performed experiments and analysed data; M.H. and A.K. helped create the NCRB6C14R mice. T.W., U.S., J.K., A.S., N.D.H., F.E.M, D.T.S., E.U., E.V. and G.T.B. provided intellectual input and tools or reagents; A.G., Y.L. and W.S. performed bioinformatics analyses. G.T.B. and E.U. conceived the ideas and interpreted data; G.T.B, E.U., F.F.A. and S.T. wrote the paper and edited it with the assistance of the other authors.
Supplemental Material
Download PDF (15.4 MB)Acknowledgments
We thank M. Camilleri, J. Janssen, S. Cree, C. O’Brien and A. Abendroth and K. Stein for expert technical support and Dr J. Babon for helpful discussions, and Prof. C. Bogdan for providing mice and helpful discussion of results. This study was made possible through Victorian State Government Operational Infrastructure Support and Australian Government NHMRC Independent Research Institute Infrastructure Support Scheme (Walter and Eliza Hall Institute of Medical Research) and the LOEWE Center for Cell and Gene therapy (to ST and EU), Frankfurt, funded by the Hessian Ministry of Higher Education, Germany (III L 4-518/17.004).
Supplemental Material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Lam VC, Lanier LL. NK cells in host responses to viral infections. Curr Opin Immunol. 2017;44:43–51. doi:10.1016/j.coi.2016.11.003.
- Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat Immunol. 2008;9(5):503–510. doi:10.1038/ni1582.
- Seidel E, Glasner A, Mandelboim O. Virus-mediated inhibition of natural cytotoxicity receptor recognition. Cell Mol Life Sci. 2012;69(23):3911–3920. doi:10.1007/s00018-012-1001-x.
- Thielens A, Vivier E, Romagne F. NK cell MHC class I specific receptors (KIR): from biology to clinical intervention. Curr Opin Immunol. 2012;24(2):239–245. doi:10.1016/j.coi.2012.01.001.
- Satoh-Takayama N, et al.. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 2008;29(6):958–970. doi:10.1016/j.immuni.2008.11.001.
- Luci C, et al.. Influence of the transcription factor RORgammat on the development of NKp46+ cell populations in gut and skin. Nat Immunol. 2009;10(1):75–82. doi:10.1038/ni.1681.
- Sanos SL, Bui VL, Mortha A, Oberle K, Heners C, Johner C, Diefenbach A. RORγt and commensal microflora are required for the differentiation of mucosal interleukin IL22-producion NKp46+ cells. Nature Immunology. 2009;10(1):83–91. doi:10.1038/ni.1684.
- Narni-Mancinelli E, et al.. Complement factor P is a ligand for the natural killer cell-activating receptor NKp46. Sci Immunol. 2017;2:10. doi:10.1126/sciimmunol.aam9628.
- Vankayalapati R, et al.. The NKp46 receptor contributes to NK cell lysis of mononuclear phagocytes infected with an intracellular bacterium. J Immunol. 2002;168(7):3451–3457. doi:10.4049/jimmunol.168.7.3451.
- Mandelboim O, et al.. Recognition of haemagglutinins on virus-infected cells by NKp46 activates lysis by human NK cells. Nature. 2001;409(6823):1055–1060. doi:10.1038/35059110.
- Draghi M, et al.. NKp46 and NKG2D recognition of infected dendritic cells is necessary for NK cell activation in the human response to influenza infection. J Immunol. 2007;178(5):2688–2698. doi:10.4049/jimmunol.178.5.2688.
- Arnon TI, et al.. Recognition of viral hemagglutinins by NKp44 but not by NKp30. Eur J Immunol. 2001;31(9):2680–2689. doi:10.1002/1521-4141(200109)31:9<2680::AID-IMMU2680>3.0.CO;2-A.
- Jarahian M, et al.. Activation of natural killer cells by newcastle disease virus hemagglutinin-neuraminidase. J Virol. 2009;83(16):8108–8121. doi:10.1128/JVI.00211-09.
- Jarahian M, et al.. Modulation of NKp30- and NKp46-mediated natural killer cell responses by poxviral hemagglutinin. PLoS Pathog. 2011;7(8):e1002195. doi:10.1371/journal.ppat.1002195.
- Chaushu S, et al.. Direct recognition of Fusobacterium nucleatum by the NK cell natural cytotoxicity receptor NKp46 aggravates periodontal disease. PLoS Pathog. 2012;8(3):e1002601. doi:10.1371/journal.ppat.1002601.
- Vitenshtein A, et al.. NK cell recognition of candida glabrata through binding of NKp46 and NCR1 to fungal ligands Epa1, Epa6, and Epa7. Cell Host Microbe. 2016;20(4):527–534. doi:10.1016/j.chom.2016.09.008.
- Gazit R, et al.. Lethal influenza infection in the absence of the natural killer cell receptor gene Ncr1. Nat Immunol. 2006;7(5):517–523. doi:10.1038/ni1322.
- Narni-Mancinelli E, et al.. Tuning of natural killer cell reactivity by NKp46 and Helios calibrates T cell responses. Science. 2012;335(6066):344–348. doi:10.1126/science.1215621.
- Sheppard S, et al.. Characterization of a novel NKG2D and NKp46 double-mutant mouse reveals subtle variations in the NK cell repertoire. Blood. 2013;121(25):5025–5033. doi:10.1182/blood-2012-12-471607.
- Glasner A, et al.. Expression, function, and molecular properties of the killer receptor Ncr1-Noe. J Immunol. 2015;195(8):3959–3969. doi:10.4049/jimmunol.1501234.
- Glasner A, Isaacson B, Mandelboim O. Expression and function of NKp46 W32R: the human homologous protein of mouse NKp46 W32R (Noe). Sci Rep. 2017;7:40944. doi:10.1038/srep40944.
- Walzer T, et al.. Identification, activation, and selective in vivo ablation of mouse NK cells via NKp46. Proc Natl Acad Sci U S A. 2007;104(9):3384–3389. doi:10.1073/pnas.0609692104.
- Gordon SM, et al.. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity. 2012;36(1):55–67. doi:10.1016/j.immuni.2011.11.016.
- Seillet C, et al.. Differential requirement for Nfil3 during NK cell development. J Immunol. 2014;192(6):2667–2676. doi:10.4049/jimmunol.1302605.
- Klose CS, et al.. A T-bet gradient controls the fate and function of CCR6-RORgammat+ innate lymphoid cells. Nature. 2013;494(7436):261–265. doi:10.1038/nature11813.
- Rankin LC, et al.. The transcription factor T-bet is essential for the development of NKp46(+) innate lymphocytes via the Notch pathway. Nat Immunol. 2013;14(4):389–395. doi:10.1038/ni.2545.
- Grundy MA, Zhang T, Sentman CL. NK cells rapidly remove B16F10 tumor cells in a perforin and interferon-gamma independent manner in vivo. Cancer Immunol Immunother. 2007;56(8):1153–1161. doi:10.1007/s00262-006-0264-1.
- Smyth MJ, et al.. Differential tumor surveillance by natural killer (NK) and NKT cells. J Exp Med. 2000;191(4):661–668. doi:10.1084/jem.191.4.661.
- Sathe P, et al.. Innate immunodeficiency following genetic ablation of Mcl1 in natural killer cells. Nat Commun. 2014;5:4539. doi:10.1038/ncomms5539.
- Noda M, et al.. CXCL12-CXCR4 chemokine signaling is essential for NK-cell development in adult mice. Blood. 2011;117(2):451–458. doi:10.1182/blood-2010-04-277897.
- Bernardini G, Antonangeli F, Bonanni V, Santoni A. Dysregulation of chemokine/chemokine receptor axes and NK cell tissue localization during diseases. Front Immunol. 2016;7:402. doi:10.3389/fimmu.2016.00402.
- Pak-Wittel MA, Yang L, Sojka DK, Rivenbark JG, Yokoyama WM. Interferon-gamma mediates chemokine-dependent recruitment of natural killer cells during viral infection. Proc Natl Acad Sci U S A. 2013;110(1):E50–59. doi:10.1073/pnas.1220456110.
- Wang H, et al.. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153(4):910–918. doi:10.1016/j.cell.2013.04.025.
- Blake JA, et al.. The Mouse Genome Database (MGD): premier model organism resource for mammalian genomics and genetics. Nucleic Acids Res. 2011;39(Database issue):D842–848. doi:10.1093/nar/gkq1008.
- Zebedee SL, Barritt DS, Raschke WC. Comparison of mouse Ly5a and Ly5b leucocyte common antigen alleles. Dev Immunol. 1991;1(4):243–254. doi:10.1155/1991/52686.
- Mercier FE, Sykes DB, Scadden DT. Single targeted exon mutation creates a true congenic mouse for competitive hematopoietic stem cell transplantation: the C57BL/6-CD45.1(STEM) mouse. Stem Cell Reports. 2016;6(6):985–992. doi:10.1016/j.stemcr.2016.04.010.
- Miletic A, et al.. NCR1-deficiency diminishes the generation of protective murine cytomegalovirus antibodies by limiting follicular helper T-cell maturation. Eur J Immunol. 2017. doi:10.1002/eji.201646763.
- Mandaric S, et al.. IL-10 suppression of NK/DC crosstalk leads to poor priming of MCMV-specific CD4 T cells and prolonged MCMV persistence. PLoS Pathog. 2012;8(8):e1002846. doi:10.1371/journal.ppat.1002846.
- Hayakawa Y, Smyth MJ. CD27 dissects mature NK cells into two subsets with distinct responsiveness and migratory capacity. J Immunol. 2006;176(3):1517–1524. doi:10.4049/jimmunol.176.3.1517.
- Meinhardt K, et al.. Identification and characterization of the specific murine NK cell subset supporting graft-versus-leukemia- and reducing graft-versus-host-effects. Oncoimmunology. 2015;4(1):e981483. doi:10.4161/2162402X.2014.981483.
- Glasner A, et al.. NKp46 receptor-mediated interferon-g production by natural killer cells increases fibronectin 1 to alter tumor architecture and control metastasis. Immunity. 2018;48:1–13. doi:10.1016/j.immuni.2017.12.007.
- Jang Y, et al.. Cutting edge: check your mice-A point mutation in the Ncr1 locus identified in CD45.1 congenic mice with consequences in mouse susceptibility to infection. J Immunol. 2018;200(6):1982–1987. doi:10.4049/jimmunol.1701676.
- Cortez VS, et al.. Transforming growth factor-beta signaling guides the differentiation of innate lymphoid cells in salivary glands. Immunity. 2016;44(5):1127–1139. doi:10.1016/j.immuni.2016.03.007.
- Liang Y, et al.. The hemagglutinin-neuramidinase protein of Newcastle disease virus upregulates expression of the TRAIL gene in murine natural killer cells through the activation of Syk and NF-kappaB. PLoS One. 2017;12(6):e0178746. doi:10.1371/journal.pone.0178746.
- Viant C, et al.. Transforming growth factor-beta and Notch ligands act as opposing environmental cues in regulating the plasticity of type 3 ILCs. Sci Signal. 2016;9(426):ra46. doi:10.1126/scisignal.aaf2176.
- Verrier T, et al.. Phenotypic and functional plasticity of murine intestinal NKp46+ group 3 innate lymphoid cells. J Immunol. 2016;196(11):4731–4738. doi:10.4049/jimmunol.1502673.
- Satoh-Takayama N, et al.. The natural cytotoxicity receptor NKp46 is dispensable for IL-22-mediated innate intestinal immune defense against Citrobacter rodentium. J Immunol. 2009;183(10):6579–6587. doi:10.4049/jimmunol.0901935.
- Rankin LC, et al.. Complementarity and redundancy of IL-22-producing innate lymphoid cells. Nat Immunol. 2016;17(2):179–186. doi:10.1038/ni.3332.
- Ferrari de Andrade L, et al.. Natural killer cells are essential for the ability of BRAF inhibitors to control BRAFV600E-mutant metastatic melanoma. Cancer Research. 2014;74(24):7298–7308. doi:10.1158/0008-5472.CAN-14-1339.
- Liao Y, Smyth GK, Shi W. The subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41(10):e108. doi:10.1093/nar/gkt214.
- Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2013.
- Law CW, Chen Y, Shi W, Smyth GK. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15(2):R29. doi:10.1186/gb-2014-15-2-r29.
- Ritchie ME, et al.. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi:10.1093/nar/gkv007.
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3. Article3. doi:10.2202/1544-6115.1027.
