ABSTRACT
The incidence and mortality rates of endometrial cancer are increasing during recent years. CA125 (gene symbol MUC16) is a well-known diagnostic and prognostic serum marker of endometrial cancer. High serum CA125 level is associated with poor prognosis. MUC16 is one of the most frequently mutated genes in endometrial cancer. However, the potential relationship and underlying mechanism between MUC16 mutations and endometrial cancer patients’ prognosis and disease progression remain unclear. In present study, we analyzed the whole exome sequencing data, RNA sequencing data and patients’ clinical information in TCGA database and demonstrated that MUC16 mutational status was an independent prognostic factor for endometrial cancer patients. Patients with somatic MUC16 mutations had a prolonged overall survival time. MUC16 mutations promoted patients’ antitumor immune responses. Cytotoxic immune cells mediated pathways were enriched in endometrial cancer samples with MUC16 mutations. Elevation of two pathways, NO2-dependent IL 12 pathway in NK cells and T cytotoxic cell surface molecules, significantly correlated with a higher rate of MUC16 mutations and a significantly favorable patients’ prognosis. An increased level of cytotoxic T lymphocytes, not NK cells, infiltration was observed in the tumor microenvironment of patients with MUC16 mutations. High expression of molecular markers of T cells and CD8+ T cells associated with a higher rate of MUC16 mutations and a better patients’ prognosis. These findings may provide deeper insight into potential endometrial cancer immunotherapy approaches.
Introduction
Endometrial cancer is the most common gynecological malignancy in developed countries and the fifth most common cancer in women worldwide, with an estimated 320,000 new cases and 76,000 deaths each year worldwide.Citation1 The incidence and mortality of endometrial cancer are increasing during recent years, likely due to rising obesity rates and an aging population.Citation2
Mucins are high molecular weight glycoproteins that are divided into two subfamilies, secreted and transmembrane mucins.Citation3 Mucins normally express on various types of epithelial cells and play numerous physiological roles ranging from protection against pathogenic infections to regulation of cellular signaling and transcription.Citation4 Many studies also demonstrated that mucins were overexpressed and aberrantly glycosylated in diverse epithelial cancers to promote cancer cell growth and invasion.Citation5–Citation8 MUC16 is the largest transmembrane mucin and its secreted counterpart is CA125, the well-known diagnostic and adverse prognostic serum marker of gynecological malignancy.Citation9,Citation10 However, its roles in carcinogenesis and progression of endometrial carcinoma are not fully clear.
The core protein of MUC16 contains a large extracellular tandem repeat domain at its N- terminus, and a transmembrane domain with a short cytoplasmic domain at its C- terminus.Citation4 Emerging evidence suggests that MUC16 might be involved in regulating immune responses in various cancer types. Gubbels JA et al reported that MUC16 protected ovarian cancer cells from NK cells targeting by inhibiting synapse formation between these tow types of cells.Citation11 Later, Belisle JA et al identified that MUC16 can also bind to NK cells, B cells and monocytes via Siglec-9, which is an inhibitory receptor that attenuates T cell and NK cell function.Citation12 A study indicated that circulating Treg proportion was related to the serum CA125 level and that MUC16 C terminal promoted Foxp3 expression and tumor-associated Treg enrichment in tumor tissues through tumor-secreted IL-6 activation of the JAK2/STAT3 pathway in pancreatic cancer.Citation13 These findings suggest that MUC16 might play important roles in inhibiting anti-cancer immune responses. There also was study showed that the pro-inflammatory cytokines TNFα and IFNγ stimulate MUC16 expression in breast, endometrial and ovarian cancers through NFκB in vitro and elevated MUC16 expression is associated with elevated cytokine levels in breast and ovarian cancer tissues.Citation14 Pro-inflammatory stimuli (oxidative stress and treatment with the cytokines IFNγ, IL-1α, and TNFα) altered MUC16 glycosylation of pancreatic cancer cells.Citation15 These indicate that MUC16 might be involved in promoting pro-inflammatory signaling in cancer.
Cancer is characterized by a sequential accumulation of genetic alterations including somatic mutations.Citation16 Cancer progression is driven by a handful of mutations in cancer-related genes (oncogenes and tumor suppressors), termed ‘driver mutations’ which facilitate uncontrolled proliferation and other hallmarks of cancer.Citation17,Citation18 Driver mutations, however, arise alongside thousands of other mutations, called “passenger mutations” that have previously been assumed to be neutral and largely ignored in cancer research.Citation19 Yet growing evidence suggests that passenger mutations can be deleterious to cancer cells and play an important role in both cancer progression and clinical outcomes. Citation20–Citation22
MUC16 is one of the most frequently mutated genes in malignant tumors of the female reproductive system through preliminarily analyzing The Cancer Genome Atlas Cancer Genome (TCGA) exome sequencing data via its online exploration tool. However, the studies on MUC16 mutational effects on endometrial cancer tumorigenesis and progression have not been performed to our knowledge. It is also not known whether MUC16 mutations have influences on endometrial cancer patients’ prognosis. In this study, we addressed these questions through a combinational analysis of whole exome sequencing data, RNA sequencing data and patients’ clinical information of the Uterine Corpus Endometrial Carcinoma (UCEC) project in TCGA database to investigate MUC16 mutational effects on endometrial cancer.
Results
MUC16 frequently mutates in endometrial cancer
530 endometrial cancer patients in TCGA-UCEC project were included in this study. 530 patients contained the whole exome sequencing data, 528 patients contained clinical and survival information and 524 patients contained RNA sequencing data.
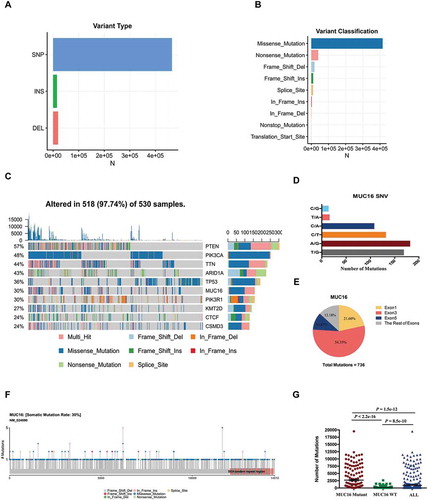
Exome sequencing of 530 endometrial cancer samples identified 498,711 somatic mutations based on consensus calls from mutect2 mutation-detection algorithm (Supplementary Table 1). The types of these mutations included short insertions (INS), short deletions (DEL) and single nucleotide polymorphisms (SNPs), which mainly were nonsynonymous and splice site SNPs (Supplementary Table 1). Synonymous, 5’UTR, 3’UTR and intronic SNPs were not included in our analysis. The major type of variants was SNPs (Number = 464454), compared with insertions and deletions (). The majority of mutations were missense (Number = 415573) and nonsense mutations (Number = 40375) (). The top ten mutated genes in endometrial cancer included PTEN (57%), PIK3CA (48%), TTN (44%), ARID1A (43%), TP53 (36%), MUC16 (30%), PIK3R1 (30%), KMT2D (27%), CTCF (24%), and CSMD3 (24%) (). MUC16 was the sixth most frequently mutated gene in endometrial cancer with a mutational frequency of 159 in 530 tumor samples (30%) and it mutated more than once in one third of samples with mutant MUC16 approximately (). We identified 744 mutations in MUC16 gene, including 621 single nucleotide variants (SNVs) that were mainly substitutions between cytosine and thymine (C/T), cytosine and adenine (C/A), adenine and guanine (A/G) and thymine and guanine (T/G) (). 736 mutations occurred in exon regions and more than 86% mutations occurred in exon1 (21.60%), exon3 (54.35%) and exon5 (10.87%) (). Most of MUC16 mutations were missense mutations and these mutations were equally distributed from N- terminal to C- terminal of MUC16 protein (, ). The total tumor mutational burden of these patients ranged from 1 mutation to 19526 mutations per sample and the mean of variants per sample was 940.964 (). Patients with MUC16 mutations had a higher tumor mutational burden compared with patients without MUC16 mutations (the means of variants per sample 2709.044 vs. 183.2156, P < 2.2e-16) ().
Figure 1. MUC16 frequently mutates in endometrial cancer.
Exome sequencing data of 530 endometrial cancer samples in TCGA-UCEC project were analyzed through the use of mutect2 mutation-detection algorithm.(A) The number of mutations based on variant type.(B) The number of mutations based on variant classification.(C) The OncoPlot of the top ten mutated genes. The upper barplot indicates the number of genetic mutations per patient, while the right barplot shows the number of genetic mutations per gene. The mutation types were added as annotations in the bottom. Variants annotated as Multi_Hit are those genes that mutated more than once in the same sample.(D) The number of mutations of MUC16 gene based on single nucleotide variant (SNV) class.(E) The pie chart of mutations of MUC16 gene occurred in indicated exons.(F) The lollipopPlot of MUC16 gene. Amino acid axis labeled for domain. The mutation types were added as annotations in the bottom.(G) The number of mutations for all patients, patients with mutant MUC16 and patients with wild type MUC16. The data are compared by unpaired two-tailed Student’s t-test. The bars represent mean and SEM.
MUC16 mutations are associated with favorable prognosis of endometrial cancer patients
The detailed clinical characteristics included age at diagnosis, histological type, histologic grade, clinical stage, race and history of neoadjuvant treatment (). The relationship between MUC16 mutational status and clinical features was evaluated through chi-square test. The results showed that MUC16 mutational status was significantly associated with patients’ age at diagnosis (P = 0.0002825) and histological type (P = 0.0002432); No significant difference was found between MUC16 mutational status and histologic grade or clinical stage (P > 0.05) (Supplementary Table 2).
Table 1. Clinical characteristics of patients with endometrial cancer. NA, not available.
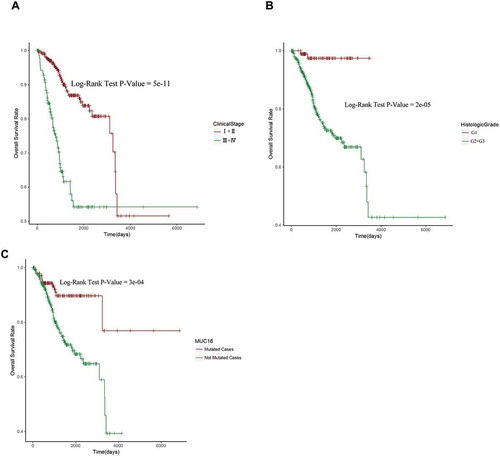
Among all clinical features and top ten mutated genes, there were seven genes (PTEN, PIK3CA, TNN, ARID1A, TP53, MUC16 and CTCF) and four clinical features (age, histological type, histologic grade and clinical stage) might correlate with patients’ survival analyzed by Kaplan-Meier method and log-rank test with P value < 0.05 ( and Supplementary Figure 1, 2). Next, we did cox proportional hazards regression analysis to further investigate the relationship between these seven genes’ mutational status, four clinical characteristics and patients’ overall survival. In univariate analysis, all the features’ P values were less than 0.05, indicating that these features may relate with patients’ overall survival in endometrial cancer (). Therefore, we included all these features in the following multivariate analysis and used a much more stringent cut-off criterion P < 0.01. In multivariate analysis, there were only histologic grade (HR = 7.5872, P = 0.00543), clinical stage (HR = 3.1203, P = 6.44E-07) and MUC16 mutational status (HR = 2.4534, P = 0.00872) that were showed to be independent prognostic factors in endometrial cancer patients (). We then plotted patients’ survival curve based on the stratifications of three independent prognostic factors separately using Kaplan-Meier curve and Log-rank test. The results showed that low histologic grade (G1), early clinical stage (I+ II) and MUC16 mutations were positively related to patients’ overall survival (). Despite its smallest P value among three independent prognostic factors, clinical stage, which had an intersect in its survival curve, was a poor stratified factor of patients’ prognosis (). Compared with clinical stage, histologic grade and MUC16 mutational status were good prognostic stratified factors, which had not intersects in their survival curves (, ).
Table 2. Univariate and multivariate Cox regression analysis in endometrial cancer patients.
Figure 2. MUC16 mutational status, histologic grade and clinical stage are independent prognostic factors for endometrial cancer patients.
(A) Kaplan-Meier curve and Log-rank test for endometrial cancer patients base on clinical stage classification.(B) Kaplan-Meier curve and Log-rank test for endometrial cancer patients base on histologic grade classification.(C) Kaplan-Meier curve and Log-rank test for endometrial cancer patients base on MUC16 mutational status classification.
MUC16 mutations are involved in antitumor immune responses
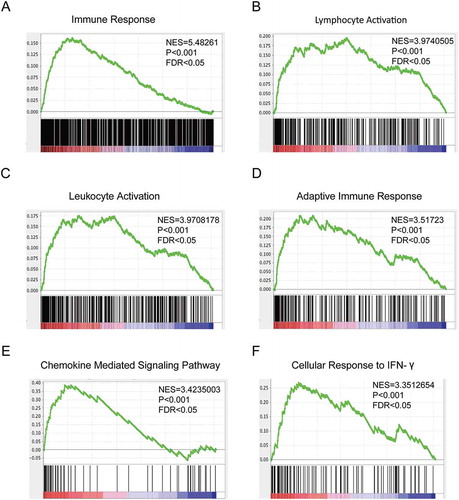
Patients with MUC16 mutations had a prolonged overall survival time, indicating that mutant MUC16 might be critically involved in the tumorigenesis and progression of endometrial cancer. To further study the roles of MUC16 mutations in endometrial cancer, we analyzed the global gene expression profiling between 158 tumor samples with mutant MUC16 and 366 tumor samples with wild type MUC16 using RNA sequencing data downloaded from TCGA database. Gene Set Enrichment Analysis (GSEA) using GO terms as the gene sets revealed enrichment in categories like immune response, lymphocyte activation, adaptive immune response, cellular response to IFN-γ and others, in endometrial cancer samples with MUC16 mutations (). Increasing evidence suggests that MUC16 might play important roles in inhibiting antitumor immune responsesCitation11–Citation13 and in promoting pro-inflammatory signaling in many cancersCitation14,Citation15. It is possible that MUC16 mutations might abrogate its inhibitory immune effects. On the other hand, we observed that patients with MUC16 mutations had a much more higher tumor mutational burden (). It is also possible that the hypermutated status might enhance antitumor immune responses. To minimize the interference of hypermutated status on antitumor immunity, we excluded hypermutated phenotypes (more than 1000 mutations per sample in Supplementary Figure 3A or more than 500 mutations per sample in Supplementary Figure 3B) and found that total mutational loads decreased significantly, but remained slightly higher in patients with MUC16 mutations. However, the exclusion of patients with hypermutated phenotypes did not alter the effects of MUC16 mutations on patients’ prognosis (Supplementary Figure 3C) and antitumor immune responses (Supplementary Figure 3D). A study reported that it was neoantigen quality, not quantity, that can predict patients’ survival and MUC16 neoantigens were enriched in long-term survivor of pancreatic cancer. Citation23 These observations indicated that MUC16 mutations might play critical roles in patents’ prognosis and in regulating antitumor immunity.
Figure 3. Mutated MUC16 are involved in endometrial cancer patients’ antitumor immune responses.
Gene Set Enrichment Analysis (GSEA) of global gene expression profiling between 158 tumor samples with mutant type MUC16 and 366 tumor samples with wild type MUC16. Gene sets annotated by GO terms were used in the analysis. NES, normalized enrichment score; P = nominal P value; FDR = false discovery rate.Gene sets representing Immune Response (A), Lymphocyte Activation (B), Leukocyte Activation (C), Adaptive Immune Response (D), Chemokine Mediated Signaling Pathway (E) and Cellular Response to IFN-γ (F) were enriched in patients with MUC16 mutations.
MUC16 mutations are involved in cytotoxic immune cell mediated antitumor responses
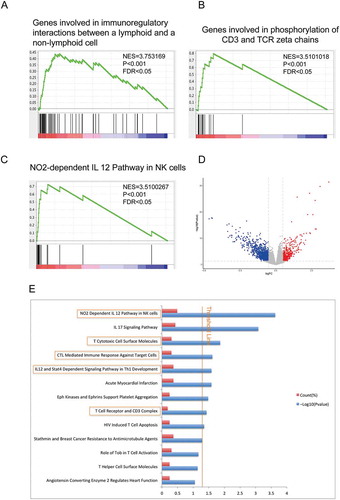
To guide our identification of immune pathways necessary for mutant MUC16 function, we performed GSEA using canonical pathways mainly from BIOCARTA, KEGG and REACTOME databases as the gene sets. The results demonstrated that cytotoxic immune cell mediated antitumor immune responses, such as genes involved in immunoregulatory interactions between a lymphoid and a non-lymphoid cell (), genes involved in phosphorylation of CD3 and TCR zeta chains () and NO2-dependent IL 12 pathway in NK cells (), were enriched in endometrial cancer samples with MUC16 mutations. Next, we employed BIOCARTA pathway analysis of the differentially expressed genes (DEGs) via DAVID database. According to the cut-off criteria (P < 0.05 and log2|FC| ≥ 0.5), a total of 1666 DEGs were identified, including 420 up-regulated and 1246 down-regulated genes ( and Supplementary Table 3). We identified that NO2-dependent IL12 pathway in NK cells was the most overrepresented pathway in tumor samples with MUC16 mutations (). The results also showed that T cell mediated antitumor immune responses like T cytotoxic cell surface molecules, cytotoxic T lymphocyte (CTL) mediated immune response against target cells and others were significantly enriched in samples with MUC16 mutations (). These findings suggested that MUC16 mutations promote antitumor immune responses through cytotoxic immune cell mediated pathways in endometrial cancer.
Figure 4. MUC16 mutations are involved in cytotoxic immune cell mediated antitumor responses.
(A)-(C) GSEA using the canonical pathways, which are annotated pathways mainly from BIOCARTA, KEGG and REACTOME databases, as the gene sets in the analysis. NES, normalized enrichment score; P = nominal P value; FDR = false discovery rate.(A) A gen set representing Genes involved in immunoregulatory interactions between a lymphoid and a non-lymphoid cell was enriched in patients with MUC16 mutations.(B) A gen set representing Genes involved in phosphorylation of CD3 and TCR zeta chains was enriched in patients with MUC16 mutations.(C) A gen set representing NO2-dependent IL 12 Pathway in NK cells was enriched in patients with MUC16 mutations.(D) Volcano plot of differentially expressed genes (DEGs) between mutant and wild type MUC16 endometrial cancer samples. Red dot represents up-regulated genes and blue dot represents down-regulated genes.(E) BIOCARTA pathway analysis of DEGs via DAVID database. Blue bars that cross the threshold line (P < 0.05) represent pathways that are significantly changed between mutant and wild type MUC16 endometrial cancer samples.
To further investigate the involvement of mutant MUC16 in antitumor immune responses through cytotoxic immune cell mediated pathways, we analyzed the expression level of 17 genes in NO2-dependent IL 12 pathway in NK cells and 12 genes in T cytotoxic cell surface molecules between tumor samples with mutant and wild type MUC16. Next, we stratified tumors samples based on the expression pattern of genes in these two pathways and examined the mutation rate of MUC16. Two main groups were observed from the unsupervised hierarchical clustering of the expression matrix from 17 genes in NO2-dependent IL 12 pathway in NK cells of the 524 samples. Cluster I had a higher gene expression pattern and exhibited a higher rate of MUC16 mutations (35.38%) ). Cluster II had a lower expression pattern and a lower rate of MUC16 mutations (20.33%) (). Similarly, two main groups were also observed in T cytotoxic cell surface molecules. Cluster I had a higher gene expression pattern and exhibited a higher rate of MUC16 mutations (33.91%) (). Cluster II had a lower expression pattern and a lower rate of MUC16 mutations (22.73%) ().
Figure 5. Hierarchical clustering analysis of two pathways, NO2-dependent IL 12 pathway in NK cells and T cytotoxic cell surface molecules.
(A) and (C) Unsupervised hierarchical clustering with Euclidean distance and ward linkage of expression matrixes from 17 genes in NO2-dependent IL 12 pathway in NK cells (A) and 12 genes in T cytotoxic cell surface molecules (C) of 524 samples (158 of which have MUC16 mutational status). Rows indicate the identity of the genes and columns indicate the identity of the patients. The MUC16 mutational status for each tumor is depicted directly above each column. Cluster I exhibits a higher expression of genes in these two pathways, and cluster II exhibits a lower expression level.(B) The Kaplan-Meier curves for the resulting clusters from the unsupervised hierarchical clustering in (A).(D) The Kaplan-Meier curves for the resulting clusters from the unsupervised hierarchical clustering in (C).
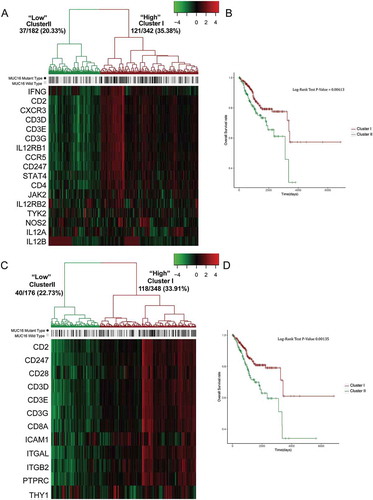
To test the biological significance of elevation of NO2-dependent IL 12 pathway in NK cells and T cytotoxic cell surface molecules in tumors with MUC16 mutations, we examined whether up-regulation of these two pathways correlated with patients’ prognosis. It was striking that cluster I, which had higher expression level of the genes in these two pathways, was correlated with a more favorable prognosis, whereas cluster II, which had lower expression pattern, was associated with a significantly poorer survival probability (, ). Therefore, not only was elevation of these two pathways significantly correlated with a higher rate of MUC16 mutations, but these patients also had a significantly favorable survival.
MUC16 mutations enhance the infiltration and antitumor immunity of cytotoxic T lymphocytes in the endometrial cancer microenvironment
To guide our identification of the specific populations and subpopulations of cytotoxic immune cells infiltration in the endometrial cancer microenvironment of patients with MUC16 mutations, we analyzed the expression level of molecular markers of T cells and NK cells between patients with and without MUC16 mutations through the use of immune cell signatures built by a research group from expression data.Citation24 Transcriptomic profiling analysis of 524 endometrial cancer samples revealed the upregulation of genes specific for T cell and CD8+ T cells in the tumor microenvironment of patients with MUC16 mutations (, left panel). Cancer samples with MUC16 mutations did not display differences in the expression of molecular markers of NK cells (, right panel). The CD56dim NK cell subpopulation displays high cytotoxic capacity, secretes cytokines as interferon (IFN)-gamma after direct contact with target cells and is thought to mediate antitumor responses.Citation25 The expression of genes specific for CD56dim NK cells was slightly higher in patients with MUC16 mutations, however, with no statistical significance (, right panel).
Figure 6. MUC16 mutations enhance the infiltration and antitumor immunity of cytotoxic T lymphocytes in the endometrial cancer microenvironment.
(A) Tumor sample transcriptomic immune profiling between patients with mutant and wild type MUC16. The data are represented as mean± SEM and compared by unpaired two-tailed Student’s t-test.(B) Unsupervised hierarchical clustering with Euclidean distance and ward linkage of expression matrixes from 16 genes in T cell signature and 36 genes in CD8+ T cell signature of 524 samples. Rows indicate the identity of the genes and columns indicate the identity of the patients. The MUC16 mutational status for each tumor is depicted directly above each column. Cluster I exhibits a higher expression of genes in these two signatures, and cluster II exhibits a lower expression level.(C) The Kaplan-Meier curves for the resulting clusters from the unsupervised hierarchical clustering in (B).
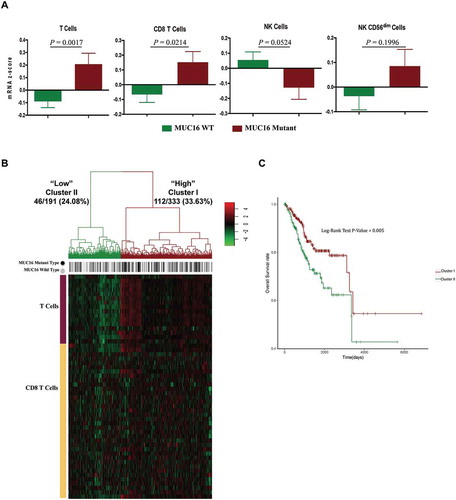
The unsupervised hierarchical clustering of the expression matrix from T cell and CD8+ T cell signatures of 524 endometrial cancer samples showed that the higher gene expression cluster (Cluster I) exhibited a higher rate of MUC16 mutations (33.63%) and the lower gene expression cluster (Cluster II) exhibited a lower rate of MUC16 mutations (24.08%) (). Consistently, Cluster I was correlated with a more favorable prognosis, whereas cluster II was associated with a significantly poorer survival probability (). Theses findings demonstrated that patients with MUC16 mutations had an elevated level of cytotoxic T lymphocytes infiltration and enhanced T cell antitumor immunity in the tumor microenvironment and exhibited prolonged overall survival time.
Discussion
As one of the most well-studied tumor biomarkers over three decades, elevated serum CA125 level suggests patients’ poor prognosis in many cancers including endometrial cancer.Citation26–Citation29 A positive significant correlation was also found between increased levels of CA125 tissue expression and elevated serum CA125 levels in ovarian cancer patients.Citation30 These findings demonstrate that CA125 expression levels are closely associated with patients’ prognosis. MUC16 mutated in various cancer types with a high mutational frequency.Citation31,Citation32 However, little is known about the relationship between MUC16 mutational status and patients’ prognosis. In our analysis, we found that although the gene expression levels of MUC16 between endometrial cancer samples with mutant and wild type MUC16 were no statistical significance (data not show), MUC16 mutational status was an independent prognostic factor for patients, indicating that patients with somatic MUC16 mutations had a prolonged overall survival time ( and ).
Accumulating evidence supports that passenger mutations correlate with slower cancer progression, enhanced antitumor immunity and improved clinical outcomes. Citation20–Citation22,Citation33,Citation34 Consistently, we observed that mutations in six genes (PTEN, PIK3CA, TNN, ARID1A, MUC16 and CTCF) were positively related to patients’ overall survival and only TP53 mutations negatively associated with patients’ prognosis (P < 0.05) (, Supplementary Figure 2). Simultaneously, we also found that many patients with MUC16 mutations also carried numerous passenger mutations resulting in higher mutational burden (), and these patients exhibited enhanced antitumor immune responses in the tumor microenvironment () and favorable prognosis (). However, the exclusion of these hypermutated cases did not alter the effects of MUC16 mutations on patients’ prognosis (Supplementary Figure 3C) and antitumor immune responses (Supplementary Figure 3D). Not only quantities, but also qualities of neoantigens generated by various mutations influence patients’ survival.Citation23 This may be one possible interpretation for our observations in Supplementary Figure 3. These findings indicated the important regulatory roles of MUC16 mutations in patients’ survival and antitumor immunity.
Increasing evidence suggests that MUC16 actively interacts with immune system. Pro-inflammatory cytokines can stimulate MUC16 expression or alter its glycosylation in normal epithelial cells or epithelial cancer cells.Citation14,Citation15,Citation35 MUC16 can repress antitumor immune responses through inhibiting NK cell or T cell function in many cancers.Citation11-Citation13 However, very little is known about the effects of MUC16 mutations on patients’ antitumor immunity. Several studies demonstrated that somatic mutations can generate cancer-specific neoantigens that are presented by HLA molecules and potentially recognized by the mature T cell repertoire as ideal targets for cancer immunotherapy and cancer vaccine.Citation36–Citation40 A study unveiled that MUC16 was the most frequently tumor specific antigen presented by HLA molecules on ovarian cancer cells and 85% of MUC16-derived HLA ligands are immunogenic and able to prime T cells, rendering MUC16 an unparalleled first-class antigen for ovarian cancer immunotherapy.Citation41 Recently, a research identified that, compared with short-term survivors, the long-term survivors of pancreatic cancer had a higher frequency of MUC16 neoantigens and had T-cell responses against mutant MUC16, suggesting that MUC16 mutations generate neoantigens that are key antitumour targets of the immune system.Citation23 Our GSEA and DEGs analyses also revealed that T cell mediated immune responses like T Cytotoxic Cell Surface Molecules and CTL mediated immune response against target cells are enriched in endometrial cancer samples with MUC16 mutations (, ). The analysis of molecular markers expression level of cytotoxic T lymphocytes indicated that increased T cells and CD8+ T cells infiltration in the tumor microenvironment of patients with MUC16 mutations (). High expression of T cytotoxic cell surface molecules pathway, T-cell and CD8+ T-cell signatures, significantly correlated with higher rates of MUC16 mutations (, ) and favorable patients’ prognosis (, ). Collectively, our data demonstrated that patients with somatic MUC16 mutations had favorable prognosis than those with wild type MUC16 and exhibited enhanced infiltration and antitumor immunity of cytotoxic T lymphocytes. More in-depth studies are needed to investigate how to exploit mutant MUC16 in endometrial cancer immunotherapy to promote patients’ survival.
Besides cytotoxic T cell, NK cell is also an important part of antitumor immune responses due to their potent cytolytic and cytokine-secreting abilities.Citation42 We observed that NO2-dependent IL 12 pathway in NK cells was the most overrepresented pathway in tumor samples with MUC16 mutations through DEGs analysis (). High expression of this pathway was associated with higher rates of MUC16 mutations and improved patients’ overall survival (, ). However, patients with MUC16 mutations did not display differences in NK cells infiltration in the endometrial cancer microenvironment (). MUC16 was reported to inhibit immune synapse formation between NK cells and cancer cells.Citation11 It can also bind to Siglec-9, an inhibitory receptor on NK cells.Citation12 There are little researches focus on how mutated tumor-specific antigens recognized by NK cells and NK cells’ efficiency in related cancer immunotherapy. Further validations were needed to identify whether MUC16 mutations correlate with antitumor function of NK cells. And the applications of mutant MUC16 proteins in NK cells associated cancer immunotherapy need further investigations.
Interleukin-12 (IL-12) promotes cell mediated immunity by activating the cytotoxic activity of both T cell and NK cell and inducing Th1 cell differentiation which is believed to represent an important link between innate and adaptive immunity.Citation43 In our analysis, we observed that two IL-12 mediated pathways in T cells and NK cells were upregulated in tumor samples with MUC16 mutations (). Studies reported that chimeric antigen receptors expressing T cells (CAR T cells), which secrete IL-12 and target retained MUC16 ectodomain, exhibited enhanced antitumor efficacy in recurrent ovarian cancer.Citation44,Citation45 The combination use of MUC16 and IL-12 provided novel and promising approachs for cancer immunotherapy. And, more researches are needed to explore appropriate protocols, the safety and efficacy of these methods in clinical application.
Methods
TCGA data downloading
The Mutation Annotation Format (MAF) data generated by mutect2 algorithm from whole exome sequencing data, RNA sequencing raw counts (HTSeq – Counts) data, RNA sequencing FPKM data (HTSeq – FPKM), clinical and survival information were downloaded from TCGA database (https://cancergenome.nih.gov/) through TCGAbiolinks R/Bioconductor package.Citation46 In brief, 530 endometrial cancer patients were included in the following analyses, all of which contained the whole exome sequencing data, 528 patients contained clinical and survival information and 524 patients contained RNA sequencing data.
Analyses and visualization of somatic mutations
We used Maftools R/Bioconductor packageCitation47 to extract detailed mutational information from MAF file. The summary data of mutect2 mutational analysis were list in Supplementary Table 1. And, the plotmafSummary function was used to plot the summary of the MAF file, which displays number of variant type and variant classification. The oncoplot functionCitation47,Citation48 and the lollipopPlot function were used to plot OncoPlot of the top ten mutated genes and lollipopPlot of MUC16 gene respectively.
Patients’ prognostic analyses
Survival curves were depicted using the Kaplan-Meier method and compared with log-rank test. Cox proportional hazards regression analysis was used for univariate and multivariate analyses to explore the association of clinical features, gene mutational status and patients’ prognosis. All the prognostic analyses were conducted by survival R package.
Differentially expressed genes analysis and biocarta pathway analysis
The differentially expressed genes (DEGs) between mutant and wild type MUC16 endometrial cancer samples were analyzed by edgeR R/Bioconductor package.Citation49 Genes with log2|FC| ≥ 0.5 and P < 0.05 were considered to be significantly DEGs. The DEGs data was list in Supplementary Table 3. BioCarta pathway analysis of DEGs was performed using DAVID database (https://david.ncifcrf.gov/).Citation50
Gene set enrichment analysis
Gene Set Enrichment Analysis (GSEA) was performed through the use of GSEAPreranked tool in GSEA software (http://www.broadinstitute. org/gsea/).Citation51 The value of log2(FC) calculated by edgeR package was used as ranking metric.
We used C5 collection that contains genes sets annotated by GO terms in the Molecular Signatures Database (MSigDB) as the gene sets in the analysis. The C5 collection, including 5917 gene sets, is divided into three sub-collections based on GO ontologies: Biological Process, Cellular Component, and Molecular Function.
We also used the canonical pathways sub-collection of C2 collection in the MSigDB as the gene sets in the analysis. This sub-collection contains 1329 gene sets, which are annotated pathways mainly from BIOCARTA, KEGG and REACTOME databases.
Hierarchical clustering
The gene expression data for T cell, CD8+ T cell signatures and the following two pathways, NO2-dependent IL 12 Pathway in NK cells and T Cytotoxic Cell Surface Molecules, were extracted from RNA sequencing FPKM data of 524 endometrial cancer samples in TCGA database. The data were subjected to log2 transformed. Unsupervised hierarchical clustering was used to discover groups based on the expression pattern of the genes in these two pathways and T cell, CD8+ T cell signatures. In total, 17 genes in NO2-dependent IL 12 Pathway in NK cells, 12 genes in T Cytotoxic Cell Surface Molecules, 16 genes in T cell signature and 36 genes in CD8+ T cell signature were used in the unsupervised hierarchical clustering respectively. Expression values of these genes of 524 samples (where rows indicate the identity of the genes, columns indicate the identity of the patients) were clustered using hierarchical clustering with Euclidean distance and ward linkage by dendextend R packageCitation52 and heatmap.2 function in gplots R package. The MUC16 mutation rates of the resulting groups were calculated. The Kaplan-Meier survival curves were plotted for the resulting groups and compared with log-rank test.
Cytotoxic t cell and NK cell populations and subpopulations analysis
T cell, CD8+ T cell, NK cell and CD56dim NK cell signatures were built by a research group, using genes specific for these immune cells.Citation24 T cell signature genes include PRKCQ, CD3D, CD3G, CD28, LCK, TRAT1, BCL11B, CD2, ITM2A, SH2D1A, CD6, CD96, NCALD, GIMAP5, CD3E and SKAP1. CD8+ T cell signature genes include CD8B, CD8A, PF4, SF1, LIME1, PRR5, GZMM, SLC16A7, SRSF7, APBA2, HAUS3, LEPROTL1, GADD45A, ZFP36L2, KAT6A, ZEB1, ZNF609, MAPKAPK5-AS1, THUMPD1, VAMP2, ZNF91, ZNF22, TMC6, DNAJB1, FLT3LG, CDKN2AIP, TSC22D3, TBCC, RBM3, ABT1, TMEM259, CAMLG, PPP1R2, AES, KLF9 and PRF1. NK cell signature genes include GAGE2, ZNF747, XCL1, XCL2, GNAS, SLC30A5, SGMS1, MCM3AP, TBXA2R, CDC5L, FGF18, MRC2, PSMD4, PRX, ZNF205, APBB2, ZNF528, MAPRE3, BCL2, KANK2, ATL2, SPN, FZR1, PDLIM4, TRPV6, LDB3, ADARB1, PPP4R3A, TCTN2, TINAGL1, IGFBP5, ALDH1B1, FUT5 and NCR1. CD56dim NK cell signature genes include SPON2, GZMB, TTC38, PMEPA1, IL21R, GTF3C1, S1PR5, KIR3DL2, KIR2DL3, KIR3DL1, KIR3DL3.
The gene expression data were extracted from RNA sequencing FPKM data of 524 endometrial cancer samples in TCGA database. mRNA z-scores were calculated via the function scale in R.
Statistical analysis
Univariate and multivariate cox proportional hazard regression analysis were carried out to compare clinical features, gene mutational status (mutated vs. not mutated) and patients’ prognosis. Survival curves were compared using log-rank test. Comparisons between two groups were performed using an unpaired two-tailed Student’s t-test (normally distributed parameters). Categorical variables were compared using a χ2 test. All comparison groups had equivalent variances. P < 0.05 was considered to be statistically significant. All statistical analysis was performed using R programming language v. 3.4.3.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Additional information
Funding
References
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86. doi:10.1002/ijc.29210. PMID: 25220842.
- Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin. 2018;68:7–30. doi:10.3322/caac.21442. PMID: 29313949.
- Baldus SE, Engelmann K, Hanisch FG. MUC1 and the MUCs: a family of human mucins with impact in cancer biology. Crit Rev Clin Lab Sci. 2004;41:189–231. doi:10.1080/10408360490452040. PMID: 15270554.
- Hattrup CL, Gendler SJ. Structure and function of the cell surface (tethered) mucins. Annu Rev Physiol. 2008;70:431–457. doi:10.1146/annurev.physiol.70.113006.100659. PMID: 17850209.
- Kufe DW. Mucins in cancer: function, prognosis and therapy. Nat Rev Cancer. 2009;9:874–885. doi:10.1038/nrc2761. PMID: 19935676.
- Bafna S, Kaur S, Batra SK. Membrane-bound mucins: the mechanistic basis for alterations in the growth and survival of cancer cells. Oncogene. 2010;29(20):2893–2904. doi:10.1038/onc.2010.87. PMID: 20348949.
- Remmers N, Anderson JM, Linde EM, DiMaio DJ, Lazenby AJ, Wandall HH, Mandel U, Clausen H, Yu F, Hollingsworth MA. Aberrant expression of mucin core proteins and o-linked glycans associated with progression of pancreatic cancer. Clin Cancer Res. 2013;19:1981–1993. doi:10.1158/1078-0432.CCR-12-2662. PMID: 23446997.
- Hollingsworth MA, Swanson BJ. Mucins in cancer: protection and control of the cell surface. Nat Rev Cancer. 2004;4:45–60. doi:10.1038/nrc1251. PMID: 14681689.
- Vuento MH, Stenman UH, Pirhonen JP, Mäkinen JI, Laippala PJ, Salmi TA. Significance of a single CA 125 assay combined with ultrasound in the early detection of ovarian and endometrial cancer. Gynecol Oncol. 1997;64:141–146. doi:10.1006/gyno.1996.4545. PMID: 8995563.
- Chao A, Tang YH, Lai CH, Chang CJ, Chang SC, Wu TI, Hsueh S, Wang CJ, Chou HH, Chang TC. Potential of an age-stratified CA125 cut-off value to improve the prognostic classification of patients with endometrial cancer. Gynecol Oncol. 2013;129:500–504. doi:10.1016/j.ygyno.2013.02.032. PMID: 23458702.
- Gubbels JA, Felder M, Horibata S, Belisle JA, Kapur A, Holden H, Petrie S, Migneault M, Rancourt C, Connor JP, et al. MUC16 provides immune protection by inhibiting synapse formation between NK and ovarian tumor cells. Mol Cancer. 2010;9:11. doi:10.1186/1476-4598-9-11. PMID: 20089172.
- Belisle JA, Horibata S, Jennifer GA, Petrie S, Kapur A, André S, Gabius HJ, Rancourt C, Connor J, Paulson JC, et al. Identification of Siglec-9 as the receptor for MUC16 on human NK cells, B cells, and monocytes. Mol Cancer. 2010;9:118. doi:10.1186/1476-4598-9-118. PMID: 20497550.
- Fan K, Yang C, Fan Z, Huang Q, Zhang Y, Cheng H, Jin K, Lu Y, Wang Z, Luo G, et al. MUC16 C terminal-induced secretion of tumor-derived IL-6 contributes to tumor-associated Treg enrichment in pancreatic cancer. Cancer Lett. 2018;418:167–175. doi:10.1016/j.canlet.2018.01.017. PMID: 29337110.
- Morgado M, Sutton MN, Simmons M, Warren CR, Lu Z, Constantinou PE, Liu J, Francis LL, Conlan RS, Bast RC Jr, et al. Tumor necrosis factor-α and interferon-γ stimulate MUC16 (CA125) expression in breast, endometrial and ovarian cancers through NFκB. Oncotarget. 2016;7:14871–14884. doi:10.18632/oncotarget.7652. PMID: 26918940.
- Wu YM, Nowack DD, Omenn GS, Haab BB. Mucin glycosylation is altered by pro-inflammatory signaling in pancreatic-cancer cells. J Proteome Res. 2009;8:1876–1886. doi:10.1021/pr8008379. PMID: 19714813.
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi:10.1126/science.1235122. PMID: 23539594.
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi:10.1016/j.cell.2011.02.013. PMID: 21376230.
- Tomasetti C, Marchionni L, Nowak MA, Parmigiani G, Vogelstein B. Only three driver gene mutations are required for the development of lung and colorectal cancers. Proc Natl Acad Sci U S A. 2015;112:118–123. doi:10.1073/pnas.1421839112. PMID: 25535351.
- Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi:10.1038/nature07943. PMID: 19360079.
- McFarland CD, Yaglom JA, Wojtkowiak JW, Scott JG, Morse DL, Sherman MY, Mirny LA. The damaging effect of passenger mutations on cancer progression. Cancer Res. 2017;77:4763–4772. doi:10.1158/0008-5472.CAN-15-3283-T. PMID: 28536279.
- McFarland CD, Mirny LA, Korolev KS. Tug-of-war between driver and passenger mutations in cancer and other adaptive processes. Proc Natl Acad Sci U S A. 2014;111(42):15138–15143. doi:10.1073/pnas.1404341111. PMID: 25277973.
- McFarland CD, Korolev KS, Kryukov GV, Sunyaev SR, Mirny LA. Impact of deleterious passenger mutations on cancer progression. Proc Natl Acad Sci U S A. 2013;110(8):2910–2915. doi:10.1073/pnas.1213968110. PMID: 23388632.
- Balachandran VP, Łuksza M, Zhao JN, Makarov V, Moral JA, Remark R, Herbst B, Askan G, Bhanot U, Senbabaoglu Y, et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature. 2017;551(7681):512–516. doi:10.1038/nature24462. PMID: 29132146.
- Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T, Lafontaine L, Berger A, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39(4):782–795. doi:10.1016/j.immuni.2013.10.003. PMID: 24138885.
- Cooper MA, Fehniger TA, Caligiuri MA. The biology of human natural killer-cell subsets. Trends Immunol. 2001;22(11):633–640. PMID: 11698225.
- Tang A, Kondalsamy-Chennakesavan S, Ngan H, Zusterzeel P, Quinn M, Carter J, Leung Y, Obermair A. Prognostic value of elevated preoperative serum CA125 in ovarian tumors of low malignant potential: a multinational collaborative study (ANZGOG0801). Gynecol Oncol. 2012;126:36–40. doi:10.1016/j.ygyno.2012.02.031. PMID: 22370601.
- Huang GS, Chiu LG, Gebb JS, Gunter MJ, Sukumvanich P, Goldberg GL, Einstein MH. Serum CA125 predicts extrauterine disease and survival in uterine carcinosarcoma. Gynecol Oncol. 2007;107:513–517. doi:10.1016/j.ygyno.2007.08.060. PMID: 17935762.
- Nakamura K, Imafuku N, Nishida T, Niwa I, Joja I, Hongo A, Kodama J, Hiramatsu Y. Measurement of the minimum apparent diffusion coefficient (ADCmin) of the primary tumor and CA125 are predictive of disease recurrence for patients with endometrial cancer. Gynecol Oncol. 2012;124:335–339. doi:10.1016/j.ygyno.2011.10.014. PMID: 22008707.
- Yang C, Cheng H, Luo G, Lu Y, Guo M, Jin K, Wang Z, Yu X, Liu C. The metastasis status and tumor burden-associated CA125 level combined with the CD4/CD8 ratio predicts the prognosis of patients with advanced pancreatic cancer: A new scoring system. Eur J Surg Oncol. 2017;43:2112–2118. doi:10.1016/j.ejso.2017.07.010. PMID: 28802662.
- Høgdall EV, Christensen L, Kjaer SK, Blaakaer J, Kjaerbye-Thygesen A, Gayther S, Jacobs IJ, Høgdall CK. CA125 expression pattern, prognosis and correlation with serum CA125 in ovarian tumor patients. From the danish “MALOVA” ovarian cancer study. Gynecol Oncol. 2007;104:508–515. doi:10.1016/j.ygyno.2006.09.028. PMID: 17113137.
- Kim N, Hong Y, Kwon D, Yoon S. Somatic mutaome profile in human cancer tissues. Genomics Inform. 2013;11:239–244. doi:10.5808/GI.2013.11.4.239. PMID: 24465236.
- Zhan H, Jiang J, Sun Q, Ke A, Hu J, Hu Z, Zhu K, Luo C, Ren N, Fan J, et al. Whole-exome sequencing-based mutational profiling of hepatitis B virus-related early-stage hepatocellular carcinoma. Gastroenterol Res Pract. 2017;2017:2029315. doi:10.1155/2017/2029315. PMID: 29333154.
- Andor N, Graham TA, Jansen M, Xia LC, Aktipis CA, Petritsch C, Ji HP, Maley CC. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat Med. 2016 Jan;22(1):105–113. doi:10.1038/nm.3984. PMID: 26618723.
- Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160(1–2):48–61. doi:10.1016/j.cell.2014.12.033. PMID: 25594174.
- Albertsmeyer AC, Kakkassery V, Spurr-Michaud S, Beeks O, Gipson IK. Effect of pro-inflammatory mediators on membrane-associated mucins expressed by human ocular surface epithelial cells. Exp Eye Res. 2010;90:444–451. doi:10.1016/j.exer.2009.12.009. PMID: 20036239.
- Matsushita H, Vesely MD, Koboldt DC, Rickert CG, Uppaluri R, Magrini VJ, Arthur CD, White JM, Chen YS, Shea LK, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482:400–404. doi:10.1038/nature10755. PMID: 22318521.
- Robbins PF, Lu YC, El-Gamil M, Li YF, Gross C, Gartner J, Lin JC, Teer JK, Cliften P, Tycksen E, et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med. 2013;19:747–752. doi:10.1038/nm.3161. PMID: 23644516.
- Gubin MM, Zhang X, Schuster H, Caron E, Ward JP, Noguchi T, Ivanova Y, Hundal J, Arthur CD, Krebber WJ, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014;515:577–581. doi:10.1038/nature13988. PMID: 25428507.
- Kreiter S, Vormehr M, van de Roemer N, Diken M, Löwer M, Diekmann J, Boegel S, Schrörs B, Vascotto F, Castle JC, et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature. 2015;520:692–696. doi:10.1038/nature14426. PMID: 25901682.
- Sahin U, Türeci Ö. Personalized vaccines for cancer immunotherapy. Science. 2018;359:1355–1360. doi:10.1126/science.aar7112. PMID: 29567706.
- Schuster H, Peper JK, Bösmüller HC, Röhle K, Backert L, Bilich T, Ney B, Löffler MW, Kowalewski DJ, Trautwein N, et al. The immunopeptidomic landscape of ovarian carcinomas. Proc Natl Acad Sci U S A. 2017;114:E9942–E9951. doi:10.1073/pnas.1707658114. PMID: 29093164.
- Paul S, Lal G. The molecular mechanism of natural killer cells function and its importance in cancer immunotherapy. Front Immunol. 2017;8:1124. doi:10.3389/fimmu.2017.01124. PMID: 28955340.
- Zundler S, Neurath MF. Interleukin-12: functional activities and implications for disease. Cytokine Growth Factor Rev. 2015;26:559–568. doi:10.1016/j.cytogfr.2015.07.003. PMID: 26182974.
- Koneru M, Purdon TJ, Spriggs D, Koneru S, Brentjens RJ. IL-12 secreting tumor-targeted chimeric antigen receptor T cells eradicate ovarian tumors in vivo. Oncoimmunology. 2015;4:e994446. doi:10.4161/2162402X.2014.994446. PMID: 25949921.
- Koneru M, O’Cearbhaill R, Pendharkar S, Spriggs DR, Brentjens RJ. A phase I clinical trial of adoptive T cell therapy using IL-12 secreting MUC-16(ecto) directed chimeric antigen receptors for recurrent ovarian cancer. J Transl Med. 2015;13:102. doi:10.1186/s12967-015-0460-x. PMID: 25890361.
- Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, Sabedot TS, Malta TM, Pagnotta SM, Castiglioni I, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44:e71. doi:10.1093/nar/gkv1507. PMID: 26704973.
- Mayakonda A, Koeffler HP. Maftools: efficient analysis, visualization and summarization of MAF files from large-scale cohort based cancer studies. bioRxiv. 2016. doi:10.1101/052662.
- Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi:10.1093/bioinformatics/btw313. PMID: 27207943.
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi:10.1093/bioinformatics/btp616. PMID: 19910308.
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi:10.1038/nprot.2008.211. PMID: 19131956.
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi:10.1073/pnas.0506580102. PMID: 16199517.
- Galili T. dendextend: an R package for visualizing, adjusting and comparing trees of hierarchical clustering. Bioinformatics. 2015;31:3718–3720. doi:10.1093/bioinformatics/btv428. PMID: 26209431.
