ABSTRACT
Cancers progress when the immune system fails to identify and eliminate malignant cells. Recognition of this, combined with advances in tumor immunology, has allowed development of therapies that induce effective anti-tumor immune responses. For incompletely-understood reasons, effective responses to immunotherapy occur in some patients and not others. Head and neck squamous cell carcinomas (HNSCC) are a common cancer type that can be divided into two subsets based on human papillomavirus (HPV) status. HPV status is a strong predictor of positive clinical outcome. Expression of exogenous viral antigens by HPV+, but not HPV-, HNSCC allows direct comparison of the immune status (immune cell presence and characteristics) between these two otherwise anatomically-similar tumors. Using TCGA data, we compared the immune landscape between HPV+ and HPV- treatment-naïve HNSCC. As compared to HPV- samples, HPV+ HNSCC exhibited a strong Th1 response characterized by increased infiltration with multiple types of immune cells and expression of their effector molecules. HPV+ HNSCC also expressed higher levels of CD39 and multiple T-cell exhaustion markers including LAG3, PD1, TIGIT, and TIM3 compared to HPV- HNSCC. Importantly, patients with higher expression of these exhaustion markers–indicative of a T-cell-inflamed tumor–correlated with markedly improved survival in HPV+, but not HPV-, HNSCC. Thus, profound differences exist between the immune landscape of HPV+ and HPV- HNSCC. These results suggest that immune checkpoint inhibitor therapy is a promising treatment strategy for HPV+ HNSCC, and that expression of immune checkpoint molecules could serve as a predictive biomarker of patient outcome in HPV+ HNSCC.
Introduction
Recent advances in understanding the basics of tumor immunology have paved the way for development of therapies that can induce effective anti-tumor immune responses. Many of these are based on immune checkpoint inhibition, in which normal negative regulatory mechanisms, which keep immune responses in check, are overcome.Citation1,Citation2 Current immuno-oncology therapies can demonstrate tremendous efficacy and induce long-term remission in patients.Citation3 However, responses to immunotherapy are often restricted to a subset of cancers, for reasons that are not entirely understood.
Head and neck squamous cell carcinomas (HNSCC) represent the 6th most common human cancer typeCitation4 and are often characterized by aggressive local invasion and overall poor prognosis.Citation5 In addition to mortality, both the disease and its treatment often result in significant patient morbidity. Indeed, treatments for HNSCC often impact the most personal characteristics of an individual, including facial appearance and the ability to eat and speak.
Infection with human papillomavirus (HPV) is a major etiological factor for tumors located in the oropharynx. Indeed, HPV positive (HPV+) HNSCC is increasing at an epidemic pace.Citation6,Citation7 Importantly, HPV negative (HPV-) and HPV+ HNSCC are molecularly distinct, with a different spectrum of mutations.Citation8 HPV+ HNSCC also constitutively express viral oncogenes that deregulate cell growth and gene expression.Citation9 In addition, clinical outcomes for HPV+ HNSCC are far superior to those in HPV- cases.Citation10,Citation11
Both HPV+ and HPV- HNSCC display a comparable frequency of somatic mutations, a major source of tumor specific neoantigens that can be recognized and targeted by anti-tumor immunity.Citation8 However, HPV+, but not HPV-, HNSCC express exogenous antigenic viral proteins that may be the source of a key difference in the tumor immune landscape between these two types of HNSCC and may contribute to the superior clinical outcomes associated with HPV+ HNSCC. Several studies have compared various immunological parameters between HPV+ and HPV- HNSCC and have commonly concluded that HPV+ HNSCC are immune “hot” tumors, with markedly more immune infiltration and higher levels of CD8+ T-cell activation than HPV- HNSCC.Citation12,Citation13 These and other studies suggest that a detailed comparison of the immunological differences between HPV+ and HPV- HNSCC provides an opportunity to identify immunological determinants that contribute to successful treatment in HNSCC that may be broadly applicable to cancer treatment in general.Citation14–Citation16
In this study, we used RNA-sequencing data from over 500 HNSCC samples from The Cancer Genome Atlas (TCGA) to compare the immune landscape between HPV+, HPV-, and normal adjacent control tissue. Importantly, these samples were treatment-naïve prior to surgical resection, avoiding any confounding effects of exposure to chemotherapy or radiation on the immune status of these tumors. Such analysis can provide scientific rationale for treating HNSCC patients with immune checkpoint inhibitors (ICIs) as first-line therapy.
We determined that HPV+ HNSCC tumors exhibit a strong Th1 response, characterized by increased infiltration with dendritic cells (DCs), CD4+ and CD8+ T-cells. HPV+ HNSCC also expressed higher levels of CD39 and multiple T-cell exhaustion markers including LAG3, PD1, TIGIT, and TIM3 compared to HPV- HNSCC. This gene expression profile is consistent with a “T-cell-inflamed” phenotype, one that is dominated by T-cell markers and chemokines associated with effector T-cell recruitment.Citation17 Importantly, higher expression of these T-cell exhaustion genes correlated with markedly improved patient survival in HPV+, but not HPV-, HNSCC. These results illustrate the profound differences between the immune landscape of HPV+ and HPV- HNSCC tumors and its association with patient outcome. Furthermore, the presence of high expression levels of multiple immune inhibitory genes in HPV+ HNSCC suggests that these cancers will exhibit strong beneficial responses to immunotherapy, providing a strong rationale for using ICIs as single treatment or combination therapies in first-line treatment of HPV+ HNSCC. This would save patients from disfiguring surgery, or the toxicities associated with conventional chemotherapy or radiation treatment.
Results
HPV+ and HPV- head & neck carcinomas exhibit differences in some leukocyte populations
To identify and understand differences in the immune landscape between HPV+ and HPV- HNSCC, we conducted a preliminary analysis using the CIBERSORT deconvolution algorithm in order to elucidate the immune cell populations present in each sample.Citation18 This computational method is based on the differential expression of 547 genes to estimate the relative fraction of 22 leukocyte populations in each of the samples for the TCGA HNSCC cohort (). Our comparison of the HPV+ and HPV- samples revealed a number of differences, including an increased percentage of CD8+ T-cells and CD4+ T-regulatory cells (Tregs) in HPV+ samples compared to HPV- samples. HPV+ samples were also estimated to contain a greatly decreased level of uncommitted (M0) and anti-inflammatory (M2) macrophages as compared to HPV- samples. Increased levels of follicular helper CD4+ T-cells, but not other types of CD4+ T-cells, were present in HPV+ versus HPV- HNSCC samples, as well increased levels of naïve and memory B-cells, but not plasma cells. These data indicate a difference between the immune landscape of HPV+ and HPV- HNSCC tumors that requires further investigation.
Figure 1. The relative population of 22 leukocyte types present in the tumor microenvironment was determined by the CIBERSORT algorithm and compared between HPV+, HPV-, and normal control samples. Four leukocyte types (neutrophils, eosinophils, naïve CD4 + T-cells, and gamma delta T-cells) are not shown, as they were undetectable in this cohort. * p ≤ 0.05; ** p ≤ 0.01; *** p ≤ 0.001; **** p ≤ 0.0001, ns (not significant).
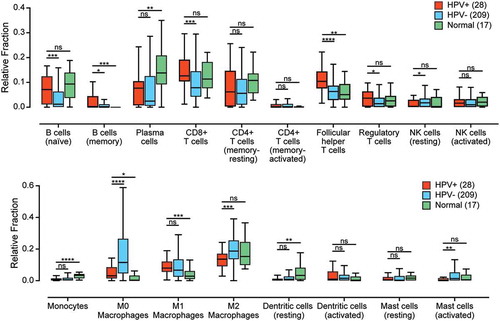
Given the observed differences in the various leukocyte populations, we also compared the total leukocyte fraction between the HPV+ and HPV- samples in the TCGA HNSCC cohort using a previously defined DNA methylation signature that is highly specific for leukocytes.Citation19 However, no significant differences were detected between the overall leukocyte fraction of HPV+ versus HPV- HNSCC (Supplementary Figure 1).
HPV+ head & neck carcinomas exhibited a Th1 CD4+ helper T-cell phenotype
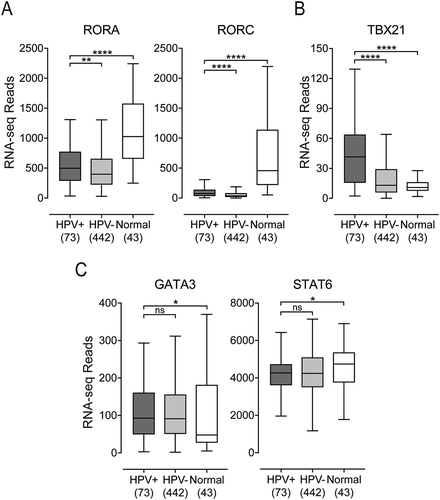
CIBERSORT identified a number of differences between HPV+ and HPV- HNSCC samples for multiple classes of T-cells, however, cancer-induced dysregulations and immune cell phenotypic plasticity can limit the ability of CIBERSORT to accurately characterize immune components of the tumor microenvironment.Citation18 Therefore, we began a detailed and mechanistically oriented comparison of the immune responses in HPV+ versus HPV- HNSCC. CD4+ helper T-cells are instrumental in establishing the type of response triggered by immune activation, whether it be a cell-mediated Th1 response, a humorally-biased Th2 response, or the proinflammatory Th17 response necessary to maintain the immunological integrity of mucosal barriers.Citation20 Each of the CD4+ subsets is characterized by a distinct transcriptional program regulated by specific master regulatory transcription factors.Citation21 To examine the tumor immune landscape of HPV+ and HPV- HNSCC, we analyzed the TCGA Illumina HiSeq RNA expression data from the HNSCC cohort for expression of TBX21 (Th1), GATA3 and STAT6 (Th2), and RORA and RORC (Th17) (). As expected, a strong Th17 signature was observed in normal control samples, and that signature expression profile was markedly reduced in HPV+ and HPV- carcinomas (). HPV+ samples had significantly increased levels of TBX21 as compared to HPV- and normal control samples, indicative of a polarization toward a Th1 response (). A modest increase in GATA3 accompanied by a reduction in STAT6 was observed in HPV+ samples versus normal control samples (). Taken together, these results indicate that a strong shift in the normal Th17 bias towards Th1 occurred in HNSCC, with a significantly accentuated Th1 signature in HPV+ versus HPV- carcinomas.
Figure 2. Expression of marker genes indicating that CD4+ helper T-cell subsets present in head & neck carcinomas stratified by human papillomavirus (HPV) status. Normalized RNA-seq data for TBX21 (Th1), GATA3 and STAT6 (Th2), and RORA and RORC (Th17) was extracted from The Cancer Genome Atlas (TCGA) database for the head & neck cancer (HNSC) cohort for HPV+, HPV−, and normal control tissues. Numbers in brackets refer to the number of samples included in each analysis. * p ≤ 0.05; ** p ≤ 0.01; *** p ≤ 0.001; **** p ≤ 0.0001, ns (not significant).
Higher levels of CD4+ and CD8+ T-cells were present in HPV+ head & neck carcinomas
We next assessed the relative proportion of CD4+ and CD8+ T-cells in these samples, based on relative expression of the lineage defining CD4 and CD8A/CD8B marker genes, respectively ( and ). HPV+ samples showed significantly increased expression of all three genes versus HPV- HNSCC or normal control samples, confirming that enhanced T-cell infiltration is a common feature of HPV+ HNSCC. We also observed significantly higher FoxP3 expression in HPV+ HNSCC compared to HPV- HNSCC and normal samples. FoxP3 is associated with Tregs infiltrating into these tumors (). Furthermore, analysis of the expression of CD137 (4-1BB), an activation-induced costimulatory molecule present primarily on CD8+ T-cells, detected significantly higher levels in HPV+ versus HPV- or normal control samples, suggesting a higher overall level of T-cell activation and effector function in HPV+ disease ().
Figure 3. Detection of tumor infiltrating T-cells and their activation status in head & neck carcinomas stratified by HPV status. Normalized RNA-seq data for genes indicative of tumor infiltrating CD8+ (CD8A and CD8B) or CD4+ (CD4) T-cells and their activation status (CD137, INFG, and TNF) was extracted from the TCGA database for the HNSC cohort for HPV+, HPV-, and normal control tissues. Numbers in brackets refer to the number of samples included in each analysis. * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001, **** p ≤ 0.0001, ns (not significant).
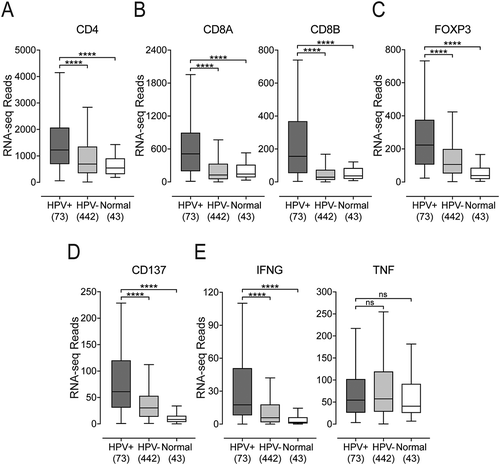
Activated cytotoxic CD8+ T-cells produce various effector molecules, including IFN-γ and TNF-α. Although expression of IFN-γ mRNA in these samples was low, it was detectable at significantly higher levels in HPV+ samples versus HPV- HNSCC or normal control tissues. No significant difference was observed for TNF (). HPV+ HNSCC tumors also expressed significantly higher levels of cytotoxic mediators, including granzyme A (GRZA), granzyme B (GRZB), and perforin (PRF1) compared to both HPV- HNSCC or normal control tissues (Supplementary Figure 2A). Taken together, these results indicate CD8+ T-cells are not only present at higher levels in HPV+ HNSCC, but are concomitantly more active and producing effector molecules such as IFN-γ, granzyme A, granzyme B, and perforin, but not TNF. We also observed low–but significantly higher than in HPV- HNSCC or normal control tissues–expression levels of genes associated with natural killer (NK) cells (SH2D1A, XCL2, NCR1, KIR2DL3, KIR3DL1, and KIR3DL2) and B-cells (IL21R and CD19) in HPV+ tumors indicating higher infiltration of these immune cells into HPV+ HNSCC (Supplementary Figures 2B and C, respectively). While many of these observations agree with the CIBERSORT estimations (), there are some differences. In some cases, this may be related to the use of different marker genes, or to the fact that CIBERSORT divides some classes of leukocytes into multiple subcategories based on a gene signature that can be affected by disease dysregulation.
Interferon-gamma-induced immunomodulatory genes are expressed at higher levels in HPV+ head & neck carcinomas
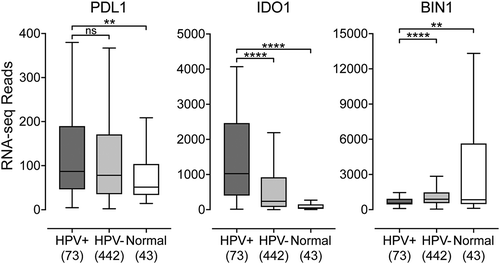
An expected consequence of higher levels of infiltrating CD8+ T-cells producing IFN-γ would be induction of expression of interferon response genes in tumor cells in close proximity to the infiltrating T-cells. Indeed, we previously reported that multiple interferon responsive components of the MHC-I antigen presentation apparatus were upregulated in HPV+ HNSCC.Citation22 We extended those studies to PDL1 and IDO, two immunomodulatory genes known to be expressed by tumor cells in response to IFN-γ (). Both PDL1 and IDO1 were significantly upregulated in HPV+ HNSCC compared to normal control. While IDO was also significantly upregulated compared to HPV- HNSCC, no significant difference was observed between PDL1 expression in HPV+ versus HPV- carcinomas. Interestingly, BIN1 expression was significantly downregulated in HPV+ compared to both HPV- HNSCC and normal control samples (). As BIN1 is a negative regulator of IDO expression and deletion of BIN1 increases IFN-γ stimulation of IDO1 gene expression, these results are consistent with the higher levels of IDO observed in HPV+ HNSCC.Citation23 Taken together, these data ( and ) indicate that HPV+ HNSCC, in treatment-naïve patients, shows a T-cell-inflamed phenotype.Citation17
Figure 4. Transcript levels of tumor-derived interferon-responsive immunomodulatory genes in head & neck carcinomas stratified by HPV status. Normalized RNA-seq data for genes associated with tumor cell mediated immunomodulation including IDO1, its negative regulator BIN1, and PDL1 was extracted from the TCGA database for the HNSC cohort for HPV+, HPV-, and normal control tissues. Numbers in brackets refer to the number of samples included in each analysis. * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001, **** p ≤ 0.0001, ns (not significant).
Dendritic cell signatures are elevated in HPV+ head & neck carcinomas
Because HPV+ HNSCC displayed a T-cell-inflamed phenotype and DCs play a key role as professional antigen-presenting cells to stimulate T-cell function, we performed an analysis of DC signatures in these tumors. Using the expression of mRNA encoding CD103 and CD11C to detect DCs in tumor samples, we found that both genes are expressed at higher levels in HPV+ HNSCC compared to normal control samples (). While CD103 was expressed at significantly higher levels in HPV+ versus HPV- carcinomas, CD11C was not. We also determined that there was a modest but significant correlation between CD103 and CD11C in individual tumors (; r = 0.56, p = 2.6e-07). This suggested that these markers tended to be coordinately expressed, although the link was not strong enough to lead to increased CD11C expression in HPV+ carcinomas in accord with increased CD103.
Figure 5. Detection of dendritic cells and their effector cytokines in head & neck carcinomas, stratified by HPV status. Normalized RNA-seq data for genes indicative of tumor infiltrating dendritic cells (CD103 and CD11C) and genes encoding cytokines indicative of dendritic cell activation status (IL12A, STAT4, IL23A, and EBI3) was extracted from the TCGA database for the HNSC cohort for HPV+, HPV-, and normal control tissues. Numbers in brackets refer to the number of samples included in each analysis. * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001, **** p ≤ 0.0001, ns (not significant).
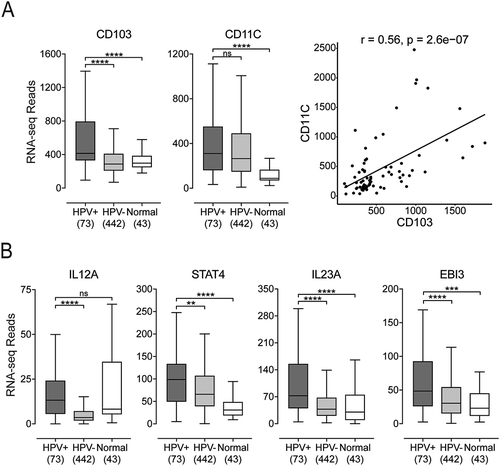
A key anti-tumor role for DCs is to express effector cytokines such as IL12 and IL23 that mediate CD8+ T-cell activation and IFN-γ production. While IL12A mRNA levels in HPV+ samples were not significantly induced versus normal control tissue, they were elevated compared to HPV- HNSCC (). IL12 activates STAT4 signaling, which is critical for anti-tumor immunity.Citation24 Notably, STAT4 expression levels were higher in HPV+ HNSCC tumors (). Activated DCs also produce IL23–a proinflammatory cytokine–that induces IFN-γ secretion from activated CD4+ T-cells.Citation25 IL23A levels were higher in HPV+ HNSCC than HPV- tumors and normal samples (). Moreover, Epstein-Barr virus-induced gene 3 (EBI3) transcripts that encode a component of IL27 (a cytokine that synergizes with IL12 and induces IFN-γ production by naïve CD4+ T-cells)Citation26 were significantly higher in HPV+ than HPV- tumors and normal samples (). We were not able to analyze the other two genes within the IL12 family of cytokines–IL12B and IL27A–because their RNA transcripts were not detectable in the majority of tumor and normal control samples. Finally, we looked at a number of inflammatory chemokines and chemokine receptors that are crucial for DC and T-cell recruitment to tumors.Citation27–Citation30 CCR5, CCL4, and CXCR3 were highly expressed in HPV+ HNSCC compared to HPV- tumors and normal samples, while CCL3 expression was elevated in HPV+ HNSCC compared to normal samples (Supplementary Figure 3). Overall, these results indicate a higher level of DC involvement in HPV+ than in HPV- HNSCC, with higher levels of effector cytokines that could be responsible for the elevated levels of T-cell infiltration of these samples.
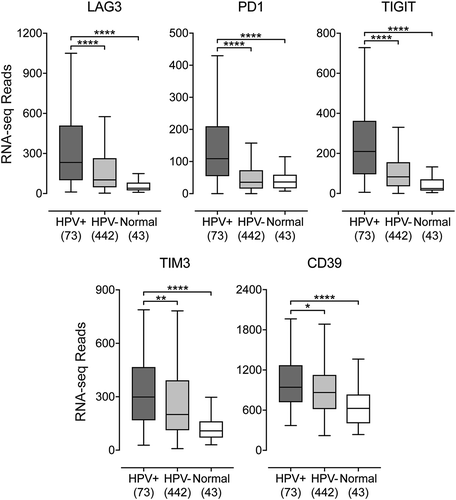
HPV+ head & neck carcinomas expressed high levels of multiple T-cell exhaustion markers
Once activated, T-cells upregulate expression of multiple cell surface receptors that negatively regulate their proliferation and moderate their level of activation. These so-called “exhaustion markers” include LAG3, PD1, TIGIT, and TIM3.Citation31 They are important targets for immune checkpoint blockade therapies that are approved or under development. Notably, all four of these checkpoint genes were significantly upregulated in HPV+ compared with HPV- HNSCC or normal control samples (). We also determined that individual tumors expressing high levels of any one of these T-cell inhibitory genes generally co-expressed high levels of the other exhaustion markers (r values ranging from 0.78 to 0.9), suggesting that these genes are coordinately expressed (Supplementary Figure 4). We also examined the expression of the immunosuppressive molecule CD39Citation32 in this cohort of patients. CD39 was recently shown to be exclusively expressed on tumor antigen-specific CD8+ T-cells and not bystander CD8+ T-cells in colorectal and lung tumors.Citation33 Notably, CD39 showed significantly higher RNA expression levels in HPV+ HNSCC tumors compared to HPV- HNSCC samples (). Interestingly, individual tumors expressing high levels of CD39 also co-expressed high levels of the four exhaustion markers (r values ranging from 0.37 to 0.62), suggesting that these genes are coordinately expressed (Supplementary Figure 5). Taken together, HPV+ HNSCC display a markedly different immune signature from HPV- HNSCC, indicative of sustained CD8+ T-cell activation in HPV+ HNSCC.
Figure 6. Transcript levels of T-cell exhaustion marker genes and CD39 in head & neck carcinomas, stratified by HPV status. Normalized RNA-seq data for genes associated with T-cell exhaustion (LAG3, PD1, TIGIT, TIM3, and CD39) was extracted from the TCGA database for the HNSC cohort for HPV+, HPV-, and normal control tissues. Numbers in brackets refer to the number of samples included in each analysis. * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001, **** p ≤ 0.0001, ns (not significant).
High levels of T-cell exhaustion markers correlated with improved survival of HPV+ head & neck carcinomas
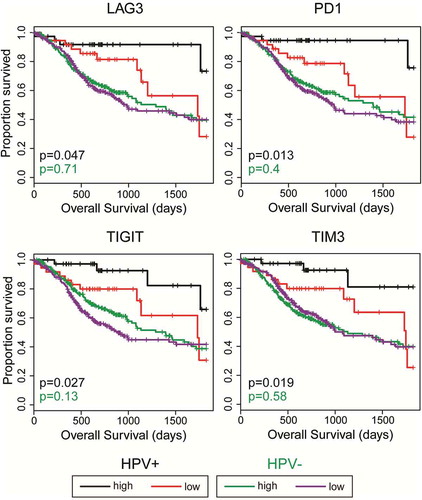
High CD8+ T-cell infiltration into HNSCC tumors has been previously linked to better patient outcome.Citation34 Moreover, expression of checkpoint molecules such as PD1 on circulating CD8+ T-cells is a biomarker of tumor antigen specificity.Citation35 We therefore dichotomized the HPV+ HNSCC data set based on median LAG3, PD1, TIGIT, or TIM3 expression and calculated the impact of high versus low expression on overall patient survival (, black and red curves). Higher than median expression of any of these genes predicted markedly improved survival in HPV+ HNSCC over those patients with tumors expressing these markers at levels below the median. This sharp overall increase in clinical outcome was also statistically significant following correction for multiple testing in all the assessed genes, with the exception of LAG3 (Supplementary Table 1). In contrast, a similar analysis did not reveal any survival advantage for HPV- samples expressing higher levels of LAG3, PD1, TIGIT, or TIM3 (, green and purple curves).
Figure 7. High LAG3, PD1, TIGIT, or TIM3 gene transcript levels are strongly associated with improved survival in treatment naïve patients with HPV+ but not HPV- head & neck carcinomas. Overall survival of patients within the HPV-positive cohort dichotomized by median expression of LAG3, PD1, TIGIT, and TIM3. Comparison between groups was made by the 2-sided log-rank test. Red = low expression of the indicated gene in HPV+ samples, Black = high expression of the indicated gene in HPV+ samples, Purple = low expression of the indicated gene in HPV- samples, Green = high expression of the indicated gene in HPV- samples.
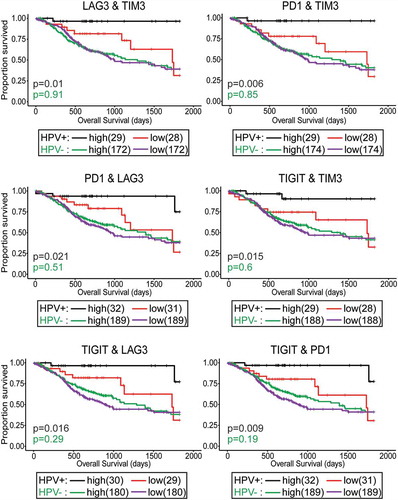
Given our observation that HPV+ tumors expressing high levels of LAG3, PD1, TIGIT, or TIM3 transcripts generally displayed higher levels of at least one more of these exhaustion markers (Supplementary Figure 4), we repeated the survival analysis by comparing HPV+ HNSCC samples that were above the median expression for each combination of two markers to those below the median for both markers (, black and red curves). Remarkably, overall survival for patients with high expression of any combination of two of the T-cell exhaustion markers identified individuals with a virtual certainty of long-term survival. No improvement was seen when a similar analysis was performed using the HPV- HNSCC cohort (, green and purple curves). These results suggest that the co-expression of high levels of LAG3, PD1, TIGIT, and TIM3 in HPV+, but not HPV-, HNSCC reflect the successful development of T-cell-based anti-tumor immunity and a T-cell-inflamed phenotype, which contributed to long-term remission. This immune signature could be used as a predictive biomarker of HPV+ HNSCC patient outcome after surgery.
Figure 8. Concordant levels of multiple markers of T-cell exhaustion is strongly associated with survival in patients with HPV+ head & neck carcinomas. Overall survival of patients within the HPV-positive cohort dichotomized by median transcript levels of the indicated pairs of T-cell exhaustion markers. Comparison between groups was made by the 2-sided log-rank test. Red = low expression of the indicated gene in HPV+ samples, Black = high expression of the indicated gene in HPV+ samples, Purple = low expression of the indicated gene in HPV- samples, Green = high expression of the indicated gene in HPV- samples.
High levels of CD39 predict improved survival of HPV+HNSCC
We also dichotomized the HPV+ and HPV- HNSCC data set based on median CD39 expression and calculated the impact of high versus low expression on overall patient survival (). Higher than median expression of CD39 predicted markedly improved survival in HPV+ HNSCC over those patients with tumors expressing this marker at levels below the median. This sharp overall increase in clinical outcome was also statistically significant. Similar to our previous analysis (), a combination of CD39 and any of the T-cell exhaustion markers identified long-term survivors in HPV+ HNSCC.
Figure 9. High CD39 gene transcript level is strongly associated with improved survival in treatment naïve patients with HPV+head & neck carcinomas. Overall survival of patients within the HPV+ and HPV- cohorts were dichotomized by median expression of CD39. Concordant levels of CD39 and multiple markers of T-cell exhaustion was strongly associated with survival in patients with HPV+ head & neck carcinomas. Overall survival of patients within the HPV-positive cohort dichotomized by median transcript levels of the indicated pairs of T-cell exhaustion markers. Comparison between groups was made by the 2-sided log-rank test. Red = low expression of the indicated gene in HPV+ samples, Black = high expression of the indicated gene in HPV+ samples, Purple = low expression of the indicated gene in HPV- samples, Green = high expression of the indicated gene in HPV- samples.
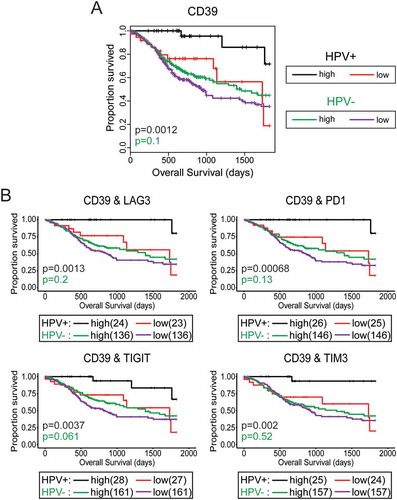
We next applied the Cox Proportional Hazards model to identify clinical variables and immunological markers predictive of overall survival (Supplementary Table 2). By univariate analysis high expression of TIGIT, TIM3, PD1, and CD39 were correlated with favorable prognosis (HR = 0.29, 0.24, 0.22 and 0.15 respectively, p < 0.05), while an HPV type other than HPV16 was associated with a markedly poorer survival (HR = 3.22, p = 0.03). Oral cavity tumors had significantly worse survival compared those in the oropharyngeal subsite (HR = 3.05, p = 0.03). There was a trend towards improved outcome with high expression of LAG3 (HR = 0.32, p = 0.06). Stepwise regression analysis was used to identify the best multivariate model of survival, and the final model included N stage, HPV type, and CD39. Notably, CD39 (HR = 0.07, p = 0.0007), and HPV type (HR = 9.23, p = 0.001 remained as significant predictors of survival in the multivariate model.
Discussion
Although evading anticancer immunity is one of the hallmarks of cancer, immunotherapy–also referred to as immuno-oncology–harnesses the power of the immune system to destroy cancer cells. Durable responses to immunotherapy among cancer patients are what sets immunotherapy apart from conventional oncology therapies, including chemotherapy, radiotherapy, and targeted therapy. Cancer immunotherapy is now widely recognized as having the potential to revolutionize cancer treatment. Despite the recent “tsunami of immunotherapy in oncology” and many reports of immunotherapy treatment resulting in near-miraculous cures in cases of highly refractory cancers, in general only a subset of patients respond to these treatments. The underlying reasons for response, or lack thereof, remain an intense area of investigation.Citation36
Based on their somatic mutation prevalence,Citation37 tumors can be categorized as immunologically “cold” or immunologically “hot”. Immunologically hot tumors such as melanoma and smoking-induced lung cancer, often respond more favorably to immune checkpoint inhibition therapy than cold tumors. Therefore, a partial explanation for immune checkpoint inhibitor response appears to be related to high mutation burden, which leads to higher levels of tumor-derived neoantigens. Indeed, a significant correlation exists between immune checkpoint inhibitor response and mutational load across cancer sites.Citation38
A variety of studies have compared various immunological parameters between HPV+ and HPV- HNSCC and generally concluded that HPV+ HNSCC are immune “hot” tumors, with markedly more immune infiltration and higher levels of CD8+ T-cell activation than HPV- HNSCC.Citation12,Citation13,Citation39,Citation40 Although this is thought to contribute to the improved prognosis of HPV+ HNSCC, the mutational loads in HPV+ and HPV- HNSCC are similar therefore the immune hot and cold model cannot explain the survival differences observed among HNSCC patients. However, HPV+, but not HPV-, HNSCC express exogenous antigenic viral oncoproteins that may represent a key difference in the tumor immune landscape between these two types of HNSCC, giving HPV+ HNSCC a T-cell-inflamed phenotype that can at least partially explain the better patient outcome in HPV+ HNSCC compared to HPV- HNSCC. Furthermore, HPV+ HNSCC tumors are different from HPV- HNSCC tumors at the epigenetic and genomic levels, but viral etiology alone may not be sufficient to confer favorable outcome in HNSCC.Citation41 Therefore, the substantial differences observed in anti-tumor immunity between HPV+ and HPV- cancers could be the major source of improved prognosis in HPV+ tumors.
In this study, for the first time, we report the detailed immune characteristics of T-cell-inflamed HPV+ HNSCC compared to non-T-cell-inflamed HPV- HNSCC. We performed a mechanistic analysis of the tumor immune landscape in treatment-naïve HNSCC using the TCGA cohort, which allows a detailed and direct comparison between HPV+ and HPV- HNSCC and normal adjacent control tissue. However, examination of only the RNA expression levels limits the ability to accurately confirm expression levels on each cell type. To address this limitation, we primarily used immune lineage-specific markers or examined co-expression correlations for markers that are expressed by more than one cell type. We found that HPV+ HNSCC has a predominant Th1 immune phenotype () with a significant increase in CD8+ and CD4+ T-cell infiltration compared to HPV- HNSCC (). Interestingly, higher T-cell infiltration into HPV+ tumors was accompanied with greater T-cell activation, as shown by high CD137 (4-1BB) gene transcript levels.
Mononuclear cells isolated from patients with advanced head and neck cancer have been reported to be defective in IFN-γ production in vitro.Citation42 Given the high CD137 RNA levels in HPV+ HNSCC samples, we expected higher effector cytokine production by T-cells in these samples. Indeed, HPV+ HNSCC expressed significantly higher levels of IFN-γ, granzyme A, granzyme B, and perforin compared to HPV- HNSCC or normal control samples, indicating that there remains some T-cell effector functionality in these tumors. Moreover, the higher levels of CD137 and multiple effector molecules in HPV+ HNSCC could be a consequence of higher infiltration of these tumors with antigen-specific tumor-infiltrating lymphocytes (TILs). In contrast, we did not observe differences in TNF transcript levels–a critical cytokine for antitumor immunityCitation43–between HPV+ and HPV- HNSCC, indicating T-cell dysfunction in these samples. Given the role of IFN-γ in inducing MHC-I expression in tumor cells, these findings explain our previous observation that HPV+ HNSCC tumors had significantly higher levels of expression of MHC-I apparatus.Citation22 Furthermore, high IFN-γ transcript levels in HPV+ HNSCC were associated with significantly higher levels of IFN-responsive gene transcripts, including IDO and PD-L1 (). These observations provide a scientific rationale for combining immunotherapy of HPV+ HNSCC with ICIs and IDO inhibitors in treatment-naïve patients. Currently the only phase II study of such a combination (NCT03325465) has been canceled in light of the recent failure of combined IDO inhibition and pembrolizumab in showing advantage in progression-free survival over pembrolizumab monotherapy in melanoma patients. However, that study (NCT03325465) was not focused specifically on HPV+ HNSCC, which we show here express higher levels of IDO and PD-1 compared to HPV- HNSCC.
The observed increased numbers of CD8+ and CD4+ T-cells in HPV+ HNSCC can likely be explained by the ability of DCs to migrate to these tumors, pick up antigens, and present those antigens to T-cells.Citation44 Indeed, we observed not only high levels of transcripts from DC-associated genes, but also molecules expressed by activated DCs, including chemokines that attract DCs and T-cells into tumors ( and Supplementary Figure 3). We also observed less M2 macrophages in HPV+ HNSCC samples () indicative of less immunosuppressive activity in these tumors compared to HPV- HNSCC. These observations provide a mechanistic insight into the processes that shape the immune structure of HPV+ HNSCC as a T-cell-inflamed tumor.
T-cell-inflamed tumors show high levels of immune regulatory genes including IDO and PD-L1, and high infiltration of FoxP3+ Tregs,Citation45 secondary to cytotoxic T-cell infiltration into those tumors. The presence of these immunosuppressive events in a T-cell-inflamed microenvironment is often accompanied by increased levels of multiple T-cell immune checkpoint molecules and T-cell anergy.Citation45 Our findings show that HPV+ HNSCC, but not HPV- HNSCC, have similarly high levels of multiple immune checkpoint molecules (LAG3, TIM3, PD1, and TIGIT; ), suggesting that combined immunotherapy with drugs that target more than one of these inhibitory molecules can be effective in overcoming resistance to anti-tumor immunity in HPV+ HNSCC tumors. There are no such trials currently under way, but these findings strongly indicate that initiation of such clinical studies is justified. CD39 is an immunosuppressive molecule that can be expressed on intratumoral TregsCitation32 and tumor-specific CD8+ TILs, but not bystander CD8+ T-cells.Citation33 CD39 RNA expression was higher in both HPV+ and HPV- HNSCC tumors compared to normal samples (). Of note, CD39 expression in HPV+ HNSCC was significantly higher than HPV- tumors. CD39 alone or in combination with any of the T-cell exhaustion markers analyzed above was also a predictive biomarker of survival in HPV+ HNSCC (). Therefore, CD39 may represent a useful biomarker of survival in HPV+ HNSCC. Our findings warrant the initiation of prospective trials in determining the predictive value of this gene as a potential genomic biomarker correlated with improved overall survival in HNSCC.
High T-cell infiltration into HNSCC, regardless of HPV status, has been reported as a positive prognostic factor.Citation46 We observed that high levels of transcripts from one or any combination of two genes expressing immune checkpoint molecules (PD1, TIM3, TIGIT, LAG3) in HPV+, but not HPV-, HNSCC can predict patient survival after surgery. These findings require confirmation by prospective clinical studies to–we anticipate–pave the way for the development of immune-predictive biomarkers for HPV+ HNSCC patients that will ultimately lead to treatment deintensification in this patient population.
Taken together, this study provides clear evidence, for the first time, that the immune landscape of HPV+ HNSCC represents a T-cell-inflamed phenotype that is very different from HPV- HNSCC. Of prime importance, we demonstrate that multiple mechanisms that negatively regulate the anti-tumor immune response are significantly upregulated in the majority of HPV+ HNSCC cases, that is secondary to CD8+ T-cell infiltration and IFN-γ production in HPV+ HNSCC. Therefore, high levels of such gene transcripts–PD1, TIM3, LAG3, and TIGIT–correlate strongly with improved clinical outcome. Crucially, many of these negative regulators of the immune response are targets of clinically-approved immune checkpoint inhibitors or inhibitors under investigation in current clinical trials. Thus, HPV+ HNSCC has many immunological features that suggest that it would be highly amenable to immune checkpoint inhibition therapy. Based on these data, we propose that HPV+ HNSCC is a T-cell-inflamed disease and should be treated differently in the clinic than HPV- HNSCC, specifically with combinations of immune checkpoint inhibitors.
Materials and methods
Data collection
Patient data from The Cancer Genome Atlas (TCGA), including Merged Clinical data (build 2016012800) and Level 3 RSEM normalized Illumina HiSeq RNA expression data (build 2016012800) for the HNSC cohort, was downloaded from the Broad Genome Data Analysis Centers Firehose server (https://gdac.broadinstitute.org/). Written informed consents were obtained from the patients (or their families) by the TCGA project. CIBERSORT immune fraction data (TCGA.Kallisto.fullIDs.cibersort.relative.tsv) and Leukocyte Fraction (TCGA_all_leuk_estimate.masked.20170107.tsv) data from the Pan-Cancer Atlas was downloaded from the National Cancer Institute Cancer Genome Commons (https://gdc.cancer.gov/about-data/publications/panimmune).
RNA expression comparisons
RSEM normalized expression data was extracted into Microsoft Excel and the HPV status was manually curated based on published datasets as described.Citation47 Primary patient samples with known HPV status were grouped as HPV+, HPV-, or normal control tissue. This classification agrees completely with work done by othersCitation41, with the exception of sample TCGA-BB-7862‐01A. That sample had minimal reads aligning to the HPV genome, none of which aligned to the HPV16 E6 or E7 oncogenes, and we classified this sample as HPV- rather than HPV+ . Patient samples with unknown HPV status were omitted from our calculations, as were samples obtained from secondary metastatic lesions. This resulted in 73 HPV+, 442 HPV-, and 43 normal control samples with data available for the HNSCC gene expression analysis. This surgically managed cohort was considered treatment-naïve, as only 9 patients received neoadjuvant radiation or chemotherapy treatment (2 HPV+, 7 HPV-). Boxplot comparison of gene expression was performed using GraphPad Prism v7.0 (Graphpad Software, Inc., San Diego, California, USA) and assembled into final form using CorelDRAW (Corel, Ottawa, Ontario, Canada). For the boxplots, center lines show the medians, box limits indicate the 25th and 75th percentiles as determined by Graphpad Prism, and whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles. Statistical significance was calculated using Graphpad Prism. P-values were assigned using a one-tailed non-parametric Mann-Whitney U test. Selected genes were compared in a pairwise fashion and concordance calculated using Spearman’s Rho analysis. Differences were considered to be statistically significant for P < 0.05.
Immune fraction estimates
The relative fractions of 22 leukocyte types as calculated using CIBERSORTCitation18 was previously reported for all TCGA samples as part of the Pan-Cancer Atlas.Citation48 Only those HNSCC samples with a CIBERSORT p < 0.01 were included in our analysis, which resulted in 28 HPV+, 209 HPV-, and 17 normal control samples used in this analysis. The total leukocyte fraction was also previously reported for all TCGA samples as part of the Pan-Cancer Atlas. The HNSCC values were extracted from this data and annotated for HPV status as described above.
Survival analysis
Five-year overall survival outcomes and disease-free progression were compared in both HPV+ and HPV- subsets of patients as dichotomized by median expression of LAG3, PD1, TIGIT, TIM3, or CD39. A second set of survival outcomes were done to compare tumors expressing high levels of each combination of two of these markers. Log-rank statistical p-values were calculated for each Cox survival model. Univariate survival analysis was done through R statistical environment (version 3.4.0) based on a Cox Proportional Hazard Model using survival package (version 2.41–3).Citation49 Stepwise bidirectional multivariate analysis was then carried out with clinical variables (gender, age, smoking history, subsite, T stage, N stage, Overall stage, and HPV type), LAG3, PD1, TIGIT, TIM3, and CD39 expression. The p-values derived from the Wald test on survival coefficients were reported for investigated variables. The derived log-rank p-values for all tested genes (listed in Supplementary Table 1) were assessed for significance after correcting for false discovery rate (FDR) using the Benjamini–Hochberg method, and an FDR threshold of 0.1 was set for significance.
Disclosure of potential conflicts of interest
Authors declare no conflicts of interest.
Authors’ contributions
Conception and design: SF Gameiro, JS Mymryk, and S Maleki Vareki
Development of methodology: SF Gameiro, F Ghasemi, JS Mymryk
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): SF Gameiro, F Ghasemi, JS Mymryk, and S Maleki Vareki
Writing, review, and/or revision of the manuscript: SF Gameiro, F Ghasemi, JW Barrett, J Koropatnick, AC Nichols, JS Mymryk, and S Maleki Vareki
Study supervision: JS Mymryk and S Maleki Vareki
Supplemental Material
Download Zip (248.9 KB)Acknowledgments
We thank Dr. Scott Bratman for providing information regarding HPV status of the TCGA HNSCC samples.
Supplementary material
Supplemental data for this article can be access on the publisher’s website.
Additional information
Funding
References
- Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–264.
- Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27:450–461.
- Del Paggio JC. Immunotherapy: cancer immunotherapy and the value of cure. Nat Rev Clin Oncol. 2018;10:253.
- Ferlay J, Shin H-R, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917.
- Baxi S, Fury M, Ganly I, Rao S, Pfister DG. Ten years of progress in head and neck cancers. J Natl Compr Canc Netw. 2012;10:806–810.
- Marur S, D’Souza G, Westra WH, Forastiere AA. HPV-associated head and neck cancer: a virus-related cancer epidemic. Lancet Oncol. 2010;11:781–789.
- Nichols AC, Palma DA, Dhaliwal SS, Tan S, Theuer J, Chow W, Rajakumar C, Um S, Mundi N, Berk S, et al. The epidemic of human papillomavirus and oropharyngeal cancer in a Canadian population. Current Oncology. 2013;20:212–219.
- The Cancer Genome Atlas Research Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582.
- Mittal S, Banks L. Molecular mechanisms underlying human papillomavirus E6 and E7 oncoprotein-induced cell transformation. Mutat Res Rev Mutat. 2017;772:23–35.
- Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tân PF, Westra WH, Chung CH, Jordan RC, Lu C, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363:24–35.
- Fakhry C, Westra WH, Li S, Cmelak A, Ridge JA, Pinto H, Forastiere A, Gillison ML. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst. 2008;100:261–269.
- Solomon B, Young RJ, Bressel M, Urban D, Hendry S, Thai A, Angel C, Haddad A, Kowanetz M, Fua T, et al. Prognostic Significance of PD-L1+and CD8+immune cells in HPV+oropharyngeal squamous cell carcinoma. Cancer Immunol Res. 2018;6:295–304.
- Mandal R, Şenbabaoğlu Y, Desrichard A, Havel JJ, Dalin MG, Riaz N, Lee K-W, Ganly I, Hakimi AA, Chan TA, et al. The head and neck cancer immune landscape and its immunotherapeutic implications. JCI Insight. 2016;1:e89829.
- Montler R, Bell RB, Thalhofer C, Leidner R, Feng Z, Fox BA, Cheng AC, Bui TG, Tucker C, Hoen H, et al. OX40, PD-1 and CTLA-4 are selectively expressed on tumor-infiltrating T cells in head and neck cancer. Clin Transl Immunol. 2016;5:e70.
- Lyford-Pike S, Peng S, Young GD, Taube JM, Westra WH, Akpeng B, Bruno TC, Richmon JD, Wang H, Bishop JA, et al. Evidence for a role of the PD-1: PD-L1pathway in immune resistance of HPV-associated head and neck squamous cell carcinoma. Cancer Res. 2013;73:1733–1741.
- Feng Z, Bethmann D, Kappler M, Ballesteros-Merino C, Eckert A, Bell RB, Cheng A, Bui T, Leidner R, Urba WJ, et al. Multiparametric immune profiling in HPV- oral squamous cell cancer. JCI Insight. 2017;2(14). pii: 93652. doi:10.1172/jci.insight.93652.
- Gajewski TF, Schreiber H, Fu Y-X. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14:1014–1022.
- Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457.
- Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ, Drill E, Shen R, Taylor AM, Cherniack AD, Thorsson V, et al. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell. 2018;173:291–296.
- Mucida D, Salek-Ardakani S. Regulation of TH17 cells in the mucosal surfaces. J Allergy Clin Immunol. 2009;123:997–1003.
- Fang D, Zhu J. Dynamic balance between master transcription factors determines the fates and functions of CD4 T cell and innate lymphoid cell subsets. J Exp Med. 2017;214:1861–1876.
- Gameiro SF, Zhang A, Ghasemi F, Barrett JW, Nichols AC, Mymryk JS. Analysis of class I major histocompatibility complex gene transcription in human tumors caused by human papillomavirus infection. Viruses. 2017;9:252.
- Muller AJ, DuHadaway JB, Donover PS, Sutanto-Ward E, Prendergast GC. Inhibition of indoleamine 2,3-dioxygenase, an immunoregulatory target of the cancer suppression gene Bin1, potentiates cancer chemotherapy. Nat Med. 2005;11:312–319.
- Lund RJ, Chen Z, Scheinin J, Lahesmaa R. Early target genes of IL-12 and STAT4 signaling in th cells. J Immunol. 2004;172:6775–6782.
- Shi Q, Yin Z, Zhao B, Sun F, Yu H, Yin X, Zhang L, Wang S. PGE2 elevates IL-23 production in human dendritic cells via a cAMP dependent pathway. Mediators Inflamm. 2015;2015:984690–984697. doi:10.1155/2015/984690.
- Pflanz S, Timans JC, Cheung J, Rosales R, Kanzler H, Gilbert J, Hibbert L, Churakova T, Travis M, Vaisberg E, et al. IL-27, a heterodimeric cytokine composed of EBI3 and p28 protein, induces proliferation of naive CD4+ T cells. Immunity. 2002;16:779–790.
- Mitchell DA, Batich KA, Gunn MD, Huang M-N, Sanchez-Perez L, Nair SK, Congdon KL, Reap EA, Archer GE, Desjardins A, et al. Tetanus toxoid and CCL3 improve dendritic cell vaccines in mice and glioblastoma patients. Nature. 2015;519:366–369.
- Castellino F, Huang AY, Altan-Bonnet G, Stoll S, Scheinecker C, Germain RN. Chemokines enhance immunity by guiding naive CD8+ T cells to sites of CD4+ T cell-dendritic cell interaction. Nature. 2006;440:890–895.
- Chen F, Yin S, Niu L, Luo J, Wang B, Xu Z, Yang G. Expression of the chemokine receptor CXCR3 correlates with dendritic cell recruitment and prognosis in gastric cancer. Genet Test Mol Biomarkers. 2018;22:35–42.
- Mikucki ME, Skitzki JJ, Frelinger JG, Odunsi K, Gajewski TF, Luster AD, Evans SS. Unlocking tumor vascular barriers with CXCR3: implications for cancer immunotherapy. Oncoimmunology. 2016;5:e1116675.
- Anderson AC, Joller N, Kuchroo VK. Lag-3, Tim-3, and TIGIT: co-inhibitory receptors with specialized functions in immune regulation. Immunity. 2016;44:989–1004.
- Jie H-B, Gildener-Leapman N, Li J, Srivastava RM, Gibson SP, Whiteside TL, Ferris RL. Intratumoral regulatory T cells upregulate immunosuppressive molecules in head and neck cancer patients. Br J Cancer. 2013;109:2629–2635.
- Simoni Y, Becht E, Fehlings M, Loh CY, Koo S-L, Teng KWW, Yeong JPS, Nahar R, Zhang T, Kared H, et al. Bystander CD8+ T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature. 2018;557:575–579.
- de Ruiter EJ, Ooft ML, Devriese LA, Willems SM. The prognostic role of tumor infiltrating T-lymphocytes in squamous cell carcinoma of the head and neck: A systematic review and meta-analysis. Oncoimmunology. 2017;6:e1356148.
- Gros A, Parkhurst MR, Tran E, Pasetto A, Robbins PF, Ilyas S, Prickett TD, Gartner JJ, Crystal JS, Roberts IM, et al. Prospective identification of neoantigen-specific lymphocytes in the peripheral blood of melanoma patients. Nat Med. 2016;22:433–438.
- Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91–97.
- Sadelain M, Rivière I, Riddell S. Therapeutic T cell engineering. Nature. 2017;545:423–431.
- Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med. 2017;377:2500–2501.
- Chen X, Yan B, Lou H, Shen Z, Tong F, Zhai A, Wei L, Zhang F. Immunological network analysis in HPV associated head and neck squamous cancer and implications for disease prognosis. Mol Immunol. 2018;96:28–36.
- Keck MK, Zuo Z, Khattri A, Stricker TP, Brown CD, Imanguli M, Rieke D, Endhardt K, Fang P, Brägelmann J, et al. Integrative analysis of head and neck cancer identifies two biologically distinct HPV and three non-HPV subtypes. Clin Cancer Res. 2015;21:870–881.
- Chakravarthy A, Henderson S, Thirdborough SM, Ottensmeier CH, Su X, Lechner M, Feber A, Thomas GJ, Fenton TR. Human papillomavirus drives tumor development throughout the head and neck: improved prognosis is associated with an immune response largely restricted to the oropharynx. J Clin Oncol. 2016;34:4132–4141.
- Conti-Freitas LC, Foss-Freitas MC, Mamede RCM, Foss NT. Interferon-gamma and interleukin-10 production by mononuclear cells from patients with advanced head and neck cancer. Clinics (Sao Paulo). 2012;67:587–590.
- Calzascia T, Pellegrini M, Hall H, Sabbagh L, Ono N, Elford AR, Mak TW, Ohashi PS. TNF-alpha is critical for antitumor but not antiviral T cell immunity in mice. J Clin Invest. 2007;117:3833–3845.
- Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature. 2015;523:231–235.
- Gajewski TF, Corrales L, Williams J, Horton B, Sivan A, Spranger S. Cancer immunotherapy targets based on understanding the t cell-inflamed versus non-T cell-inflamed tumor microenvironment. Adv Exp Med Biol. 2017;1036:19–31.
- Nguyen N, Bellile E, Thomas D, McHugh J, Rozek L, Virani S, Peterson L, Carey TE, Walline H, Moyer J, et al. Tumor infiltrating lymphocytes and survival in patients with head and neck squamous cell carcinoma. Head Neck. 2016;38:1074–1084.
- Gameiro SF, Kolendowski B, Zhang A, Barrett JW, Nichols AC, Torchia J, Mymryk JS. Human papillomavirus dysregulates the cellular apparatus controlling the methylation status of H3K27 in different human cancers to consistently alter gene expression regardless of tissue of origin. Oncotarget. 2017;8(42):27564–725761.
- Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang T-H, Porta-Pardo E, Gao GF, Plaisier CL, Eddy JA, et al. The immune landscape of cancer. Immunity. 2018;48:812–814.
- Mayakonda A, Koeffler HP. Maftools: efficient analysis, visualization and summarization of MAF files from large-scale cohort based cancer studies. bioRxiv. 2016;052662. doi:10.1101/052662.
