ABSTRACT
Osteosarcoma metastasizes to the lung, and there is a link between the predominance of tumor-promoting immunosuppressive M2 macrophages in the metastases and poor patient survival. By contrast, M1 macrophage predominance correlates with longer survival. M2 macrophages can be induced by various stimuli in the tumor microenvironment, including exosomes, which are 40- to 150-nm vesicles that are involved in intercellular communication and contribute to tumor progression and immune evasion. Recognizing that tumor cells can influence the tumor microenvironment to make it more permissive and because of the link between M2 dominance and curtailed patient survival, we evaluated the effect of exosomes from non-metastatic K7 and Dunn osteosarcoma cells and the metastatic sublines K7M3 and DLM8 on macrophage phenotype and function. Incubating MHS mouse alveolar macrophages with K7M3 and DLM8 exosomes induced expression of IL10, TGFB2, and CCL22 mRNA (markers of M2 macrophages) and decreased phagocytosis, efferocytosis, and macrophage-mediated tumor cell killing. In contrast, exosomes from non-metastatic K7 or Dunn cells did not inhibit phagocytosis, efferocytosis, and macrophage-mediated cytotoxicity or induce increased expression of IL10, TGFB2 or CCL22 mRNA. In addition, metastatic osteosarcoma cell exosomes significantly increased the secretion of TGFB2, a key signaling pathway associated with tumor- mediated immune suppression. Finally, the inhibition of TGFB2 reversed the suppressive activity of alveolar macrophages exposed to metastatic osteosarcoma cell exosomes. Our data suggest that the exosomes from metastatic osteosarcoma cells can modulate cellular signaling of tumor-associated macrophages, thereby promoting the M2 phenotype and creating an immunosuppressive, tumor-promoting microenvironment through the production of TGFB2.
Introduction
Osteosarcoma is the most common primary malignancy of bone in children, adolescents, and young adults.Citation1 Metastasis occurs almost exclusively to the lung, and 5-year survival rates drop drastically, to less than 30%, once this occurs.Citation2,Citation3 The success of current chemotherapies has plateaued, highlighting an increasing clinical need to better understand this disease in order to identify new therapeutic targets and improve patient outcomes. Characterizing and understanding the biological properties that both permit and support osteosarcoma cells to grow and thrive in the lung microenvironment may allow for the development of new therapies for metastatic osteosarcoma. The tumor microenvironment is complex, comprising of surrounding stromal cells, innate and adaptive immune cells, cancer cells, and mesenchymal stem cells.Citation4 Recently, macrophages, an integral phagocyte of the innate immune system, have been shown to be important to patient survival in osteosarcoma and other cancers.Citation5,Citation6
Macrophages are professional phagocytes and are highly specialized to remove dead or dying cells and cellular debris. They have also been shown to be critical components of the tumor microenvironment.Citation6–Citation8 There are two classically accepted macrophage phenotypes: an antitumorigenic M1 phenotype and a tumor-promoting M2 phenotype. M1 macrophages have been shown to support the adaptive immune response, targeting dead or dying cells as well as infectious agents, and they are characterized by increased expression of inflammatory chemokines and cytokines such as interleukins IL1 and IL6 and tumor necrosis factor alpha (TNFA).Citation7 Additionally, M1 macrophages within osteosarcoma lung metastases have been shown to correlate with better patient outcome.Citation6,Citation9 M2 macrophages are associated with the development of an immunosuppressive microenvironment through the expression of IL10 and transforming growth factor beta 2 (TGFB2).Citation7 M2 macrophages have also been shown to promote cancer by enhancing tumor invasiveness, angiogenesis, and metastasis.Citation8 Growing evidence suggests that a high density of M2-like tumor-associated macrophages is associated with poor prognosis and poor patient survival in many cancers.Citation10 In both primary and metastatic osteosarcoma, macrophages located within the tumor microenvironment are typically polarized to the M2-like subtype, which further allows the tumor to grow and proliferate without check.Citation6 The M2 phenotype has been suggested to play a role in tumor progression and metastasis and have also been shown to be modulated by various stimuli in the tumor microenvironment, including exosomes.Citation11
Exosomes are small, 40- to 150-nm vesicles that have been shown to be an integral part of intercellular communication, resulting in increased tumor progression and immune evasion as well as prolonged tumor survival.Citation12–Citation14 Exosomes have also been shown to be released by a wide variety of cells, including cancer cells, both in vitro and in vivo.Citation13 Like cells, exosomes possess a lipid bilayer membrane that protects their contents, which include proteins, mRNAs, microRNAs, and genomic DNA, and have been shown to play a critical role in intercellular signaling.Citation13 Exosomes have also been shown to modulate macrophage phenotype in other cancers such as glioblastoma, gastric cancer, melanoma, colorectal cancer, and breast cancer.Citation15–Citation21 Exosomes from non-metastatic melanoma cells have been shown to stimulate an innate immune response via the recruitment of natural killer cells and monocytes and to suppress metastasis in the lung in vivo.Citation22 In osteosarcoma, exosomes have been shown to be related to migration, invasion, and tumor progression, but the complex interplay between osteosarcoma cells and macrophages has yet to be investigated.Citation23–Citation25
Because a tumor-promoting microenvironment contains M2 macrophages, and M2 macrophages are linked to poor outcomes in patients with osteosarcoma lung metastases, we compared the extracellular communications between metastatic and non-metastatic osteosarcoma cells and alveolar macrophages. It was our hypothesis that metastatic osteosarcoma cells help to create a tumor-permissive environment by exosomal communication. Our results highlight a potential extracellular mechanism by which metastatic osteosarcoma cells communicate with alveolar macrophages to suppress the innate immune response and reveal a potential new therapeutic opportunity to improve the response of relapsed osteosarcoma in the lung to various immunotherapies.
Materials and methods
Cell culture
Murine alveolar macrophage cell line MHS and human monocyte THP1 cells (American Type Culture Collection, ATCC) were maintained in RPMI 1640 medium (Gibco, 11875–093) supplemented with 10% heat-inactivated fetal bovine serum (FBS) in a 37°C, 5% CO2 atmosphere. Murine osteosarcoma cell lines (non-metastatic K7 and Dunn; metastatic K7M3 and DLM8) and murine fibroblast cell line NIH-3T3 were maintained in Dulbecco modified essential medium (Corning, SH30243.01) supplemented with 10% heat-inactivated FBS in a 37°C, 5% CO2 atmosphere. Human osteosarcoma cell lines (non-metastatic SAOS2 and metastatic LM7) were maintained in Dulbecco modified essential medium (Corning, SH30243.01) supplemented with 10% heat-inactivated FBS in a 37°C, 5% CO2 atmosphere.
Isolation of exosomes
Osteosarcoma and fibroblast cell cultures were serum starved for 48–72 h and the conditioned media were collected and subjected to centrifugation at 800 × g at room temperature for 5 min and then to centrifugation at 2,000 × g at room temperature for 10 min. The supernatant was then filtered through a 0.22-μm vacuum filter (Corning, 431153). The filtered supernatant was then subjected to ultracentrifugation at 100,000 × g at 4°C for 2 h using an Optima XE-90 preparative ultracentrifuge (Beckman Coulter, Pasadena, CA). The exosome pellet was washed in 35 mL of phosphate-buffered saline solution (PBS) and underwent a second ultracentrifugation at 100,000 × g at 4°C for 2 h. Next, the exosome pellet was resuspended in 210 μL of PBS. Exosome concentration was determined by the BCA assay (Pierce, 23227) with bovine serum albumin as a standard. All experiments were conducted with an exosome treatment concentration of 50 µg/mL. Exosome size was quantified by nanoparticle tracking analysis using the NanoSight NS300 (Malvern Panalytical, Malvern, United Kingdom).
Real-time qPCR
Total RNA was collected from cells using the RNeasy Mini Kit (Qiagen, 74104). RNA was reverse-transcribed into cDNA using qScript cDNASuperMix (Quanta Biosciences, 95047–100). Quantitative RT-PCR analyses of murine IL10 (forward: 5´- GCTCTTACTGACTGGCATGAG- 3´; reverse: 5´- CGCAGCTCTAGGAGCATGTG- 3´); TGFB2 (forward: 5´- CTTCGACGTGACAGACGCT- 3´; reverse: 5´- GCAGGGGCAGTGTAAACTTATT- 3´); CCL22 (forward: 5´- AGGTCCCTATGGTGCCAATGT- 3´; reverse: 5´- CGGCAGGATTTTGAGGTCCA- 3´); CXCL9 (forward: 5´- TCCTTTTGGGCATCATCTTCC- 3´; reverse: 5´- TTTGTAGTGGATCGTGCCTCG- 3´); CXCL10 (forward: 5´- CCAAGTGCTGCCGTCATTTTC- 3´; reverse: 5´- GGCTCGCAGGGATGATTTCAA- 3´); IL1B (forward: 5´- CCAGCTTCAAATCTCACAGCAG- 3´; reverse: 5´- CTTCTTTGGGTATTGCTTGGGAT- 3´); TNFa (forward: 5´- GCCTCTTCTCATTCCTGCTTG- 3´; reverse: 5´- CTGATGAGAGGGAGGCCATT- 3´); and histone HF3FA (forward: 5´- TGTGGCCCTCCGTGAAATC-3´; reverse: 5´-GCGTGCTAGCTGGATGTCTT-3´) were carried out in triplicate in a 384-well plate using a Lightcycler 480 (Roche Applied Sciences, Indianapolis, IN). The data were normalized to the housekeeping gene histone Hf3fa and then the fold-differences were calculated using the formula f = 2(d), where f = fold-difference in specific gene expression and d = cycle number difference between compared sources of mRNA (i.e., corrected for differences in histone). Melting curves were also analyzed for specificity of PCR product amplification.
Reagents, antibodies and immunoblot analysis
Monoclonal antibodies were purchased from Abcam (Boston, MA) for Calreticulin (ab92516), HSP90B1 (ab3674), CD9 (ab92726) and Beta-actin (ab8226). A monoclonal antibody for CD81 was purchased from Santa Cruz Biotechnology (sc-166029). For immunoblotting, cells were lysed in RIPA buffer (ChemCruz, sc-24948) contained protease pellet (Roche, 04693159001) while exosomes were lysed in 8 M urea 2.5% SDS buffer contained protease pellet. Protein concentrations were determined using the BCA assay (Pierce, 23225) with BSA as a standard. Thirty micrograms of total cellular or exosomal protein were loaded per lane and separated by SDS-PAGE. After transfer at 4 C, the nitrocellulose membrane (Invitrogen, Carlsbad, CA) was blocked with either 5% nonfat dry milk or 5% BSA in Tris-buffered saline (pH 8.0) prior to the addition of primary antibodies and followed with peroxidase-conjugated anti-mouse IgG or anti-rabbit IgG. Protein bands were detected with using a Bio-Rad Chemi-Doc image station with UV-light box (Hercules, CA). An ELISA kit for mouse IL10 was purchased from R&D Systems (M1000B) and performed per the manufacturer’s instructions. A Bio-Plex Pro™ TGF-β 3-plex Assay (171W4001M) was purchased from Bio-rad Technologies and performed according to the manufacturer’s instructions. A neutralizing TGFB2/1.2 Antibody was purchased from R&D Systems (AF-302-NA) and used at a concentration recommended by the manufacturer.
Immunogold labeling of whole mount exosomes
Samples were placed on formvar-carbon coated mesh nickel grids and treated with poly-L-lysine for 1 h. Excess sample was blotted with filter paper and allowed to dry. Grids were washed with PBS and then incubated with CD9 antibody overnight. Grids were washed and then incubated with secondary gold antibody for 2 h at room temperature. The grids were washed and then negatively stained with Millipore paper-filtered aqueous 1% uranyl acetate for 1 min. The stain was blotted dry with filter paper and the samples were allowed to dry. Samples were then examined in a JEM 1010 transmission electron microscope (JEOL, USA Inc., Peabody MA) at an accelerating voltage of 80 kV. Digital images were obtained using the AMT imaging system (Advance Microscopy Techniques Corp., Danvers, MA).
Confocal microscopy
Osteosarcoma and fibroblast exosomes were labeled with Cell Tracker CM-DiI red dye (Invitrogen, C7000). Briefly, exosomes were incubated with 1 micromole of dye at 37°C for 5 min. Exosomes were then incubated at 4°C for 15 min. The labeled exosomes were diluted in 35 mL of PBS and subjected to ultracentrifugation at 100,000 × g at 4°C for 2 h. The exosome pellet was washed in 35 mL of PBS and a second ultracentrifugation was performed at 100,000 × g at 4°C for 2 h. Next, the exosome pellet was resuspended in 210 μL of PBS. MHS cells were plated on cell culture slides (Corning, 53106–304) and treated with labeled osteosarcoma or fibroblast exosomes. The slides were imaged after 24 h using the Nikon Eclipse Ti de-convolution inverted bright field and fluorescent microscope (Nikon Instruments, Melville, New York). PBS treated MHS cells were used as control.
IncuCyte exosome uptake assay
Exosomes were prepared exactly as for confocal microscopy. MHS cells were seeded in a 96-well plate and treated with labeled exosomes. The plate was imaged using the IncuCyte S3 Live-Cell Analysis System (Essen Biosciences, Ann Arbor, MI). PBS treated MHS cells were used as control.
IncuCyte phagocytosis/efferocytosis assay
MHS cells or THP1 cells were seeded in a 96-well-plate and cultured overnight. THP1 cells were activated with PMA (150 ng/mL) for twenty-four hours. To evaluate phagocytosis, osteosarcoma cells and fibroblasts were cultured separately and then labeled with the IncuCyte pHrodo red labeling reagent (Essen Biosciences, 4649) per the manufacturer’s instructions. The pHrodo-labeled cells were added to the MHS cells and then imaged and analyzed using the IncuCyte S3 Live-Cell Analysis System. To evaluate efferocytosis, the osteosarcoma cells and fibroblasts were treated with gemcitabine (SAGENT Pharmaceuticals, 234–10), which induces apoptosis. The doses of gemcitabine used were cell line–specific and chosen from analysis of live-cell imaging over a time course of 48 h using the IncuCyte S3 Live-Cell Analysis System. Additionally, viability was measured using the Vi-Cell XR automated cell viability analyzer (Beckman Coulter, Pasadena, CA). Doses of gemcitabine were chosen so that treated cells were still viable at 24 h but apoptotic at 48 h; doses were as follows: K7M3, 25 µg/mL; K7, 0.78 µg/mL; DLM8, 0.31 µg/mL; Dunn, 0.08 µg/mL; 3T3, 0.31 µg/mL.
IncuCyte macrophage-mediated tumor cell killing assay
Osteosarcoma cells and fibroblasts were seeded in a 96-well-plate and cultured overnight. The cells were labeled with IncuCyte Caspase 3/7 Green Apoptosis Reagent (Essen Biosciences, 4440). MHS or THP1 cells were added to the labeled osteosarcoma cells or fibroblasts, and the cells were imaged and analyzed using the IncuCyte S3 Live-Cell Analysis System.
Statistical analysis
The Student t-test was used to compare the ratios of gene expression, protein secretion and the measures of macrophage phagocytosis, efferocytosis, and macrophage-mediated tumor cell killing. Each experiment was conducted at least twice using three replicates per sample. A p value of > 0.05 was considered significant. Microsoft Excel was used to analyze the above data.
Results
Osteosarcoma cell exosomes have common exosome characteristics and can be taken up by alveolar macrophages
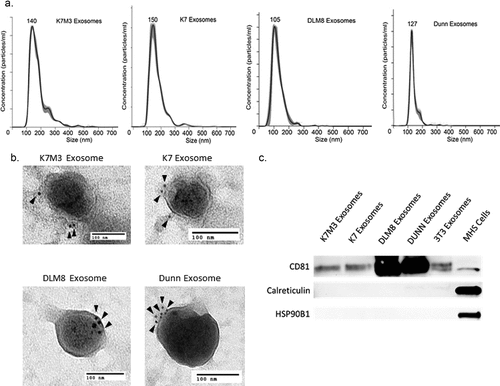
Non-metastatic and metastatic melanoma cell exosomes can have varying effects on the cells of the immune system.Citation22 We therefore elected to compare the effects of exosomes from murine osteosarcoma cells that form lung metastases and those that do not. K7 osteosarcoma cells are non-metastatic and form only a primary tumor. K7M3 osteosarcoma cells were derived from K7 cellsCitation26 but, unlike K7 cells, metastasize to the lung following intratibial injection. Dunn cells are also non-metastatic, while DLM8 is the metastatic subline.Citation27 Exosomes were extracted from all four cell types via differential centrifugation, size exclusion filtration, followed by ultracentrifugation. Exosomes from a normal fibroblast cell line were included as a control (Supplementary Figure 1). Exosome size was evaluated and analyzed using nanoparticle tracking analysis and transmission electron microscopy. Exosomes were found to be within the acceptable size range (), were positive for CD9 expression (), and also featured a membrane bilayer characteristic consistent with the defining features of exosomes. Exosomes were also shown to express exosomal marker CD81, but were not shown to express HSP90B1 or Calreticulin (), both markers of cytoplasmic content.
Figure 1. Osteosarcoma cell exosomes exhibit exosome morphology and the exosome markers CD9 and CD81. Exosomes were extracted from K7M3, K7, DLM8, and Dunn osteosarcoma cells via ultracentrifugation. (a) Exosome size and concentration were assessed via Nanosight analysis. (b) Transmission electron microscopy and immunogold staining was used to analyze CD9 expression. Arrows indicated positive CD9 expression. (c) Western blot analysis was used to evaluate CD81, Calreticulin and HSP90B1 expression in K7M3, K7, DLM8, Dunn, and 3T3 cell exosomes. MHS cells were used as a positive control
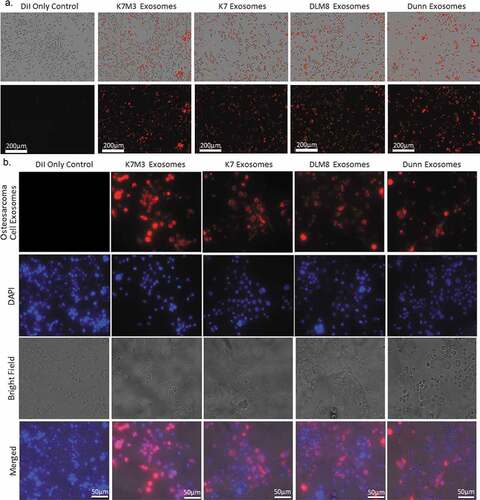
We next investigated whether mouse-derived alveolar macrophages could take up exosomes derived from osteosarcoma cells and fibroblasts. Murine alveolar macrophages (MHS cells) were cultured and incubated for 24 h with exosomes extracted from K7, K7M3, Dunn, and DLM8 cells () or from 3T3 cells and then labeled with CM-DiI red fluorescent dye. (Supplemental Figure 1C). Cells were then imaged using an IncuCyte S3 Live-Cell Analysis System. Uptake of exosomes from all osteosarcoma cells and fibroblasts was demonstrated and confirmed by using labeled exosomes and fluorescent microscopy (, Supplemental Figure 1D).
Figure 2. Alveolar macrophages can uptake osteosarcoma cell exosomes. Osteosarcoma cell exosomes were extracted from non-metastatic K7 and Dunn cells and from metastatic K7M3 and DLM8 cells via ultracentrifugation and then labeled with Cell Tracker CM-DiI red fluorescent dye. Alveolar macrophages were cultured overnight and then treated with labeled osteosarcoma cell exosomes for 24 h. Both fluorescent and bright field images were collected using the (a) IncuCyte Live-Cell System at 20× and the (b) Nikon Eclipse Ti de-convolution inverted bright field and fluorescent microscope at 60× under oil immersion. Osteosarcoma cell exosomes are shown in red and DAPI in blue. Bright field images are also included, as well as the merged images from all three channels
Exosomes from metastatic osteosarcoma cells decreased macrophage tumoricidal activity
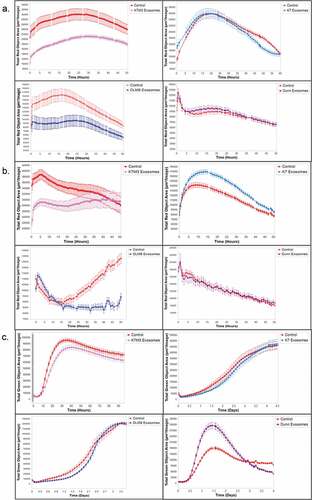
Because macrophages are professional phagocytes and play an important role in tumor cell recognition and killing through the phagocytic process, we investigated the effect of exosomal uptake on three different macrophage functions :Citation7,Citation8,Citation28 phagocytosis, efferocytosis, and cytotoxicity. Uptake of exosomes from K7M3 and DLM8 cells led to a significant decrease in the phagocytic function of MHS alveolar macrophages as compared to untreated control macrophages (). Significant inhibition was seen at twenty four hours for MHS cells treated with K7M3 or DLM8 exosomes. By contrast, there was no significant change in phagocytosis following uptake of exosomes from non-metastatic K7 or Dunn cells () or from 3T3 cells (Supplemental Figure 2A). When MHS cells were incubated with metastatic K7M3 or DLM8 cell exosomes, efferocytosis was significantly decreased at twelve and thirty hours respectively as compared to controls (). By contrast, exosomes from non-metastatic osteosarcoma cells () as well as exosomes derived from normal fibroblasts (Supplemental Figure 2B) did not decrease efferocytosis. Exosomes from K7 cells significantly increased MHS cell efferocytosis at twenty-four hours while there was no significant difference in MHS cells exposed to either Dunn cell or 3T3 cell exosomes. Finally, uptake of exosomes from metastatic osteosarcoma cells (K7M3 or DLM8) significantly decreased macrophage-mediated tumor cell killing (). By contrast, MHS cells incubated with K7 or Dunn cell exosomes () or with 3T3 cell exosomes (Supplemental Figures 2C and 2D) showed no decrease in macrophage-mediated tumor cell killing. Indeed, macrophage-mediated tumor cell killing was significantly increased following uptake of Dunn or 3T3 exosomes at twenty-four and forty-eight hours respectively. MHS cells incubated with 3T3 exosomes showed enhanced cytotoxic activities against both DLM8 and K7M3 cells (Supplemental Figure 2C) as well as against K7 and Dunn cells (Supplemental Figure 2D), and against normal 3T3 cells (Supplemental Figure 2E). Taken together, these data suggest that metastatic osteosarcoma cell exosomes inhibit the phagocytic and cytotoxic functions of alveolar macrophages, while non-metastatic osteosarcoma cell and normal fibroblast-derived exosomes do not.
Figure 3. Metastatic osteosarcoma cell exosomes inhibit alveolar macrophage phagocytosis and efferocytosis and macrophage-mediated tumor cell killing. (a) Alveolar macrophages (MHS cells) were cultured overnight and then incubated with exosomes from osteosarcoma cells (non-metastatic K7 and Dunn; metastatic K7M3 and DLM8) for 24 h. Osteosarcoma cells, labeled with IncuCyte pHrodo labeling reagent, were then added to the MHS cell culture. Phagocytic activity was determined using the IncuCyte S3 Live-Cell Analysis system and the data analyzed by IncuCyte software. Significant inhibition (p < .05) was seen at twenty hours for K7M3 and DLM8. (b) MHS cells were cultured overnight and then incubated with exosomes from the same four osteosarcoma cell lines for 24 h. Effercytosis is the process of clearing dying apoptotic cells. Therefore, osteosarcoma cells, previously treated with gemcitabine for 24 h at a dose that induced apoptosis were labeled with IncuCyte pHrodo labeling reagent, were then added to the MHS cell culture. Efferocytosis was determined by using the IncuCyte S3 Live-Cell Analysis system, and the data were analyzed by the IncuCyte software. Significant inhibition (p < .05) was seen at twelve and thirty hours for K7M3 and DLM8 respectively. Significant enhancement (p < .05) was seen at twenty-four hours for K7. (c) To assess the effect of exosomes on macrophage-mediated cytotoxicity, osteosarcoma cells were cultured overnight and then labeled with the IncuCyte Caspase 3/7 green apoptosis reagent. MHS cells were cultured overnight with exosomes from the four osteosarcoma cell lines and were then added to the labeled osteosarcoma cells. Cytotoxicity was determined using the IncuCyte S3 Live-Cell Analysis system and the data were analyzed by the IncuCyte software. MHS cells treated with PBS was used as a control. Significant inhibition (p < .05) was seen at twenty-four hours for K7M3 and DLM8. Significant enhancement (p < .05) was seen at twenty-four hours for Dunn
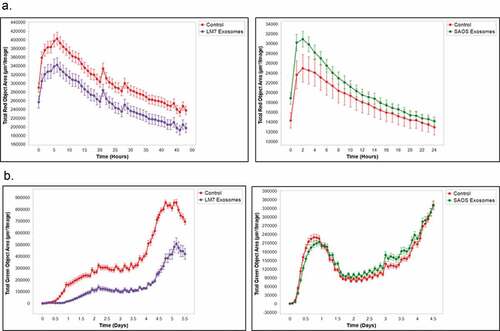
To further validate these findings, we investigated the effect of exosomes from non-metastatic and metastatic human osteosarcoma cells on macrophage function using THP1, a human-derived monocyte cell line. THP1 cells were derived from the peripheral blood and can be activated by PMA (phorbol 12-mystristate 13-acetate) into macrophages. SAOS2 human osteosarcoma cells do not form lung metastases when injected in vivo. LM7 cells were derived from SAOS2 cells, but are metastatic to the lung. SAOS2 and LM7 cell exosomes display common exosome characteristics such as size (Supplemental Figure 1A), the exosome marker CD81, as well as low levels of Calreticulin and HSP90B1 (Supplemental Figure 1B). Additionally, THP1 cells take up SAOS2 and LM7 cell exosomes within twenty-four hours after activation (Supplemental Figure 1 C). Similar to our findings using K7M3 and DLM8 exosomes, when THP1 cells were activated and then exposed to LM7 cell exosomes, there was a significant decrease in phagocytic function as well as macrophage-mediated tumor cell killing (). In contrast, when THP1 cells were activated and exposed to SAOS2 exosomes, there was no effect on phagocytosis and macrophage-mediated tumor cell killing (). These data confirm that metastatic osteosarcoma cell exosomes inhibit the critical functions of macrophages which may have an impact on tumor survival and metastatic growth in the lung.
Figure 4. Metastatic human osteosarcoma cell exosomes inhibit macrophage phagocytosis and macrophage-mediated tumor cell killing.. (a) Human monocytes (THP1 cells) were cultured and activated with PMA (150 ng/mL) overnight and then incubated with exosomes from human osteosarcoma cells (non-metastatic SAOS2; metastatic LM7) for 24 h. Osteosarcoma cells, labeled with IncuCyte pHrodo labeling reagent, were then added to the THP1 cell culture. Phagocytic activity was determined using the IncuCyte S3 Live-Cell Analysis system and the data analyzed by IncuCyte software. The exosomes from LM7 cells induced a significant inhibition (p < .05) in phagocytic activity at twenty hours. (b) Osteosarcoma cells were cultured overnight and then labeled with the IncuCyte Caspase 3/7 green apoptosis reagent. THP1 cells were activated overnight with PMA (150 ng/mL) and then exposed to exosomes from the two osteosarcoma cell lines and were then added to the labeled osteosarcoma cells. Cytotoxicity was determined using the IncuCyte S3 Live-Cell Analysis system and the data were analyzed by the IncuCyte software. THP1 cells treated with PBS was used as a control. Significant inhibition (p < .05) in cytotoxic activity was seen at twenty-four hours for the THP1 cells incubated with LM7 exosomes
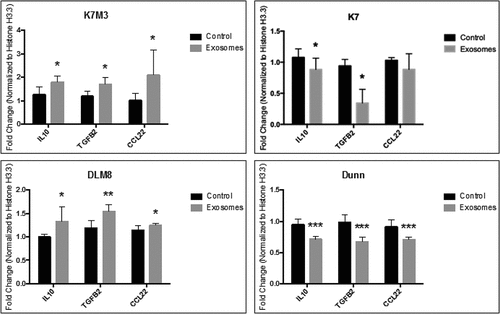
Metastatic osteosarcoma cell exosomes increased expression of M2-related cytokines and chemokines
We next quantified markers of M2-like macrophages following exposure to osteosarcoma cell exosomes. Exosomes from K7M3 and DLM8 cells resulted in significant increases in expression levels of IL10, TGFB2, and CCL22 (), cytokines and chemokines associated with the M2 phenotype, compared to control. MHS cells treated with exosomes from non-metastatic K7 or Dunn cells () or 3T3 cells (Supplemental Figure 2F) did not show increased expression of IL10, TGFB2, and CCL22. We also assessed CXCL9, CXCL10, IL1B and TNFa, markers of M1 macrophages (Supplementary Figure 4). There was a significant decrease in CXCL9 expression in MHS cells exposed to DUNN and K7M3 exosomes compared to normal exosome control. There was also a significant decrease in CXCL10 expression in response to DUNN exosome uptake. Additionally, there was a significant decrease in IL1B expression in response to DUNN and DLM8 exosomes, as well as a significant increase in expression in response to K7 exosome uptake. Lastly, there was significant decrease in TNFa expression in response to DUNN, K7 and K7M3 exosome uptake. Taken together, these data show that exosomes from metastatic cells had little to no effect on the expression of M1 markers. These data indicate that metastatic osteosarcoma cell exosomes induce an M2-like phenotype.
Figure 5. Metastatic osteosarcoma cell exosomes significantly increase mRNA expression of M2 macrophage–related cytokines and chemokines. Alveolar macrophages (MHS cells) were cultured overnight and then incubated with exosomes from osteosarcoma cells (non-metastatic K7 and Dunn; metastatic K7M3 and DLM8) for 24 h. The cells were harvested, total RNA was collected, and cDNA conversion was performed. Hf3fa was used as a loading control. Expression of IL10, TGFB2, and CCL22 mRNAs was quantitated by qPCR. * indicates a p value of < 0.05. ** indicates a p value < .005. *** indicates a p vale < 0.0005
Exosomes from metastatic osteosarcoma cells decreased phagocytosis in alveolar macrophages through the induction of TGFB2
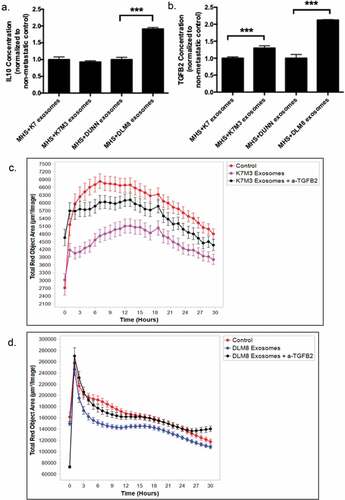
Because both TGFB2 and IL10 expression were elevated in MHS cells following uptake of metastatic osteosarcoma cell exosomes, we next investigated the secretion rate of both cytokines in response to osteosarcoma exosome uptake. When MHS cells were exposed to DLM8 exosomes, there was a significant increase in IL10 levels compared to MHS cells exposed to Dunn cell exosomes (). However, there was no significant difference in IL10 levels secreted by MHS cells exposed to K7 or K7M3 cell exosomes. By contrast, the uptake of K7M3 and DLM8 exosomes, induced a significant increase in TGFB2 levels as compared to MHS cells exposed to K7 or Dunn exosomes (). To determine whether TGFB2 mediated the decreased macrophage activity induced by the metastatic K7M3 exosomes, () or DLM8 (), MHS cells were treated with K7M3 or DLM8 exosomes with or without neutralizing a-TGFB2 antibody. Significant inhibition (p < .05) in phagocytic activity was seen at twelve hours for the MHS cells incubated with both K7M3 and DLM8 exosomes as compared to control. By contrast, MHS cells incubated with K7M3 or DLM8 exosomes plus anti-TGFB2 were similar to control. The baseline phagocytic activity was not statistically different (p > .05) in the cells treated with anti-TGFB2 (). Treatment with anti-TGFB2 prevented the decreased phagocytosis induced by the metastatic exosomes. Taken together, these data suggest that metastatic osteosarcoma cell exosomes induce an immunosuppressive phenotype through the secretion of TGFB2.
Figure 6. Induction of TGFB2 by exosomes from K7M3 and DLM8 metastatic cells plays a role in inhibiting macrophage function. Alveolar macrophages (MHS cells) were cultured overnight and then incubated with exosomes from osteosarcoma cells (non-metastatic K7 and Dunn; metastatic K7M3 and DLM8) for 48 h. IL10 (a) and TGFB2 (b) secretion in culture medium was measured by ELISA. *** indicates a p vale < 0.0005 (c,d) Alveolar macrophages (MHS cells) were cultured overnight and then incubated with exosomes from metastatic K7M3 (c) cells and DLM8 (d) cells with or without TGFB2 antibody (0.1ug/mL) for 24 h. Osteosarcoma cells, labeled with IncuCyte pHrodo labeling reagent, were then added to the MHS cell culture. Phagocytic activity was determined using the IncuCyte S3 Live-Cell Analysis system and the data analyzed by IncuCyte software. Significant inhibition (p < .05) in phagocytic activity was seen at twelve hours for the MHS cells incubated with both K7M3 and DLM8 exosomes as compared to control. There was no significant differences observed at twelve hours between control and K7M3 or DLM8 exosomes plus anti-TGFB2
Discussion
The data presented demonstrated that murine alveolar macrophages take up exosomes from both non-metastatic and metastatic murine osteosarcoma cells and from normal human fibroblasts. Uptake of metastatic osteosarcoma cell exosomes resulted in decreased phagocytosis, efferocytosis, and macrophage-mediated tumor cell killing. In contrast, non-metastatic osteosarcoma cell exosomes did not decrease phagocytosis, efferocytosis, or macrophage-mediated tumor cell killing. The inhibitory effect of exosomes from the murine metastatic cells was also seen when human macrophages were incubated with exosomes from human LM7 metastatic cells but not with non-metastatic parental SAOS2 cells.
The control curves in the experiments with DLM8 and Dunn cells () were noted to be different. This may be due to the differences in the baseline activity of control untreated MHS cells to phagocytose Dunn versus DLM8 cells. The critical finding however is that regardless of the baseline activity, decreased phagocytosis of DLM8 cells was seen when MHS cells were treated with DLM8 exosomes for 24 h and then incubated with labeled DLM8 cells. There was no difference in phagocytic activity when MHS cells were incubated with Dunn exosomes. Thus, uptake of exosomes from the metastatic DLM8 cells compromised macrophage function as defined by phagocytosis, efferocytosis and cytotoxic activity. We also noted an increase in cytotoxic function following incubation of MHS cells with exosomes from the non-metastatic Dunn cells (). We have no explanation for why Dunn cells increased MHS-mediated cytotoxicity at this time. However, once again this data shows that the compromised macrophage function is being specifically induced by the metastatic exosomes and not merely due to the uptake of exosomes from any osteosarcoma cells.
Metastatic osteosarcoma cell exosomes induced increased mRNA expression of IL10, TGFB2, and CCL22, which are cytokines and chemokines associated with M2 macrophages. This was not seen using the non-metastatic osteosarcoma cell exosomes. In addition, metastatic osteosarcoma cell exosomes induced increased production of TGFB2 protein, which has been shown to be a key modulator of immune suppression within the tumor microenvironment.Citation29 Finally, inhibition of TGFB2 reversed the suppressive phagocytic activity in alveolar macrophages induced by the metastatic osteosarcoma cell exosomes, suggesting that TGFB2 plays a key role in the inhibition of macrophage function in response to metastatic osteosarcoma cell exosome uptake. These data taken together support the concept that metastatic osteosarcoma cells can induce an immunosuppressive tumor microenvironment through the secretion of exosomes.
It is now established that macrophage polarization toward an M2 phenotype and M2 macrophage predominance in osteosarcoma lung metastases contribute to the pathogenesis and poor prognosis of osteosarcoma.Citation5,Citation6 Consequently, identifying mechanisms that trigger M2 macrophage polarization has the potential to reveal a novel treatment target for patients with relapsed osteosarcoma in the lung. Since M2 macrophages have been shown to hinder or inhibit responses to CAR T-cells and checkpoint inhibitors, understanding and targeting M2 polarization may also improve the efficacy of immunotherapy. Despite the translational relevance, however, our understanding of how osteosarcoma cells interact with the lung microenvironment to make it tumor permissive is limited. In this study, we identified a potential mechanism by which metastatic osteosarcoma cells control the lung microenvironment to inhibit the immune response. We demonstrated that exosomes from metastatic osteosarcoma cells are different than exosomes from non-metastatic or normal cells in that they induced an M2 phenotype characterized by increased expression of IL10, TGFB2, and CCL22 inhibited macrophage-mediated phagocytosis, efferocytosis, and cytotoxicity, functions that are critical to the successful elimination of tumor cells. In alveolar macrophages exposed to exosomes from non-metastatic K7 or Dunn cells or from normal fibroblasts, we saw no induction of IL10, TGFB2, and CCL22 expression, and no inhibition of phagocytosis, efferocytosis, or macrophage-mediated tumor cell killing. Additionally, the metastatic exosomes induced an increase in the secretion of TGFB2, a molecular mediator of tumor-induced immunosuppression.Citation29 Finally, inhibition of TGFB2 in alveolar macrophages reversed the immunosuppressive activity induced by metastatic osteosarcoma cell exosomes.
We recently showed the critical nature of macrophage polarization to the M1 phenotype in the anti-tumor response to anti-PD-1 therapy.Citation30 Anti-PD-1 therapy induced regression of microscopic osteosarcoma lung metastases and redirected macrophages within the metastases from an M2 to an M1 phenotype.Citation30 Therapeutic efficacy was shown to be mediated by macrophages and not natural killer cells or T cells.Citation30 Other investigators have also demonstrated the therapeutic potential of targeting M2 macrophages within the tumor microenvironment. Combining anti-PD-1 with a selective HDAC6 inhibitor significantly decreased the M2 macrophage population and reduced tumor growth.Citation31 It has also been shown that an agonist anti-CD40 antibody redirected macrophages toward the M1 phenotype, leading to reestablishment of immune surveillance and reduction in pancreatic tumor volume.Citation10 Furthermore, anti-CD47 therapy resulted in functional skewing of macrophages to an M1 phenotype, which enhanced the antitumor responses.Citation32 Blocking the CSF-1/CSF-1 R signaling axis resulted in reprogramming of M2 macrophages, which stimulated antitumor immunity in pancreatic ductal adenocarcinoma.Citation8 Finally, liposome-encapsulated muramyl tripeptide, a synthetic analogue of muramyl dipeptide, which activates macrophages and augments the M1 phenotype, had a beneficial impact on osteosarcoma patient survival.Citation33
It is important to note that alveolar macrophages specifically function to keep airspaces sterile, despite the exposure of the alveolar microenvironment to a wide array of potentially infectious and inflammatory particulates. Alveolar macrophages play a crucial role in preserving the balance between inflammatory and immunosuppressive responses in the lung in order to maintain tissue homeostasis.Citation34 The lung alveolar membrane is the largest surface of the body in contact with the outside environment and is thus exposed to a diverse array of microbes and organic and inorganic particulates.Citation34 Complex cell signaling pathways are responsible for the recognition of potentially harmful materials and subsequent activation or suppression of the immune system. The innate immune system is active and immediately responsive to harmful stimuli. Alveolar macrophages account for approximately 95% of airspace leukocytes, while the other 5% comprises neutrophils and lymphocytes.Citation34 Alveolar macrophages thus act as the sentinel phagocytic cell of the innate immune system in the lung and are avidly phagocytic in response to inhaled particulates that reach the alveolar spaces of the lung.Citation34 In addition to phagocytosis, alveolar macrophages also function in the process of efferocytosis or the phagocytosing of dead or dying cells.Citation35 Efferocytosis allows dead cells to be removed from the microenvironment before their membrane integrity is breached and their contents leak into the surrounding tissue.Citation35 This process prevents exposure of tissue to toxic enzymes, oxidants, and other intracellular components such as caspases and proteases.Citation35 These inflammatory processes may not be beneficial to tumor cell survival and growth. Macrophages also are able to selectively and effectively kill a wide variety of neoplastic cells in a contact-dependent, non-phagocytic manner.Citation36 Additionally, macrophages secrete different cytokines and chemokines depending upon their phenotype, which can kill tumor cells or support their growth. M1 macrophages secrete a diverse array of inflammatory cytokines and chemokines, whereas M2 macrophages secrete immunosuppressive cytokines and chemokines, such as IL10 and CCL22, as well as TGFB2.Citation37 Therefore the capacity of tumor cells in the lung to control the polarization of alveolar macrophages away from the M1 phenotype to the M2 phenotype is beneficial to the tumor’s successful survival.
TGFB is a potent homeostatic regulator of the anti-inflammatory response and has been shown to display immunosuppressive activities toward various components of the immune system.Citation29 The major physiological function of TGFB is to constrain and inhibit the expansion and function of many components of the immune system, including both adaptive and innate immunity. TGFB has also been shown to be a crucial component within the tumor microenvironment, which has revealed roles in both immune evasion and poor responses to cancer immunotherapy.Citation29 Malfunctions in TGFB signaling result in immune dysfunction and has been associated with cancer progression. TGFB has been shown to control adaptive immunity by inhibiting the generation and function of antigen presenting dendritic cells and effector T cells. TGFB has also been shown to modulate innate immunity by inhibiting natural killer cell function as well as the complex behavior of neutrophils and macrophages.Citation29 In mice lacking TGFB type II receptor expression, activation of macrophage to the M2 phenotype was inhibited, suggesting a critical role for TGFB signaling in alveolar macrophage polarization. There are three isoforms of TGFB: TGFB1, TGFB2, TGFB3, but the functions of these isoforms in macrophages are not well understood.Citation29,Citation38 When TGFB2 was inhibited through the use of an antisense oligonucleotide, lung metastases of breast and kidney tumors were reduced in part through the reeducation of M2 macrophages to the M1 antitumor phenotype.Citation39 Furthermore, TGFB1 and TGFB2 were found to be differentially regulated in central nervous system and peripheral nerve injury, and TGFB2 was the principal cytokine that controlled microglia and macrophage phagocytic function, rather than TGFB1.Citation40 Finally, exosomes produced by mesenchymal stem cells have been shown to contain TGFB and drive differentiation of myeloid cells to an immunosuppressive M2 phenotype in breast cancer.Citation41 Our data suggest that metastatic osteosarcoma cell exosomes or factors within the metastatic osteosarcoma cell exosomes induce TGFB2 signaling within alveolar macrophages, which may redirect M1 alveolar macrophages to the M2 phenotype and thereby modulate macrophage-mediated phagocytosis, efferocytosis, and tumor cell killing.
In summary, our results show that metastatic osteosarcoma cells can communicate with innate immune cells such as macrophages within the tumor microenvironment using exosomes, which in turn create a more tumor-permissive environment protecting the tumor cells from immune-mediated killing. The exact mechanism by which exosomes control this dynamic change is unclear, but it appears to involve the upregulation of TGFB2 signaling. While we understand that this data is derived in vitro and does not necessarily equate to what may occur in vivo, to our knowledge, this is the first study of its kind to demonstrate that communication within the osteosarcoma tumor microenvironment may differ depending on the metastatic status of a tumor cell. Understanding how tumor cells communicate with and control the microenvironment will assist in discovering why patients with osteosarcoma have shown little or no response to immunotherapy and may help in designing combination approaches to block this pathway during immunotherapy. The data from this study together with our previously published data showing that M1 macrophages are critical in the antitumor response to anti-PD-1Citation30 as well as previously published data indicating that the inhibition of TGFB2 can reeducate M1 macrophages to an M2 phenotypeCitation39 suggest that targeting TGFB2 may improve macrophage M1 function and response to treatment by redirecting tumor-associated macrophages from the M2 to the M1 phenotype.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Download ()Acknowledgments
We thank Kenneth Dunner, Jr., and the High-resolution Electron Microscopy Facility at The University of Texas MD Anderson Cancer Center for assistance with imaging of the CD9 immunogold transmission electron microscopy experiments. We also thank Anchit Bhagat and Simin Kiany for assistance and advice in designing some experiments. We would also like to thank Kathryn Hale and Scientific Publications Services, Research Medical Library for their assistance in editing this manuscript.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
Additional information
Funding
References
- Saraf AJ, Fenger JM, Roberts RD. Osteosarcoma: accelerating progress makes for a hopeful future. Front Oncol. 2018;8:4. doi:10.3389/fonc.2018.00004.
- Harting MT, Blakely ML. Management of osteosarcoma pulmonary metastases. Semin Pediatr Surg. 2006;15(1):25–13. doi:10.1053/j.sempedsurg.2005.11.005.
- Longhi A, Errani C, De Paolis M, Mercuri M, Bacci G. Primary bone osteosarcoma in the pediatric age: state of the art. Cancer Treat Rev. 2006;32(6):423–436. doi:10.1016/j.ctrv.2006.05.005.
- Kalluri R. The biology and function of fibroblasts in cancer. Nat Rev Cancer. 2016;16(9):582–598. doi:10.1038/nrc.2016.73.
- Dumars C, Ngyuen J-M, Gaultier A, Lanel R, Corradini N, Gouin F, Heymann D, Heymann M-F. Dysregulation of macrophage polarization is associated with the metastatic process in osteosarcoma. Oncotarget. 2016;7(48):78343–78354. doi:10.18632/oncotarget.13055.
- Heymann MF, Lezot F, Heymann D. The contribution of immune infiltrates and the local microenvironment in the pathogenesis of osteosarcoma. Cell Immunol. 2019 Sep;343:103711. doi: 10.1016/j.cellimm.2017.10.011.
- Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol. 2010;11(10):889–896. doi:10.1038/ni.1937.
- Brown JM, Recht L, Strober S. The promise of targeting macrophages in cancer therapy. Clin Cancer Res. 2017;23(13):3241–3250. doi:10.1158/1078-0432.CCR-16-3122.
- Xiao Q, Zhang X, Wu Y, Yang Y. Inhibition of macrophage polarization prohibits growth of human osteosarcoma. Tumour Biol. 2014;35(8):7611–7616. doi:10.1007/s13277-014-2005-y.
- Mantovani A, Marchesi F, Malesci A, Laghi L, Allavena P. Tumour-associated macrophages as treatment targets in oncology. Nat Rev Clin Oncol. 2017;14(7):399–416. doi:10.1038/nrclinonc.2016.217.
- Chen W, Jiang J, Xia W, Huang J. Tumor-related exosomes contribute to tumor-promoting microenvironment: an immunological perspective. J Immunol Res. 2017;2017:1073947. doi:10.1155/2017/1073947.
- Keller S, Sanderson MP, Stoeck A, Altevogt P. Exosomes: from biogenesis and secretion to biological function. Immunol Lett. 2006;107(2):102–108. doi:10.1016/j.imlet.2006.09.005.
- Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200(4):373–383. doi:10.1083/jcb.201211138.
- Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. 2012;1820(7):940–948. doi:10.1016/j.bbagen.2012.03.017.
- Gabrusiewicz K, Li X, Wei J, Hashimoto Y, Marisetty AL, Ott M, Wang F, Hawke D, Yu J, Healy LM, et al. Glioblastoma stem cell-derived exosomes induce M2 macrophages and PD-L1 expression on human monocytes. Oncoimmunology. 2018;7(4):e1412909. doi:10.1080/2162402X.2017.1412909.
- de Vrij J, Maas SLN, Kwappenberg KMC, Schnoor R, Kleijn A, Dekker L, Luider TM, de Witte LD, Litjens M, van Strien ME, et al. Glioblastoma-derived extracellular vesicles modify the phenotype of monocytic cells. Int J Cancer. 2015;137(7):1630–1642. doi:10.1002/ijc.29521.
- Wang F, Li B, Wei Y, Zhao Y, Wang L, Zhang P, Yang J, He W, Chen H, Jiao Z, et al. Tumor-derived exosomes induce PD1(+) macrophage population in human gastric cancer that promotes disease progression. Oncogenesis. 2018;7(5):41. doi:10.1038/s41389-018-0049-3.
- Marton A, Vizler C, Kusz E, Temesfoi V, Szathmary Z, Nagy K, Szegletes Z, Varo G, Siklos L, Katona RL, et al. Melanoma cell-derived exosomes alter macrophage and dendritic cell functions in vitro. Immunol Lett. 2012;148(1):34–38. doi:10.1016/j.imlet.2012.07.006.
- Edin S, Wikberg ML, Rutegård J, Oldenborg P-A, Palmqvist R. Phenotypic skewing of macrophages in vitro by secreted factors from colorectal cancer cells. PLoS One. 2013;8(9):e74982. doi:10.1371/journal.pone.0074982.
- Chow A, Zhou W, Liu L, Fong MY, Champer J, Van Haute D, Chin AR, Ren X, Gugiu BG, Meng Z, et al. Macrophage immunomodulation by breast cancer-derived exosomes requires Toll-like receptor 2-mediated activation of NF-kappaB. Sci Rep. 2014;4:5750. doi:10.1038/srep05750.
- Ham S, Lima LG, Chai EPZ, Muller A, Lobb RJ, Krumeich S, Wen SW, Wiegmans AP, Möller A. Breast cancer-derived exosomes alter macrophage polarization via gp130/STAT3 signaling. Front Immunol. 2018;9:871. doi:10.3389/fimmu.2018.00871.
- Plebanek MP, Angeloni NL, Vinokour E, Li J, Henkin A, Martinez-Marin D, Filleur S, Bhowmick R, Henkin J, Miller SD, et al. Pre-metastatic cancer exosomes induce immune surveillance by patrolling monocytes at the metastatic niche. Nat Commun. 2017;8(1):1319. doi:10.1038/s41467-017-01433-3.
- Jerez S, Araya H, Thaler R, Charlesworth MC, López-Solís R, Kalergis AM, Céspedes PF, Dudakovic A, Stein GS, van Wijnen AJ, et al. Proteomic analysis of exosomes and exosome-free conditioned media from human osteosarcoma cell lines reveals secretion of proteins related to tumor progression. J Cell Biochem. 2017;118(2):351–360. doi:10.1002/jcb.25642.
- Brady JV, Troyer RM, Ramsey SA, Leeper H, Yang L, Maier CS, Goodall CP, Ruby CE, Albarqi HAM, Taratula O, et al. A preliminary proteomic investigation of circulating exosomes and discovery of biomarkers associated with the progression of osteosarcoma in a clinical model of spontaneous disease. Transl Oncol. 2018;11(5):1137–1146. doi:10.1016/j.tranon.2018.07.004.
- Gong L, Bao Q, Hu C, Wang J, Zhou Q, Wei L, Tong L, Zhang W, Shen Y. Exosomal miR-675 from metastatic osteosarcoma promotes cell migration and invasion by targeting CALN1. Biochem Biophys Res Commun. 2018;500(2):170–176. doi:10.1016/j.bbrc.2018.04.016.
- Khanna C, Prehn J, Yeung C, Caylor J, Tsokos M, Helman L. An orthotopic model of murine osteosarcoma with clonally related variants differing in pulmonary metastatic potential. Clin Exp Metastasis. 2000;18(3):261–271. doi:10.1023/A:1006767007547.
- Asai T, Ueda T, Itoh K, Yoshioka K, Aoki Y, Mori S, Yoshikawa H. Establishment and characterization of a murine osteosarcoma cell line (LM8) with high metastatic potential to the lung. Int J Cancer. 1998;76(3):418–422. doi:10.1002/(SICI)1097-0215(19980504)76:3<418::AID-IJC21>3.0.CO;2-5.
- Buscher K, Ehinger E, Gupta P, Pramod AB, Wolf D, Tweet G, Pan C, Mills CD, Lusis AJ, Ley K, et al. Natural variation of macrophage activation as disease-relevant phenotype predictive of inflammation and cancer survival. Nat Commun. 2017;8:16041. doi:10.1038/ncomms16041.
- Batlle E, Massague J. Transforming growth factor-beta signaling in immunity and cancer. Immunity. 2019;50(4):924–940. doi:10.1016/j.immuni.2019.03.024.
- Dhupkar P, Gordon N, Stewart J, Kleinerman ES. Anti-PD-1 therapy redirects macrophages from an M2 to an M1 phenotype inducing regression of OS lung metastases. Cancer Med. 2018;7(6):2654–2664. doi:10.1002/cam4.1518.
- Knox T, Sahakian E, Banik D, Hadley M, Palmer E, Noonepalle S, Kim J, Powers J, Gracia-Hernandez M, Oliveira V, et al. Selective HDAC6 inhibitors improve anti-PD-1 immune checkpoint blockade therapy by decreasing the anti-inflammatory phenotype of macrophages and down-regulation of immunosuppressive proteins in tumor cells. Sci Rep. 2019;9(1):6136. doi:10.1038/s41598-019-42237-3.
- Xu J-F, Pan X-H, Zhang S-J, Zhao C, Qiu B-S, Gu H-F, Hong J-F, Cao L, Chen Y, Xia B, et al. CD47 blockade inhibits tumor progression human osteosarcoma in xenograft models. Oncotarget. 2015;6(27):23662–23670. doi:10.18632/oncotarget.4282.
- Meyers PA, Schwartz CL, Krailo MD, Healey JH, Bernstein ML, Betcher D, Ferguson WS, Gebhardt MC, Goorin AM, Harris M, et al. Osteosarcoma: the addition of muramyl tripeptide to chemotherapy improves overall survival–a report from the children’s oncology group. J Clin Oncol. 2008;26(4):633–638. doi:10.1200/JCO.2008.14.0095.
- Martin TR, Frevert CW. Innate immunity in the lungs. Proc Am Thorac Soc. 2005;2(5):403–411. doi:10.1513/pats.200508-090JS.
- Korns D, Frasch SC, Fernandez-Boyanapalli R, Henson PM, Bratton DL. Modulation of macrophage efferocytosis in inflammation. Front Immunol. 2011;2:57. doi:10.3389/fimmu.2011.00057.
- Keller R, Keist R, Wechsler A, Leist TP, Van Der Meide PH. Mechanisms of macrophage-mediated tumor cell killing: a comparative analysis of the roles of reactive nitrogen intermediates and tumor necrosis factor. Int J Cancer. 1990;46(4):682–686. doi:10.1002/ijc.2910460422.
- Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011;11(11):723–737. doi:10.1038/nri3073.
- Haque S, Morris JC. Transforming growth factor-beta: A therapeutic target for cancer. Hum Vaccin Immunother. 2017;13(8):1741–1750. doi:10.1080/21645515.2017.1327107.
- Huber-Ruano I, Raventós C, Cuartas I, Sánchez-Jaro C, Arias A, Parra JL, Wosikowski K, Janicot M, Seoane J. An antisense oligonucleotide targeting TGF-beta2 inhibits lung metastasis and induces CD86 expression in tumor-associated macrophages. Ann Oncol. 2017;28(9):2278–2285. doi:10.1093/annonc/mdx314.
- Stoll G, Schroeter M, Jander S, Siebert H, Wollrath A, Kleinschnitz C, Brück W. Lesion-associated expression of transforming growth factor-beta-2 in the rat nervous system: evidence for down-regulating the phagocytic activity of microglia and macrophages. Brain Pathol. 2004;14(1):51–58. doi:10.1111/j.1750-3639.2004.tb00497.x.
- Biswas S, Mandal G, Roy Chowdhury S, Purohit S, Payne KK, Anadon C, Gupta A, Swanson P, Yu X, Conejo-Garcia JR, et al. Exosomes produced by mesenchymal stem cells drive differentiation of myeloid cells into immunosuppressive M2-polarized macrophages in breast cancer. J Immunol. 2019;203(12):3447–3460. doi:10.4049/jimmunol.1900692.
