ABSTRACT
The outcome of patients with cutaneous melanoma has been strongly modified by recent advances obtained with Immune Checkpoint Inhibitors (ICIs). However, despite this breakthrough, durable response to ICIs is limited to a subset of patients. We investigated whether the expression of TRF2, which preserves telomere integrity, and have an effect on tumor immunosurveillance notably by directly recruiting and activating myeloid-derived suppressor cells (MDSCs), could be a prognostic biomarker in patients with relapsed or metastatic melanoma based on different treatment regimens. We evaluated retrospectively the association of TRF2 expressed in melanoma cells in combination with intratumoral CD33+ CD15+ CD14- MDSCs, as detected by immunohistochemistry and quantified by digital analysis, to clinicopathological features and overall survival (OS) among 48 patients treated with ICIs and 77 patients treated with other treatment options. The densities/mm2 of TRF2+ cells (P=.003) and CD33+ cells (P=.004) were individually significantly related to poor OS. In addition, only the combined expression of CD33+/CD15+/CD14- cells/mm2 was significantly correlated to poor OS (P=.017) in the whole study population as well as in patients treated by ICIs (P=.023). There was no significant difference in OS when analyzing the other markers individually or in combination according to the treatment regimen. The pre-treatment assessment of TRF2 expression and CD33+ cells/mm2 along with the density of CD33+/CD15+/CD14- cells/mm2 could assess OS and better predict clinical response of patients with melanoma treated by ICIs.
Introduction
Metastatic or advanced melanoma is a fatal skin cancer, with a 5-y survival rate of less than 30%.Citation1 The development of the Immune Checkpoint Inhibitors (ICIs) targeting Programmed Death-1 (PD-1) and its ligand PD-L1 represents a true paradigm shift with a 52% increase in the 5-y median overall survival.Citation2–5 However, durable response to ICIs is limited to only a subset of patients, whereas 40% of the patients do not respond to ICIs in monotherapy. In most clinical trials, the expression of PD-L1 alone did not allow optimal selection of responding patients.Citation3 Only a recent study on resected high-risk stage III melanoma demonstrated a benefit from the PD-L1 positivity of the 3-y recurrence-free survival rate being superior to pembrolizumab compared with placebo.Citation6
While PD-L1 alone is currently inadequate as prognostic and predictive marker in metastatic melanoma, other potential promising biomarkers are currently emerging.Citation7 A recent study showed that an increase in CD8 + T cells from baseline to post-treatment biopsy may be significantly associated with a decrease in tumor size in patients with metastatic melanoma treated with ICIs.Citation8 Notably, the CD4+ regulatory T cells (Tregs) expressing Foxp3 have immune suppressive functions and promote tumor progression by suppressing effective anti-tumor immunity.Citation9 Moreover, patients with increased levels of CD4+ and CD8 + T cells have better response than those with low levels, and potentially the ratio of T effector cells to Tregs may be a good predictor of response to ICIs.Citation10 In addition, circulating PD-1+ Tregs rapidly declines after the initiation of the anti-PD-1 treatment, which is associated with better clinical outcome.Citation11
High baseline eosinophil count and low LDH count were associated with improved survival in melanoma patients treated with pembrolizumab.Citation8 A recent study of patients with metastatic melanoma had 65 cytokines profiled as part of a 65-plex discovery assay. Eleven cytokines were found to be significantly upregulated in patients who experienced severe immune-related adverse events; these 11 cytokines (G-CSF, GM-CSF, Fractalkine, FGF-2, IFNα2, IL12p70, IL1a, IL1B, IL1RA, IL2, IL13) were integrated into a single cytokine toxicity score (CYTOX) and validated its ability to predict immune-related adverse events.Citation12
The shelterin protein TRF2 (telomere repeats binding factor 2) is at the center of the molecular events that preserve telomere integrity.Citation13,Citation14 TRF2 binds to telomeric repeated sequences TTAGGG and its main role is to protect telomeres from being recognized as double stranded breaks in order to maintain genome stability by inhibiting the DNA Damage Response (DDR).Citation15–18 Notably, in various mouse models, TRF2 inhibition has been shown to impair tumorigenesis independently of its functions in telomere protection and maintenance, but via cell extrinsic effects on immunosurveillance and angiogenesis.Citation19–21 Consistent with these various oncogenic properties, an increased level of TRF2 expression is observed in a large panel of carcinomas and has been reported to be associated with poor outcomes.Citation15–18 However, the prognostic impact of TRF2 remains unknown in relapsed or metastatic melanoma.
An efficient antitumor immune response is of vital importance in preventing cancer progression and metastasis, as well as in successful chemotherapy or immunotherapy.Citation22 Among the immunosuppressive properties of tumors, the recruitment and activation of myeloid-derived suppressor cells (MDSCs) facilitate cancer progression.Citation23 A recent study demonstrated that cancer cells recruited and directly activated MDSCs in a TRF2-dependent manner, dampening NK and CD8 + T cell cytotoxicity.Citation18 Moreover, patients with several types of carcinomas (including, breast, gastric, ovarian, and lung) with high TRF2 expression also exhibited marked MDSCs infiltration and reduced overall survival.Citation18 CD11b and CD33 are mainly used as markers for human MDSCs.Citation24 However, these markers are expressed by cells of the myelocytic lineage and by NK cells, so they are not specific enough to identify human neutrophils.Citation25,Citation26 Instead, neutrophils (or G-MDSCs) are found to be CD14 low and CD15 high, whereas the monocytes (or Mo-MDSCs) are CD14 high and CD15 low.Citation27 Some studies showed that patients with melanoma have significantly high levels of blood circulating CD33+ CD11b+CD15 + G-MDSCs with immune suppressive phenotype, while low levels of G-MDSCs before anti-CTLA-4 therapy could correlate with an objective clinical response, long-term survival, and an improved clinical status.Citation28–30
The objective of our study was to correlate the expression of TRF2 in melanoma cells combined with the quantification of intratumoral MDSCs, to overall survival (OS) and response to treatment in order to determine whether the combination of two proteins evaluation could be an effective prognostic biomarker in patients with relapsed or metastatic melanoma.
Patients and methods
Study population
This retrospective cohort included 125 patients with consecutive primary cutaneous malignant melanoma diagnosed between July 2013 and February 2017 and treated at the Department of Dermatology, University of Nice, Archet 2 Hospital (Nice, France) (). The patients initially diagnosed with stage I–II melanoma, were enrolled in the study at the time of the regional or distant metastatic relapse. The availability of histological material from the metastasis as well as the presence of an informed signed consent was required criteria to include a case in the study.
Table 1. Clinical and histomolecular characteristics of the metastatic melanoma cohorts treated by chemotherapy, targeted therapy, or immunotherapy. *χ2-test or Student’s t-test were used to investigate difference between groups.
Out of the 125 patients, 91 (73%) presented with regional metastases (35 in transit and 56 lymph node metastases) and 34 (27%) with distant metastases (19 lung and 15 subcutaneous).
Two groups of patients were distinguished in this study: a group of 48 patients (38%) who received at least one treatment of immunotherapy (anti-PD-1 inhibitors-pembrolizumab/nivolumab and/or anti-CTLA4) and a group of 77 patients (62%) who did not receive immunotherapy treatment, albeit some had other treatments (chemotherapy or targeted therapies with anti-BRAF and anti-MEK agents) ().
Among the patients treated with immunotherapy, 35 (73%) had exclusively immunotherapy, while 13 (27%) received an immunotherapy treatment before or after having other treatments (chemotherapy or targeted therapies).
All tumor specimens were used with the informed signed consent from the patients. The study was approved by the local ethics committee (Human Research Ethics Committee, Nice University Hospital Center/hospital-related Biobank BB-0033-00025; http://www.biobank-cotedazur.fr/) and was performed in accordance with the guidelines of the Declaration of Helsinki.
Immunohistochemistry and digital image analysis
Formalin-fixed paraffin-embedded (FFPE) serial 4 μm tissue sections were freshly cut, deparaffinized, pre-treated, and stained with monoclonal antibodies (Abs) directed against CD33 (clone SP266, ready-to-use, Roche, Tucson, AZ, USA), CD14 (clone EP128, dilution 1/200, Epitomics, Burlingame, CA, USA), CD15 (clone MMA, ready-to-use, Roche, Tucson, AZ, USA), and TRF2 (clone 4A794.15, dilution 1/500, OriGene, Rockville, MA, USA) on a BenchMark ULTRA autostainer (Ventana Medical Systems, Tucson, AZ, USA).
Stains were detected using anti-immunoglobulin-coupled horseradish peroxidase with
3,3-diaminobenzidine (DAB, OptiView Kit, Roche Diagnostics, Ventana, catalog no. 760–700) as substrate. Nuclear counterstaining was performed with Mayer hematoxylin. Each IHC run contained a positive control (tonsil) and a negative Ab control (buffer, no primary Ab).
Slides were scanned at high resolution 200x on a Nanozoomer 2.0-HT Scanner (Hamamatsu photonics, Hamamatsu, Japan). Digital image analysis was carried out by two senior pathologists (M.I. and P.H.) using the HALOTM image analysis software, v2.3.2089.52 (Indica Labs, London, UK).Citation31 The AI classifier in HALO was used to separate the image into two classes: tumor and other components (stroma, glass slide, artifacts). The classifier mask is shown overlaying the IHC image where classified tumor regions are shown in red, and the other components on the slide in yellow (Supplementary Fig. S1). Once the chosen classifier has been created and saved, it was used in the Multiplex IHC v2.0.3 module in HALO to automatically analyze the biomarkers included in the study.
BRAF molecular analysis
The BRAF mutational status was determined on tumor DNA isolated from FFPE tissue samples of melanoma metastases using the QIAamp DNA FFPE tissue kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. Pyrosequencing of BRAF exon 15 using the Therascreen BRAF Pyro Kit (Qiagen) was performed as previously described.Citation32
Statistical analysis
Data are reported as the median ± S.D., extremes, absolute frequencies, percentages, 95% confidence intervals, and missing data percentages, as specified. All statistical analyzes were performed at alpha risk = 5% under bilateral assumption using R.3.2.3 software on Windows. The data were compared using the Χ2 test and the Fisher test in the case of noncompliance with Χ2 application conditions or the Student’s T-test or the Mann–Whitney test in the case of noncompliance with the student test conditions.
Overall survival (OS) since primary was defined as the interval between the time of the biopsy/resection of the metastasis and the date of death of the patient or of the last follow up. Patients lost to follow-up were censored on the date of last contact. The survival curves were compared by the Log-Rank test. Kaplan-Meier survival curves were determined to assess the prognostic significance of single or combined biomarkers on OS. The cutoff predicting OS was defined as the median density of expressing cells/mm2. Multivariate analyzes were performed using Cox regression models with corresponding adjusted Hazard Ratio (HR) calculations. P-values <0.05 indicated statistical significance.
Results
Expression patterns of the analyzed tissue biomarkers
Examples of the digital analysis with the multiplex IHC module in HALO are shown in .
Figure 1. Representative images of immune biomarkers and TRF2 staining, and their cell detection mask overlays used in the digital image analysis. Original magnification, x 200
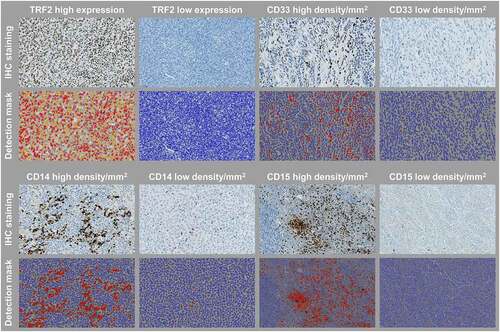
Collinearity was used to assess the association between the putative biomarkers within the tumor areas (Supplementary Fig. S2). All biomarkers demonstrated some degree of positive correlation with each other. Of the biomarkers assessed, the most significant relationships were observed between CD14 and CD33 expressing cells (rho = 0.8; P <.0001), CD14 and TRF2 expression (rho = 0.8; P <.0001), CD15 and CD33 expressing cells (rho = 0.77; P <.0001), and CD14 and CD15 expressing cells (rho = 0.75; P <.0001), whereas a moderate correlation was found between CD33 expressing cells and TRF2 expression (rho = 0.55; P <.0001). For further analyses, patient stratification was defined by density using median value cutoffs.
Patients characteristics
The main clinical and histo-molecular characteristics of this cohort are shown in .
Of the 125 patients included for analysis, 41 (32.8%) were female and 84 (67.2%) were male patients. Overall median age was 64.2 y (range, 23–92 y). A majority of patients had an ECOG status equal to 0 (97, 77.6%). Of the 125 cases, superficial spreading malignant melanoma accounted for 44.8%, nodular melanoma 25.6%, acral lentiginous melanoma 4.8%, invasive lentigo maligna melanoma 4%, and 20.8% of the cases were not classified. 32% of the cases harbored a BRAF mutation on exon 15.
Correlations with the clinicopathological characteristics
The density of CD14+ cells was significantly associated with BRAF mutational status (P=.02; ). The density of CD15+ cells was significantly correlated to the ulceration (P=.005) and the Breslow depth (P=.02), whereas the TRF2+ expression was significantly associated with the histological subtype (P<.001; ).
Table 2. Correlative analysis between the clinical and histomolecular characteristics of the patients and the analyzed biomarkers in the metastatic melanoma cohort. *χ2-test, Student’s t-test or ANOVA test were used to investigate difference between groups.
Survival analysis
The median follow-up of the study was of 53 months (95% CI, 44–70). According to the univariate analysis, the ECOG status, the pTNM stage, and the presence of ulceration were significantly associated to poor OS in our study cohort (P=.022, P=.044, and P<.001, respectively).
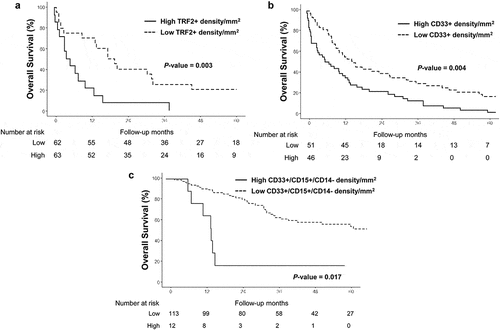
The densities of TRF2+ cells (HR, 2.4; 95% CI, 1.1–5.1; P=.003) and CD33+ cells (HR, 1.46; 95% CI, 0.7–3.1; P=.004) were individually significantly associated with poor OS (), except the density of CD15+ cells (HR, 1.7; 95% CI, 0.94–3; P=.078) and CD14+ cells (HR, 0.99; 95% CI, 0.57–1.7; P=.386; not shown). In addition, based on the results for their individual survival analysis and collinearity of the biomarkers assessed, combination of dichotomized densities was explored for OS outcome. Of these analyses, only the combined expression of CD33+/CD15+/CD14- cells/mm2 was significantly correlated to poor OS (median OS, 3.6 months versus 12.6 months; HR, 3.6; 95% CI, 1.1–12; P=.017; ).
Figure 2. Kaplan-Meier overall survival curves according to TFR2, CD33, and CD33+/CD15+/CD14- status in the whole study population (n = 125)
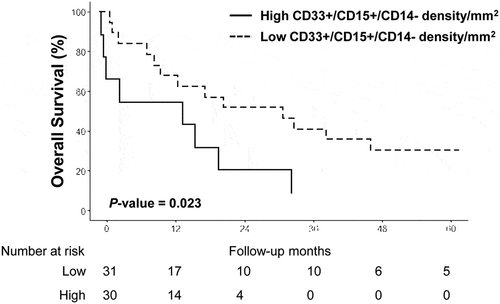
Moreover, in the population of patients treated by immunotherapy, the combined expression of CD33+/CD15+/CD14- cells/mm2 was significantly associated with poor OS (median OS, 13.3 months versus 20.7 months; HR, 3.2; 95% CI, 1.1–8; P=.023; ). There was no significant difference in OS when analyzing the other markers individually or in combination according to the treatment regimen.
Figure 3. Kaplan-Meier overall survival curve according to CD33+/CD15+/CD14- status in patients treated by immunotherapy (n = 48)
In the multivariate analysis, the ECOG status and the combined expression of CD33+/CD15+/CD14- cells/mm2 were significant and independent prognostic factors associated with OS compared to the other groups ().
Table 3. Multivariate analysis for overall survival in the cohort population
Discussion
Treatment with ICIs in patients with advanced or metastatic melanoma can demonstrate impressive response rates.Citation33,Citation34 However, although the benefit is restricted to approximately 40% of the patients treated with anti-PD-1 therapy, there are no approved stratification strategies for ICIs in melanoma.Citation33,Citation34 Thus, despite active research and development for having robust prognostic or predictive biomarkers for responsiveness of ICIs in melanoma in routine clinical practice, there is an urgent need for robust and easy to use biomarkers in daily practice to guide the clinical decision-making.Citation35,Citation36
In the current study, the high TRF2 expression and high density of CD33+ cells were found to represent baseline biomarkers significantly affecting OS of melanoma patients.
The TRF2 protein is a key factor in telomere protection, which contributes to oncogenesis.Citation37,Citation38 While elevated TRF2 expression is observed in a large number of solid malignancies, notably carcinomas,Citation16,Citation39–41 little is known about its oncogenic and clinical role in melanoma. The in vitro TRF2 inhibition in human melanoma cells can impair their tumorigenicity, whereas a basal level of telomere instability favors an efficient response to TRF2 inhibition and the combined anti-TRF2 and G4-ligand therapy would have synergistic inhibitory effects on tumor cell growth.Citation42 In addition, we previously demonstrated that high expression of TRF2 in circulating tumor microemboli detected in metastatic melanoma patients had potential impact for the assessment of disease aggressiveness.Citation17
Recent findings have also raised the possibility that overexpression of TRF2 may be a critical step in human oncogenesis by contributing to bypass tumor immune surveillance. Based on the upregulation of TRF2, tumor cells recruit and activate MDSCs, acting as a general suppressor of the immune response by inhibiting NK and T cell responses, thus establishing a direct link between cancer-associated telomere modifications and the immunosuppressive tumor microenvironment.Citation18
In our study, the high density of CD33+ cells was significantly correlated with worse OS. Only one other recent study evaluated the relationship between the expression of CD33+ MDSCs and the outcome of patients with cutaneous melanoma, showing that high expression of CD33 was associated with poor clinicopathological variables and was an independent prognostic factor.Citation43 Moreover, CD33+ MDSCs are increased in the peripheral blood of advanced melanoma patients, being an indicator of worse survival at baseline and following treatment with ipilimumab.Citation28,Citation44 MDSCs have been shown to exert immunosuppressive function on T cells, thereby possibly counteracting the beneficial effect of ICIs.Citation45 However, CD33 is found in maturing granulocytes, monocytes, and multipotent myeloid precursors and is also expressed in subsets of activated T cells, natural killer cells, and B cells.Citation25,Citation26 Instead neutrophils (or G-MDSCs) besides expressing CD33, are found to be CD14 low and CD15 high, whereas monocytes (or Mo-MDSCs) are CD14 high and CD15 low.
In the present study, whereas the density of CD15+ cells or CD14+ cells was not correlated to survival, only the combined expression of CD33+/CD15+/CD14- cells/mm2 was significantly predictive of poor OS in both the whole population as well as in patients treated by immunotherapy. Thus, it seems that the G-MDSCs and not Mo-MDSCs may be related to the outcome, suggesting that the blockade of G-MDSCs immunosuppressive mechanisms could be explored as a therapeutic approach to reestablishing T-cells activity and immunotherapy success in melanoma patients.Citation46
Recent reports have suggested the significance of G-MDSCs in patients with advanced melanoma treated using ICIs.Citation47 Increased microRNAs in the plasma of melanoma patients are associated with the generation of G-MDSCs mediated by melanoma extracellular vesicles, and are even associated with resistance to treatment with ICIs in melanoma patients.Citation47
Nevertheless, there are a few limitations to our study that need to be considered. This is a heterogeneous patient population. The number of patients treated with ICIs was limited (n = 48). The question whether the suggested biomarkers are prognostic in general or prognostic for outcome after specific ICIs cannot be answered. As tumors often exhibit significant cellular and spatial heterogeneity, it would be important to be able to perform high-resolution multiplexed IHC analysis across whole-sections of tumors to analyze the different putative biomarkers in relationship with survival. In addition, we were not able to validate the prognostic relevance of our findings in an independent cohort. Thus, further validation is strongly warranted.
In conclusion, the pre-treatment evaluation of TRF2 expression and CD33+ cells/mm2 along with the density of CD33+/CD15+/CD14- cells/mm2 are significantly correlated with poor OS and could predict clinical response of patients with recurrent or metastatic melanoma treated by ICIs, and so be a promising, easy to use new biomarkers in patients with melanoma.
Disclosure of Potential Conflicts of Interest
M. Ilié has received honoraria for travel support and consulting/advisory roles for AstraZeneca, Bristol-Myers Squibb, Roche, Boehringer-Ingelheim and Merck & Co. outside the submitted work.
P. Hofman has received honoraria for travel support and consulting/advisory roles for AstraZeneca, Roche, Bristol-Myers Squibb, Novartis, Pfizer, Bayer, Illumina, Ed Lilly, MSD, Qiagen, Thermofisher, Biocartis, and Merck & Co. outside the submitted work.
The remaining authors have declared no conflict of interests.
Supplemental Material
Download ()Supplementary material
Supplemental data for this article can be accessed on the publisher’swebsite.
Additional information
Funding
References
- Gershenwald JE, Scolyer RA, Hess KR, Sondak VK, Long GV, Ross MI, Lazar AJ, Faries MB, Kirkwood JM, McArthur GA, et al. Melanoma staging: evidence-based changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J Clin. 2017;67(6):472–9. doi:10.3322/caac.21409.
- Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366(26):2443–2454. doi:10.1056/NEJMoa1200690.
- Robert C, Long GV, Brady B, Dutriaux C, Maio M, Mortier L, Hassel JC, Rutkowski P, McNeil C, Kalinka-Warzocha E, et al. Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med. 2015;372(4):320–330. doi:10.1056/NEJMoa1412082.
- Wolchok JD, Chiarion-Sileni V, Gonzalez R, Rutkowski P, Grob JJ, Cowey CL, Lao CD, Wagstaff J, Schadendorf D , Ferrucci PF, et al. Overall survival with combined nivolumab and ipilimumab in advanced melanoma. N Engl J Med. 2017;377:1345–1356. doi:10.1056/NEJMoa1709684.
- Larkin J, Chiarion-Sileni V, Gonzalez R, et al. Five-year survival with combined nivolumab and ipilimumab in advanced melanoma. N Engl J Med. 2019;381:1535–1546. doi:10.1056/NEJMoa1910836.
- Eggermont AMM, Blank CU, Mandala M, et al. Longer follow-up confirms recurrence-free survival benefit of adjuvant pembrolizumab in high-risk stage III melanoma: updated results from the EORTC 1325-MG/KEYNOTE-054 trial. J Clin Oncol. 2020;38:3925–3936. doi:10.1200/JCO.20.02110.
- Adam T, Becker TM, Chua W, et al. The multiple potential biomarkers for predicting immunotherapy response-finding the needle in the haystack. Cancers. 2021;13. doi:10.3390/cancers13020277
- Fujii T, Naing A, Rolfo C, et al. Biomarkers of response to immune checkpoint blockade in cancer treatment. Crit Rev Oncol Hematol. 2018;130:108–120. doi:10.1016/j.critrevonc.2018.07.010.
- Li B, Severson E, Pignon JC, Zhao H, Li T, Novak J, Jiang P, Shen H, Aster JC, Rodig S, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17(1):174. doi:10.1186/s13059-016-1028-7.
- Pirozyan MR, McGuire HM, Emran AA, Tseng H-Y, Tiffen JC, Lee JH, Carlino MS, Menzies AM, Long GV, Scolyer RA, et al. Pretreatment innate cell populations and CD4 T cells in blood are associated with response to immune checkpoint blockade in melanoma patients. Front Immunol. 2020;11:372. doi:10.3389/fimmu.2020.00372.
- Gambichler T, Schroter U, Hoxtermann S, Susok L, Stockfleth E, Becker JC. Decline of programmed death-1-positive circulating T regulatory cells predicts more favourable clinical outcome of patients with melanoma under immune checkpoint blockade. Br J Dermatol. 2020;182(5):1214–1220. doi:10.1111/bjd.18379.
- Lim SY, Lee JH, Gide TN, Menzies AM, Guminski A, Carlino MS, Breen EJ, Yang JYH, Ghazanfar S, Kefford RF, et al. Circulating cytokines predict immune-related toxicity in melanoma patients receiving anti-PD-1-based immunotherapy. Clin Cancer Res. 2019;25(5):1557–1563. doi:10.1158/1078-0432.CCR-18-2795.
- Broccoli D, Smogorzewska A, Chong L, De Lange T. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat Genet. 1997;17(2):231–235. doi:10.1038/ng1097-231.
- Bilaud T, Brun C, Ancelin K, Koering CE, Laroche T, Gilson E. Telomeric localization of TRF2, a novel human telobox protein. Nat Genet. 1997;17(2):236–239. doi:10.1038/ng1097-236.
- Nakanishi K, Kawai T, Kumaki F, Hiroi S, Mukai M, Ikeda E, Koering CE, Gilson E. Expression of mRNAs for telomeric repeat binding factor (TRF)-1 and TRF2 in atypical adenomatous hyperplasia and adenocarcinoma of the lung. Clin Cancer Res. 2003;9:1105–1111.
- Diehl MC, Idowu MO, Kimmelshue KN, York TP, Jackson-Cook CK, Turner KC, Holt SE, Elmore LW. Elevated TRF2 in advanced breast cancers with short telomeres. Breast Cancer Res Treat. 2011;127(3):623–630. doi:10.1007/s10549-010-0988-7.
- Long E, Ilie M, Bence C, Butori C, Selva E, Lalvée S, Bonnetaud C, Poissonnet G, Lacour J-P, Bahadoran P, et al. High expression of TRF2, SOX10, and CD10 in circulating tumor microemboli detected in metastatic melanoma patients. A potential impact for the assessment of disease aggressiveness. Cancer Med. 2016;5(6):1022–1030. doi:10.1002/cam4.661.
- Cherfils-Vicini J, Iltis C, Cervera L, Pisano S, Croce O, Sadouni N, Győrffy B, Collet R, Renault VM, Rey‐Millet M, et al. Cancer cells induce immune escape via glycocalyx changes controlled by the telomeric protein TRF 2. EMBO J. 2019;38(11). doi:10.15252/embj.2018100012.
- Biroccio A, Cherfils-Vicini J, Augereau A, Pinte S, Bauwens S, Ye J, Simonet T, Horard B, Jamet K, Cervera L, et al. TRF2 inhibits a cell-extrinsic pathway through which natural killer cells eliminate cancer cells. Nat Cell Biol. 2013;15(7):818–828. doi:10.1038/ncb2774.
- El Mai M, Wagner KD, Michiels JF, Ambrosetti D, Borderie A, Destree S, Renault V, Djerbi N, Giraud-Panis M-J, Gilson E, et al. The telomeric protein TRF2 regulates angiogenesis by binding and activating the PDGFRbeta promoter. Cell Rep. 2014;9(3):1047–1060. doi:10.1016/j.celrep.2014.09.038.
- Zizza P, Dinami R, Porru M, Cingolani C, Salvati E, Rizzo A, D’Angelo C, Petti E, Amoreo CA, Mottolese M, et al. TRF2 positively regulates SULF2 expression increasing VEGF-A release and activity in tumor microenvironment. Nucleic Acids Res. 2019;47(7):3365–3382. doi:10.1093/nar/gkz041.
- Fridman WH, Zitvogel L, Sautes-Fridman C, Kroemer G. The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol. 2017;14(12):717–734. doi:10.1038/nrclinonc.2017.101.
- Groth C, Hu X, Weber R, Fleming V, Altevogt P, Utikal J, Umansky V. Immunosuppression mediated by myeloid-derived suppressor cells (MDSCs) during tumour progression. Br J Cancer. 2019;120(1):16–25. doi:10.1038/s41416-018-0333-1.
- Aarts CEM, Kuijpers TW. Neutrophils as myeloid-derived suppressor cells. Eur J Clin Invest. 2018;48(Suppl 2):e12989. doi:10.1111/eci.12989.
- Hernandez-Caselles T, Martinez-Esparza M, Perez-Oliva AB, Quintanilla-Cecconi AM, García-Alonso A, Alvarez-López DMR, García-Peñarrubia P. A study of CD33 (SIGLEC-3) antigen expression and function on activated human T and NK cells: two isoforms of CD33 are generated by alternative splicing. J Leukoc Biol. 2006;79(1):46–58. doi:10.1189/jlb.0205096.
- Pillay J, Tak T, Kamp VM, Koenderman L. Immune suppression by neutrophils and granulocytic myeloid-derived suppressor cells: similarities and differences. Cell Mol Life Sci. 2013;70(20):3813–3827. doi:10.1007/s00018-013-1286-4.
- Dumitru CA, Moses K, Trellakis S, Lang S, Brandau S. Neutrophils and granulocytic myeloid-derived suppressor cells: immunophenotyping, cell biology and clinical relevance in human oncology. Cancer Immunol Immunother. 2012;61(8):1155–1167. doi:10.1007/s00262-012-1294-5.
- Sade-Feldman M, Kanterman J, Klieger Y, Ish-Shalom E, Olga M, Saragovi A, Shtainberg H, Lotem M, Baniyash M. Clinical Significance of Circulating CD33+ CD11b+ HLA-DR− Myeloid Cells in Patients with Stage IV Melanoma Treated with Ipilimumab. Clin Cancer Res. 2016;22(23):5661–5672. doi:10.1158/1078-0432.CCR-15-3104.
- Stanojevic I, Miller K, Kandolf-Sekulovic L, Mijuskovic Z, Zolotarevski L, Jovic M, Gacevic M, Djukic M, Arsenijevic N, Vojvodic D. A subpopulation that may correspond to granulocytic myeloid-derived suppressor cells reflects the clinical stage and progression of cutaneous melanoma. Int Immunol. 2016;28(2):87–97. doi:10.1093/intimm/dxv053.
- Martens A, Wistuba-Hamprecht K, Geukes Foppen M, Yuan J, Postow MA, Wong P, Romano E, Khammari A, Dreno B, Capone M, et al. Baseline peripheral blood biomarkers associated with clinical outcome of advanced melanoma patients treated with ipilimumab. Clin Cancer Res. 2016;22(12):2908–2918. doi:10.1158/1078-0432.CCR-15-2412.
- Hofman P, Badoual C, Henderson F, Berland L, Hamila M, Long-Mira E, Lassalle S, Roussel H, Hofman V, Tartour E, et al. Multiplexed immunohistochemistry for molecular and immune profiling in lung cancer-just about ready for prime-time? Cancers. 2019;11(3):283. doi:10.3390/cancers11030283.
- Ilie MI, Lassalle S, Long-Mira E, Bonnetaud C, Bordone O, Lespinet V, Lamy A, Sabourin J-C, Haudebourg J, Butori C, et al. Diagnostic value of immunohistochemistry for the detection of the BRAF V600E mutation in papillary thyroid carcinoma: comparative analysis with three DNA-based assays. Thyroid. 2014;24(5):858–866. doi:10.1089/thy.2013.0302.
- Ribas A, Hamid O, Daud A, Hodi FS, Wolchok JD, Kefford R, Joshua AM, Patnaik A, Hwu W-J, Weber JS, et al. Association of pembrolizumab with tumor response and survival among patients with advanced melanoma. JAMA. 2016;315(15):1600–1609. doi:10.1001/jama.2016.4059.
- Bai R, Lv Z, Xu D, Cui J. Predictive biomarkers for cancer immunotherapy with immune checkpoint inhibitors. Biomark Res. 2020;8(1):34. doi:10.1186/s40364-020-00209-0.
- Tray N, Weber JS, Adams S. Predictive biomarkers for checkpoint immunotherapy: current status and challenges for clinical application. Cancer Immunol Res. 2018;6(10):1122–1128. doi:10.1158/2326-6066.CIR-18-0214.
- Pilla L, Alberti A, Di Mauro P, Gemelli M, Cogliati V, Cazzaniga ME, Bidoli P, Maccalli C. Molecular and immune biomarkers for cutaneous melanoma: current status and future prospects. Cancers. 2020;12(11):3456. doi:10.3390/cancers12113456.
- D’adda Di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi:10.1038/nature02118.
- Rudolph KL, Millard M, Bosenberg MW, DePinho RA. Telomere dysfunction and evolution of intestinal carcinoma in mice and humans. Nat Genet. 2001;28(2):155–159. doi:10.1038/88871.
- Dinami R, Porru M, Amoreo CA. TRF2 and VEGF-A: an unknown relationship with prognostic impact on survival of colorectal cancer patients. J Exp Clin Cancer Res. 2020;39:111. doi:10.1186/s13046-020-01612-z.
- Hsu CP, Ko JL, Shai SE, Lee L-W. Modulation of telomere shelterin by TRF1 [corrected] and TRF2 interacts with telomerase to maintain the telomere length in non-small cell lung cancer. Lung Cancer. 2007;58:310–316. doi:10.1016/j.lungcan.2007.06.019.
- Oh BK, Kim YJ, Park C. Up-regulation of telomere-binding proteins, TRF1, TRF2, and TIN2 is related to telomere shortening during human multistep hepatocarcinogenesis. Am J Pathol. 2005;166:73–80. doi:10.1016/S0002-9440(10)62233-X.
- Biroccio A, Rizzo A, Elli R, Koering CE, Belleville A, Benassi B, Leonetti C, Stevens MFG, D'Incalci M, Zupi G, et al. TRF2 inhibition triggers apoptosis and reduces tumourigenicity of human melanoma cells. Eur J Cancer. 2006;42:1881–1888. doi:10.1016/j.ejca.2006.03.010.
- Choi JW, Kim YJ, Yun KA, Won CH, Lee MW, Choi JH, Chang SE, Lee WJ. The prognostic significance of VISTA and CD33-positive myeloid cells in cutaneous melanoma and their relationship with PD-1 expression. Sci Rep. 2020;10(1):14372. doi:10.1038/s41598-020-71216-2.
- Jordan KR, Amaria RN, Ramirez O, Callihan EB, Gao D, Borakove M, Manthey E, Borges VF, McCarter MD. Myeloid-derived suppressor cells are associated with disease progression and decreased overall survival in advanced-stage melanoma patients. Cancer Immunol Immunother. 2013;62(11):1711–1722. doi:10.1007/s00262-013-1475-x.
- Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9(3):162–174. doi:10.1038/nri2506.
- De Cicco P, Ercolano G, Ianaro A. The new era of cancer immunotherapy: targeting myeloid-derived suppressor cells to overcome immune evasion. Front Immunol. 2020;11:1680.
- Huber V, Vallacchi V, Fleming V, Hu X, Cova A, Dugo M, Shahaj E, Sulsenti R, Vergani E, Filipazzi P. Tumor-derived microRNAs induce myeloid suppressor cells and predict immunotherapy resistance in melanoma. J Clin Invest. 2018;128(12):5505–5516. doi:10.1172/JCI98060.
