ABSTRACT
Immunotherapy targeting the CD274 (PD-L1)/PDCD1 (PD-1) immune checkpoint axis has emerged as a promising treatment strategy for various cancers. Experimental evidence suggests that phosphatidylinositol-4,5-bisphosphonate 3-kinase (PI3K) signaling may upregulate CD274 expression. Thus, we hypothesized that PIK3CA mutation, PTEN loss, or their combined status might be associated with CD274 overexpression in colorectal carcinoma. We assessed tumor CD274 and PTEN expression by immunohistochemistry and assessed PIK3CA mutation by pyrosequencing in 753 patients among 4,465 incident rectal and colon cancer cases that had occurred in two U.S.-wide prospective cohort studies. To adjust for potential confounders and selection bias due to tissue availability, inverse probability weighted multivariable ordinal logistic regression analyses used the 4,465 cases and tumoral data including microsatellite instability, CpG island methylator phenotype, KRAS and BRAF mutations. PIK3CA mutation and loss of PTEN expression were detected in 111 of 753 cases (15%) and 342 of 585 cases (58%), respectively. Tumor CD274 expression was negative in 306 (41%), low in 195 (26%), and high in 252 (33%) of 753 cases. PTEN loss was associated with CD274 overexpression [multivariable odds ratio (OR) 1.83; 95% confidence interval (CI), 1.22–2.75; P = .004]. PIK3CA mutation was statistically-insignificantly (P = .036 with the stringent alpha level of 0.005) associated with CD274 overexpression (multivariable OR, 1.54; 95% CI, 1.03–2.31). PIK3CA-mutated PTEN-lost tumors (n = 33) showed higher prevalence of CD274-positivity (82%) than PIK3CA-wild-type PTEN-lost tumors (n = 204; 70% CD274-positivity) and PTEN-expressed tumors (n = 147; 50% CD274-positivity) (P = .003). Our findings support the role of PI3K signaling in the CD274/PDCD1 pathway.
Introduction
Over the past few decades, cancer immunotherapies have changed the landscape of cancer treatment. Among them, immune checkpoint inhibitors targeting PDCD1 (programmed cell death 1, PD-1) and its ligand, CD274 (programmed cell death 1 ligand 1, PD-L1) on tumor cells, are increasingly used for a variety of cancers, including colorectal cancer.Citation1,Citation2 Recent studies suggest that oncogenic activation of signaling pathways play an important role in CD274 upregulation in cancer cells.Citation3–5
Phosphatidylinositol-4,5-bisphosphonate 3-kinase (PI3K) signaling plays a central role in several cellular functions that are influential in oncogenesis and metastasis.Citation6,Citation7 PIK3CA encodes the catalytic subunit of PI3K that is involved in cell growth, proliferation, survival, and apoptosis, through induction of AKT phosphorylation and, subsequently, MTOR activation.Citation7 Conversely, PTEN counteracts this mechanism by dephosphorylating phosphatidylinositol-3,4,5-triphosphonate to phosphatidylinositol-4,5-biphosphonate.Citation7–9
Evidence indicates that PIK3CA mutation is associated with KRAS mutation in colorectal cancer and the prevalence of PIK3CA mutation gradually increases from rectum to cecum.Citation10–12 While PIK3CA mutation in colorectal cancer may not be a prognostic biomarker,Citation7,Citation10,Citation13–17 it may be a predictive biomarker for response to aspirin.Citation18–22 With regard to CD274 (PD-L1) overexpression in colorectal cancer, while it may not be a prognostic biomarker,Citation23–27 it has been shown to predict resistance to aspirin,Citation28 suggesting a possible interplay between the PTGS2 and PDCD1 pathways.Citation29 With regard to PTEN, evidence indicates that loss of PTEN expression is a potential therapeutic target in colorectal cancer.Citation30 Experimental studies suggest that activation of PI3K signaling may upregulate CD274 expression in certain experimental models,Citation4,Citation31,Citation32 including colon cancer cell lines.Citation26 However, as evidence indicates that tumor microenvironment can substantially change cellular gene expression profiles,Citation33,Citation34 experimental findings under artificial or non-human conditions need to be tested in human tumor tissue research. We therefore tested the hypothesis that PIK3CA mutation and PTEN loss might be associated with tumor CD274 overexpression in human colorectal cancer specimens.
To test our hypothesis, we used a database of 4,465 incident colorectal cancer cases, including 753 cases with available molecular data, from two large U.S.-wide prospective cohort studies. This comprehensive dataset allowed us to examine the association of PIK3CA mutation, PTEN loss, and their combined status with CD274 expression in tumor tissue after adjustment for potential confounders and selection bias due to tissue availability.
Materials and methods
Study population
We used two large prospective cohort studies in the U.S., the Nurses’ Health Study (NHS, 121,701 women aged 30–55 years followed since 1976) and the Health Professionals Follow-up Study (HPFS, 51,529 men aged 40–75 years followed since 1986).Citation35 Every two years, follow-up questionnaires were obtained to update information, such as lifestyle factors and newly-diagnosed diseases, including colorectal cancer. The overall response rate for these questionnaires was more than 90% in each follow-up questionnaire cycle. The National Death Index was used to confirm deaths of participants and identify unreported lethal colorectal cancer cases. We documented 4,465 colorectal cancer cases that had occurred in the two cohort studies during the follow-up until 2012. Participating physicians, who were blinded to exposure data, reviewed medical records of colorectal cancer patients to collect data on tumor characteristics including tumor size, tumor location, and disease stage based on the American Joint Committee on Cancer (AJCC) tumor, node and metastases classification, and identified causes of death for participants. We obtained formalin-fixed paraffin-embedded (FFPE) tumor tissue samples from the hospitals where participants underwent tumor resection. A single pathologist (S.O.), blinded to other data, reviewed all hematoxylin and eosin-stained tissue sections of colorectal cancer cases and recorded pathological features.Citation36 In this study, we utilized a molecular pathological epidemiology database of 753 colorectal cancer cases with available data on PIK3CA mutation status and CD274 (PD-L1) expression and 3,712 colorectal cancer cases without tissue data (). Comparison of clinical characteristics between cases with available tissue data and those without available tissue data is shown in Table S1. We included both colon and rectal cancers based on based on the colorectal continuum theory: i.e., a gradual change of clinical and tumor characteristics throughout the colorectum.Citation11,Citation37
Figure 1. Flow diagram of study population in the nurses’ health study and the health professionals follow-up study
Informed consent was obtained from all study participants at study enrollment. The study protocol was approved by the institutional review boards of the Brigham and Women’s Hospital and Harvard T.H. Chan School of Public Health (Boston, MA, USA), and those of participating registries as required.
Tumor tissue analyses
Tumor DNA was extracted from archival FFPE tissue sections with QIAamp DNA FFPE Tissue Kit. Microsatellite instability (MSI) status was examined using 10 microsatellite markers (D2S123, D5S346, D17S250, BAT25, BAT26, BAT40, D18S55, D18S56, D18S67, and D18S487), and MSI-high was defined as the presence of instability in ≥30% of the markers.Citation38 Methylation status of eight CpG island methylator phenotype (CIMP)-specific promoters (CACNA1G, CDKN2A, CRABP1, IGF2, MLH1, NEUROG1, RUNX3, and SOCS1) and long interspersed nucleotide element-1 (LINE-1) were assessed as previously described.Citation39–41 CIMP-high was defined as the presence of methylated promoters in at least six of the eight markers. Polymerase chain reaction and pyrosequencing were targeted for KRAS (codons 12, 13, 61, and 146), BRAF (codon 600), and PIK3CA (exons 9 and 20) to detect mutations.Citation22,Citation42 The PCR products were sequenced by Pyrosequencing PSQ96 HS System (Biotage AB) following the manufacturer’s instructions.Citation43
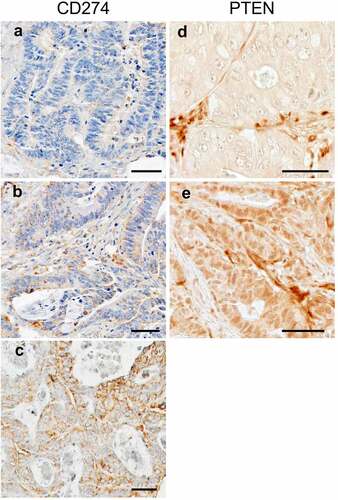
Tissue microarrays were constructed from FFPE tissue.Citation44 Immunohistochemical analyses of CD274 (PD-L1) and PTEN expression in tumor cells were performed using anti-CD274 antibody (Clone MIH1, dilution, 1:50; eBioscience) and anti-PTEN antibody (Clone 6H2.1, dilution 1:200; Abcam), respectively (), following standardized protein nomenclature recommended by the expert panel.Citation45 Blind to other data, immunohistochemical expression was recorded by a single investigator for each marker (CD274 by Y.M.; PTEN by K.N.). Tumor CD274 expression was evaluated based on immunostaining in the cytoplasm and membrane of tumor cells, as previously described.Citation24,Citation46 Tumor CD274 expression was interpreted as negative, low, or high. Appropriate positive and negative controls were included in each run of immunohistochemistry. PTEN expression was evaluated as intact in the presence of moderate or strong nuclear and cytoplasmic staining in tumor cells as previously described.Citation47 Loss of PTEN expression was defined as the absence of staining or only weak nuclear and/or cytoplasmic staining of tumor cells.Citation47 A second investigator (A.dS. for CD274; Y.B. for PTEN) independently reviewed 148 cases for CD274 expression and 109 cases for PTEN expression, and the weighted kappa values between the two independent investigators were 0.65 for CD274 expression (P < .001) and 0.45 for PTEN expression (P < .001).
Figure 2. Tumor CD274 and PTEN expression in colorectal cancer. Tumor CD274 expression was evaluated based on immunostaining in the cytoplasm and membrane of tumor cells. Tumor CD274 expression was interpreted as negative (a), low (b), or high (c) according to membranous and cytoplasmic intensity. Tumor PTEN expression was evaluated based on immunostaining in the cytoplasm and nuclear of tumor cells. Cytoplasmic and nuclear PTEN expression level was classified as lost (d) or intact (e) according to cytoplasmic and nuclear intensity. Scale bars represent 50 μm
Statistical analysis
All statistical analyses were performed using SAS software (version 9.4, SAS Institute, Cary, NC, USA). All P values were two-sided, and we used the stringent two-sided α level of 0.005 for our hypothesis testing, as recommended by a panel of expert statisticians.Citation48 Our primary hypothesis testing was an assessment of a statistical association of PIK3CA mutation status (wild-type and mutant; as a predictor variable) and PTEN expression (intact and lost; as an predictor variable) with CD274 expression score (negative, low, and high; as an ordinal outcome variable). All other analyses were secondary analyses. We used multivariable ordinal logistic regression models for our primary hypothesis testing. The proportional odds assumption was generally satisfied in ordinal logistic regression models (P > .05). The chi-square test was used to compare categorical data between PIK3CA mutation and PTEN expression categories.
To adjust for selection bias due to the availability of tumor tissue, we integrated the inverse probability weighting (IPW) method into multivariable ordinal logistic regression analyses using covariate data of the 4,465 incident colorectal cancer cases.Citation49,Citation50 Multivariable ordinal logistic regression analyses initially included sex (female vs. male), age at diagnosis (continuous), year of diagnosis (continuous), family history of colorectal cancer in any first-degree relatives (present vs. absent), tumor location (proximal colon vs. distal colon vs. rectum), tumor differentiation (well to moderate vs. poor), disease stage (I to II vs. III to IV), MSI status (MSI-high vs. non-MSI-high), CIMP (high vs. low/negative), LINE-1 methylation level (continuous), BRAF mutation (mutant vs. wild-type), and KRAS mutation (mutant vs. wild-type). To select variables for the final models, a threshold of P = .05 was used in a backward stepwise elimination procedure. The variables which remained in the final models are shown in . In this analysis, cases with missing data (family history of colorectal cancer in any first-degree relatives, 0.9%; tumor location, 0.3%; tumor differentiation, 0.3%; disease stage, 6.0%; MSI status, 2.4%; CIMP status, 1.7%; BRAF mutation, 1.4%; and KRAS mutation, 2.2%) were imputed as the most common category of the given variable to avoid overfitting of the models. For cases with missing data on LINE-1 methylation level (2.7%), we substituted the mean value and assigned a separate indicator variable. It was confirmed that no results were substantially altered after excluding the cases with missing information in any of the covariates. We categorized LINE-1 methylation level as low vs. high based on the median level. We conducted stratified analyses by LINE-1 methylation level (high vs. low) and CIMP status (high vs. low/negative), and assessed a statistical interaction using the Wald test for the cross-product term of PIK3CA mutation/PTEN expression and CIMP status or LINE-1 methylation level in the logistic regression model.
Results
PIK3CA mutation was detected in 111 (15%) of 753 cases, whereas loss of PTEN expression was detected 342 (58%) of 585 cases. Among 753 cases, tumor CD274 (PD-L1) expression was negative in 306 (41%), low in 195 (26%), and high in 252 (33%) cases. We summarized the clinical, pathological, and molecular characteristics of colorectal cancer cases according to PIK3CA mutation and PTEN expression in tumor tissue (). Loss of PTEN was associated with tumor CD274 overexpression (P < .001).
Table 1. Clinical, pathological, and molecular characteristics of colorectal cancer cases according to PIK3CA mutation and PTEN expression in tumor tissue
We used multivariable logistic regression model (to adjust for confounding) combined with inverse probability weighting (IPW) method on all 4,465 incident colorectal cancer cases to adjust for selection bias due to tissue availability (). Loss of PTEN was statistically significantly associated with higher CD274 expression [multivariable odds ratio (OR) 1.83; 95% confidence interval (CI), 1.22–2.75; P = .004]. PIK3CA mutation was statistically-insignificantly (P = .036 with the stringent alpha level of 0.005) associated with CD274 overexpression (multivariable OR, 1.54; 95% CI, 1.03–2.31). We confirmed that similar results were obtained by sensitivity analyses without IPW adjustment (Tables S2 and S3). We also stratified analyses by LINE-1 methylation level and CIMP status and did not observe significant effect modification (Table S4).
Table 2. Inverse probability weighting-adjusted ordinal logistic regression analysis to assess the association of tumor PIK3CA mutation or PTEN expression (predictor) with CD274 (PD-L1) expression (outcome)
We evaluated the association of the combined status of PIK3CA mutation and PTEN expression with CD274 overexpression (). PIK3CA-mutated PTEN-lost tumors (n = 33) showed higher prevalence of CD274-positivity (82%) than PIK3CA-wild-type PTEN-lost tumors (n = 204; 70% CD274-positivity), PIK3CA-mutated PTEN-expressed tumors (n = 24; 50% CD274-positivity), and PIK3CA-wild-type PTEN-expressed tumors (n = 123; 50% CD274-positivity) (P = .003). In the multivariable ordinal logistic regression model, the coexistence of PIK3CA mutation and loss of PTEN expression was significantly associated with CD274 overexpression (multivariable OR, 3.70; 95% CI, 1.69–8.19, compared to PIK3CA wild-type PTEN-intact tumors; P = .001) ().
Table 3. CD274 (PD-L1) expression score according to PIK3CA mutation and PTEN expression in tumor tissue
Table 4. Inverse probability weighting-adjusted ordinal logistic regression analysis to assess the association of the combination of PIK3CA mutation and PTEN expression (predictor) with CD274 (PD-L1) expression (outcome)
Discussion
We conducted this study to test the hypothesis that tumor PIK3CA mutation or PTEN loss might be associated with tumor CD274 (PD-L1) expression levels in colorectal cancer. We found that loss of PTEN was statistically significantly associated with higher CD274 expression, independent of other molecular features, including MSI status, CIMP status, and LINE-1 methylation level. In addition, the combination of PIK3CA mutation and loss of PTEN expression was more strongly associated with higher CD274 expression than loss of PTEN expression alone, suggesting that PIK3CA mutation may have additional influences on CD274 expression.
To our best knowledge, only one prior study has examined the association between PTEN loss and CD274 expression in human colorectal cancer specimens (from 314 cases).Citation26 Although another study has examined PIK3CA mutation in relation to CD274 expression in 66 colorectal cancer patients, the association could not be assessed because the sample size for this comparison was small.Citation51 The current study is the largest study that assessed PIK3CA mutation and PTEN loss (and the first study that assessed their combined status) in relation to CD274 expression in colorectal cancer. It is important to further investigate the consequences of activated PI3K signaling, as our findings suggest a potential role for the PI3K signaling pathway in CD274 (PD-L1) upregulation in colorectal cancer.
The PI3K signaling pathway is crucial in numerous cellular processes, including metabolism, cell survival, differentiation, proliferation, motility, and angiogenesis.Citation52 Evidence suggests that aberrant alterations of the PI3K pathway by either PIK3CA mutation or PTEN loss are potential predictive biomarkers for adjuvant therapy in colorectal cancer.Citation15,Citation22,Citation30 Recent studies have shown that activation of the PI3K pathway regulates tumor-intrinsic and immune-intrinsic features of the immunosuppressive tumor microenvironment.Citation53,Citation54 In particular, PTEN abrogation generates an immune-suppressive microenvironment by altering cytokine secretion patterns.Citation8 Moreover, a few studies have indicated that activation of PI3K signaling may promote immune escape through regulating PDCD1 (PD-1)/CD274 expression.Citation3,Citation55 In triple-negative breast cancer, a study reported that knockdown of PTEN genes led to high CD274 levels and decreased T-cell proliferation and increased apoptosis.Citation4 Other studies have shown that the inhibition of the PI3K pathway results in CD274 downregulation in various cancer cell lines.Citation3,Citation5,Citation56 We previously assessed the association between tumor CD274 expression and T cell infiltration.Citation46 We found that tumor CD274 expression level was not associated with overall T cell density but inversely associated with FOXP3+ cell densities,Citation46 suggesting that the PDCD1 immune checkpoint pathway and regulatory T cells infiltration may be generally mutually exclusive mechanisms of immune evasion. Although more research is needed to clarify the downstream signaling of PDCD1, our findings, based on a large U.S. nationwide sample of human colorectal cancers, support the association of loss of PTEN expression with higher CD274 expression, spurring subsequent studies to assess whether PI3K pathway inhibition can be exploited as a new treatment strategy to supplement immune checkpoint inhibition in colorectal cancer.Citation8,Citation57
The mechanisms by which the PI3K signaling pathway upregulate CD274 expression remain to be fully characterized. Although direct transcriptional upregulation by the PI3K pathway may be responsible for higher CD274 expression in breast cancer cell lines,Citation4 post-translational mechanisms have also been implicated in colorectal cancer or other cancer cell lines.Citation26,Citation31,Citation32 It is possible that the PI3K signaling pathway upregulate CD274 expression via transcriptional and/or post-transcriptional mechanisms depending on tumor type. Evidence also indicates that tumor CD274 expression differs by colorectal cancer molecular subtypes.Citation46,Citation58 More evidence from in vitro and in vivo studies of different tumor types is needed to clarify the mechanisms. Mounting evidence suggests that epigenetic aberrations contribute to cancer development.Citation59 LINE-1 hypomethylation, which reflects the global DNA hypomethylation, has been associated with poor clinical outcomes in colorectal cancer.Citation60,Citation61 CIMP-high colorectal cancer represents a subset of colorectal cancer developing through epigenetic instability.Citation62–65 We conducted stratified analyses by LINE-1 methylation level and CIMP status, and assessed the effect of those markers on our results. However, there was little evidence for substantial effect modification.
We acknowledge several limitations in this study. First, our study examined PIK3CA mutation and PTEN loss but did not examine somatic mutations in PTEN gene.Citation66 However, the majority of cases with PTEN loss in colorectal cancer have been attributed to epigenetic causes such as hypermethylation,Citation30 and immunohistochemistry used in this study is able to detect loss of protein expression irrespective of cause. Second, measurement errors may exist in molecular tissue data. However, such errors would likely be nearly randomly distributed and drive our results toward the null hypothesis. Third, our study was an observational, cross-sectional analysis, and further in vivo and in vitro experimental studies are needed to elucidate the mechanisms underlying our findings. Lastly, in this study, separate single-color immunohistochemistry assays did not allow the examination of co-expression patterns of PTEN and CD274 at the single cell level. Therefore, multiplex immunohistochemistry or immunofluorescence assays should be considered in future studies.
This study has notable strengths. First, the integrated molecular pathological epidemiologyCitation67,Citation68 database of clinical, pathological, and tumor molecular characteristics allowed us to rigorously investigate the potential interactive association of the PI3K pathway and tumor PDCD1/CD274 axis in colorectal cancer. Moreover, our prospective cohort studies enabled us to adjust for selection bias due to tissue availability utilizing the 4,465 incident colorectal cancer cases.Citation50 We have conducted a separate analysis that examined lymphocytic reaction patterns in relation to colorectal cancer survival, using the same cohort studies.Citation69 In the current study, we tested the hypothesis on CD274 expression in relation to PIK3CA mutation and loss of PTEN expression. As illustrated by these studies, because the large integrated database of clinical, pathological, and tumor molecular characteristics has been established, we can utilize it to test different hypotheses in an efficient and robust manner. In addition, cases and specimens in our study were drawn from a large number of hospitals located throughout the U.S., which increased the generalizability of our findings.
In conclusion, our data indicate that PI3K pathway activation by PTEN loss and/or PIK3CA mutation is associated with CD274 (PD-L1) overexpression in colorectal tumor tissue, supporting the role of PI3K signaling in the CD274 upregulation.
Abbreviations
AJCC, American Joint Committee on Cancer; CI, confidence interval; CIMP, CpG island methylator phenotype; FFPE, formalin-fixed paraffin-embedded; HPFS, Health Professionals Follow-up Study; IPW, inverse probability weighting; LINE-1, long interspersed nucleotide element-1; MSI, microsatellite instability; NHS, Nurses’ Health Study; OR, odds ratio; PD-1, programmed cell death 1; PD-L1, programmed cell death 1 ligand 1; PI3K, phosphatidylinositol-4,5-bisphosphonate 3-kinase.
Role of the sponsors
The funders had no role in design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript.
Use of standardized official symbols
We use HUGO (Human Genome Organization)-approved official symbols (or root symbols) for genes and gene products, including AKT, BRAF, CACNA1G, CD274, CDKN2A, CRABP1, IGF2, KRAS, MLH1, MTOR, NEUROG1, PDCD1, PIK3CA, PTEN, PTGS2, RUNX3, and SOCS1; all of which are described at www.genenames.org. Gene symbols are italicized whereas symbols for gene products are not italicized.
Supplemental Material
Download ()Acknowledgments
We would like to thank the participants and staff of the Nurses’ Health Study and the Health Professionals Follow-up Study for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. The authors assume full responsibility for analyses and interpretation of these data.
Availability of data and materials
The datasets generated and/or analyzed in this study are not publicly available. Further information including the procedures to obtain and access data from the Nurses’ Health Studies and the Health Professionals
Follow-up Study are described at https://www.nurseshealthstudy.org/researchers/ and
Disclosure statement
A.T.C. previously served as a consultant for Bayer Healthcare and Pfizer Inc. M.G. has received research funding from Bristol-Myers Squibb, Merck and Servier. J.A.M. has received institutional research funding from Boston Biomedical, has served as an advisor/consultant to Ignyta and COTA Healthcare, and served on a grant review panel for the National Comprehensive Cancer Network funded by Taiho Pharmaceutical. This study was not funded by any of these commercial entities.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
Additional information
Funding
References
- Yearley JH, Gibson C, Yu N, Moon C, Murphy E, Juco J, Lunceford J, Cheng J, Chow LQM, Seiwert TY, et al. PD-L2 expression in human tumors: relevance to Anti-PD-1 therapy in cancer. Clin Cancer Res. 2017;23(12):3158–11. doi:10.1158/1078-0432.CCR-16-1761.
- Tunger A, Sommer U, Wehner R, Kubasch AS, Grimm MO, Bachmann MP, Platzbecker U, Bornhäuser M, Baretton G, Schmitz M. The evolving landscape of biomarkers for anti-PD-1 or ANTI-PD-L1 therapy. J Clin Med. 2019;8(10):1534.
- Lastwika KJ, Wilson W, Li QK, Norris J, Xu H, Ghazarian SR, Kitagawa H, Kawabata S, Taube JM, Yao S, et al. Control of PD-L1 expression by oncogenic activation of the AKT-mTOR pathway in non-small cell lung cancer. Cancer Res. 2016;76(2):227–238. doi:10.1158/0008-5472.CAN-14-3362.
- Mittendorf EA, Philips AV, Meric-Bernstam F, Qiao N, Wu Y, Harrington S, Su X, Wang Y, Gonzalez-Angulo AM, Akcakanat A, et al. PD-L1 expression in triple-negative breast cancer. Cancer Immunol Res. 2014;2(4):361–370. doi:10.1158/2326-6066.CIR-13-0127.
- Jiang X, Zhou J, Giobbie-Hurder A, Wargo J, Hodi FS. The activation of MAPK in melanoma cells resistant to BRAF inhibition promotes PD-L1 expression that is reversible by MEK and PI3K inhibition. Clin Cancer Res. 2013;19(3):598–609. doi:10.1158/1078-0432.CCR-12-2731.
- Koveitypour Z, Panahi F, Vakilian M, Peymani M, Seyed Forootan F, Nasr Esfahani MH, Ghaedi K. Signaling pathways involved in colorectal cancer progression. Cell Biosci. 2019;9(1):97. doi:10.1186/s13578-019-0361-4.
- Papadatos-Pastos D, Rabbie R, Ross P, Sarker D. The role of the PI3K pathway in colorectal cancer. Crit Rev Oncol Hematol. 2015;94(1):18–30. doi:10.1016/j.critrevonc.2014.12.006.
- Cretella D, Digiacomo G, Giovannetti E, Cavazzoni A. PTEN Alterations As A Potential Mechanism For Tumor Cell Escape from PD-1/PD-L1 inhibition. Cancers (Basel). 2019;11(9):1318.
- Molinari F, Frattini M. Functions and regulation of the PTEN gene in colorectal cancer. Front Oncol. 2013;3:326.
- Rosty C, Young JP, Walsh MD, Clendenning M, Sanderson K, Walters RJ, Parry S, Jenkins MA, Win AK, Southey MC, et al. PIK3CA activating mutation in colorectal carcinoma: associations with molecular features and survival. PLoS One. 2013;8(6):e65479. doi:10.1371/journal.pone.0065479.
- Yamauchi M, Morikawa T, Kuchiba A, Imamura Y, Qian ZR, Nishihara R, Liao X, Waldron L, Hoshida Y, Huttenhower C, et al. Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut. 2012;61(6):847–854. doi:10.1136/gutjnl-2011-300865.
- Liao X, Morikawa T, Lochhead P, Imamura Y, Kuchiba A, Yamauchi M, Nosho K, Qian ZR, Nishihara R, Meyerhardt JA, et al. Prognostic role of PIK3CA mutation in colorectal cancer: cohort study and literature review. Clin Cancer Res. 2012;18(8):2257–2268. doi:10.1158/1078-0432.CCR-11-2410.
- Ogino S, Liao X, Imamura Y, Yamauchi M, McCleary NJ, Ng K, Niedzwiecki D, Saltz LB, Mayer RJ, Whittom R, et al. Predictive and prognostic analysis of PIK3CA mutation in stage III colon cancer intergroup trial. J Natl Cancer Inst. 2013;105(23):1789–1798. doi:10.1093/jnci/djt298.
- Vogelaar FJ, Erning FNV, Reimers MS, Van Der Linden H, Pruijt H, Van Den Brule AJC, Bosscha K. The prognostic value of microsatellite instability, KRAS, BRAF and PIK3CA mutations in stage II colon cancer patients. Mol Med. 2016;21(1):1038–1046. doi:10.2119/molmed.2015.00220.
- Mei ZB, Duan CY, Li CB, Cui L, Ogino S. Prognostic role of tumor PIK3CA mutation in colorectal cancer: a systematic review and meta-analysis. Ann Oncol. 2016;27(10):1836–1848. doi:10.1093/annonc/mdw264.
- Manceau G, Marisa L, Boige V, Duval A, Gaub M-P, Milano G, Selves J, Olschwang S, Jooste V, le Legrain M, et al. PIK3CA mutations predict recurrence in localized microsatellite stable colon cancer. Cancer Med. 2015;4(3):371–382. doi:10.1002/cam4.370.
- Ogino S, Nosho K, Kirkner GJ, Shima K, Irahara N, Kure S, Chan AT, Engelman JA, Kraft P, Cantley LC, et al. PIK3CA mutation is associated with poor prognosis among patients with curatively resected colon cancer. J Clin Oncol. 2009;27(9):1477–1484. doi:10.1200/JCO.2008.18.6544.
- Zumwalt TJ, Wodarz D, Komarova NL, Toden S, Turner J, Cardenas J, Burn J, Chan AT, Boland CR, Goel A, et al. Aspirin-Induced chemoprevention and response kinetics are enhanced by PIK3CA mutations in colorectal cancer cells. Cancer Prev Res (Phila). 2017;109(3):208–218. doi:10.1158/1940-6207.CAPR-16-0175.
- Gu M, Nishihara R, Chen Y, Li W, Shi Y, Masugi Y, Hamada T, Kosumi K, Liu L, Da Silva A, et al. Aspirin exerts high anti-cancer activity in PIK3CA-mutant colon cancer cells. Oncotarget. 2017;8(50):87379–87389. doi:10.18632/oncotarget.20972.
- Paleari L, Puntoni M, Clavarezza M, DeCensi M, Cuzick J, DeCensi A. PIK3CA mutation, aspirin use after diagnosis and survival of colorectal cancer. a systematic review and meta-analysis of epidemiological studies. Clin Oncol (R Coll Radiol). 2016;28(5):317–326. doi:10.1016/j.clon.2015.11.008.
- Domingo E, Church DN, Sieber O, Ramamoorthy R, Yanagisawa Y, Johnstone E, Davidson B, Kerr DJ, Tomlinson IPM, Midgley R, et al. Evaluation of PIK3CA mutation as a predictor of benefit from nonsteroidal anti-inflammatory drug therapy in colorectal cancer. J Clin Oncol. 2013;31(34):4297–4305. doi:10.1200/JCO.2013.50.0322.
- Liao X, Lochhead P, Nishihara R, Morikawa T, Kuchiba A, Yamauchi M, Imamura Y, Qian ZR, Baba Y, Shima K, et al. Aspirin use, tumor PIK3CA mutation, and colorectal-cancer survival. N Engl J Med. 2012;367(17):1596–1606. doi:10.1056/NEJMoa1207756.
- Berntsson J, Eberhard J, Nodin B, Leandersson K, Larsson AH, Jirström K. Expression of programmed cell death protein 1 (PD-1) and its ligand PD-L1 in colorectal cancer: relationship with sidedness and prognosis. Oncoimmunology. 2018;7:e1465165. doi:10.1080/2162402X.2018.1465165.
- Masugi Y, Nishihara R, Hamada T, Song M, Da Silva A, Kosumi K, Gu M, Shi Y, Li W, Liu L, et al. Tumor PDCD1LG2 (PD-L2) expression and the lymphocytic reaction to colorectal cancer. Cancer Immunol Res. 2017;5(11):1046–1055. doi:10.1158/2326-6066.CIR-17-0122.
- Li Y, Liang L, Dai W, Cai G, Xu Y, Li X, Li Q, Cai S. Prognostic impact of programed cell death-1 (PD-1) and PD-ligand 1 (PD-L1) expression in cancer cells and tumor infiltrating lymphocytes in colorectal cancer. Mol Cancer. 2016;15(1):55. doi:10.1186/s12943-016-0539-x.
- Song M, Chen D, Lu B, Wang C, Zhang J, Huang L, Wang X, Timmons CL, Hu J, Liu B, et al. PTEN loss increases PD-L1 protein expression and affects the correlation between PD-L1 expression and clinical parameters in colorectal cancer. PLoS One. 2013;8(6):e65821. doi:10.1371/journal.pone.0065821.
- Droeser RA, Hirt C, Viehl CT, Frey DM, Nebiker C, Huber X, Zlobec I, Eppenberger-Castori S, Tzankov A, Rosso R, et al. Clinical impact of programmed cell death ligand 1 expression in colorectal cancer. Eur J Cancer. 2013;49(9):2233–2242. doi:10.1016/j.ejca.2013.02.015.
- Hamada T, Cao Y, Qian ZR, Masugi Y, Nowak JA, Yang J, Song M, Mima K, Kosumi K, Liu L, et al. Aspirin use and colorectal cancer survival according to tumor CD274 (Programmed Cell Death 1 Ligand 1) expression status. J Clin Oncol. 2017;35(16):1836–1844. doi:10.1200/JCO.2016.70.7547.
- Hutchinson L. Immunotherapy: evading immune escape: synergy of COX and immune-checkpoint inhibitors. Nat Rev Clin Oncol. 2015;12:622.
- Salvatore L, Calegari MA, Loupakis F,Fassan M, Di Stefano B, Bensi M, Bria E, Tortora G. PTEN in colorectal cancer: shedding light on its role as predictor and target. Cancers (Basel). 2019;11(11):1765.
- Crane CA, Panner A, Murray JC, Wilson SP, Xu H, Chen L, Simko JP, Waldman FM, Pieper RO, Parsa AT, et al. PI(3) kinase is associated with a mechanism of immunoresistance in breast and prostate cancer. Oncogene. 2009;28(2):306–312. doi:10.1038/onc.2008.384.
- Parsa AT, Waldron JS, Panner A, Crane CA, Parney IF, Barry JJ, Cachola KE, Murray JC, Tihan T, Jensen MC, et al. Loss of tumor suppressor PTEN function increases B7-H1 expression and immunoresistance in glioma. Nat Med. 2007;13(1):84–88. doi:10.1038/nm1517.
- Oczko-Wojciechowska M, Pfeifer A, Jarzab M, Swierniak M, Rusinek D, Tyszkiewicz T, Kowalska M, Chmielik E, Zembala-Nozynska E, Czarniecka A, et al. Impact of the tumor microenvironment on the gene expression profile in papillary thyroid cancer. Pathobiology. 2020;87(Suppl. 2):143–154. doi:10.1159/000507223.
- Gil J, Ramsey D, Pawlowski P, Szmida E, Leszczynski P, Bebenek M, Sasiadek MM. The influence of tumor microenvironment on ATG4D gene expression in colorectal cancer patients. Med Oncol. 2018;35(12):159. doi:10.1007/s12032-018-1220-6.
- Nishihara R, Wu K, Lochhead P, Morikawa T, Liao X, Qian ZR, Inamura K, Kim SA, Kuchiba A, Yamauchi M, et al. Long-term colorectal-cancer incidence and mortality after lower endoscopy. N Engl J Med. 2013;369(12):1095–1105. doi:10.1056/NEJMoa1301969.
- Ogino S, Nosho K, Irahara N, Meyerhardt JA, Baba Y, Shima K, Glickman JN, Ferrone CR, Mino-Kenudson M, Tanaka N, et al. Lymphocytic reaction to colorectal cancer is associated with longer survival, independent of lymph node count, microsatellite instability, and CpG island methylator phenotype. Clin Cancer Res. 2009;15(20):6412–6420. doi:10.1158/1078-0432.CCR-09-1438.
- Yamauchi M, Lochhead P, Morikawa T, Huttenhower C, Chan AT, Giovannucci E, Fuchs C, Ogino S. Colorectal cancer: a tale of two sides or a continuum?: figure 1. Gut. 2012;61(6):794–797. doi:10.1136/gutjnl-2012-302014.
- Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Meyerhardt JA, Loda M, Giovannucci EL, Fuchs CS. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58(1):90–96. doi:10.1136/gut.2008.155473.
- Irahara N, Nosho K, Baba Y, Shima K, Lindeman NI, Hazra A, Schernhammer ES, Hunter DJ, Fuchs CS, Ogino S. Precision of pyrosequencing assay to measure LINE-1 methylation in colon cancer, normal colonic mucosa, and peripheral blood cells. J Mol Diagn. 2010;12(2):177–183. doi:10.2353/jmoldx.2010.090106.
- Nosho K, Irahara N, Shima K, Kure S, Kirkner GJ, Schernhammer ES, Hazra A, Hunter DJ, Quackenbush J, Spiegelman D, et al. Comprehensive biostatistical analysis of CpG island methylator phenotype in colorectal cancer using a large population-based sample. PLoS One. 2008;3(11):e3698. doi:10.1371/journal.pone.0003698.
- Ogino S, Kawasaki T, Brahmandam M, Cantor M, Kirkner GJ, Spiegelman D, Makrigiorgos GM, Weisenberger DJ, Laird PW, Loda M, et al. Precision and performance characteristics of bisulfite conversion and real-time PCR (MethyLight) for quantitative DNA methylation analysis. J Mol Diagn. 2006;8(2):209–217. doi:10.2353/jmoldx.2006.050135.
- Imamura Y, Lochhead P, Yamauchi M, Kuchiba A, Qian Z, Liao X, Nishihara R, Jung S, Wu K, Nosho K, et al. Analyses of clinicopathological, molecular, and prognostic associations of KRAS codon 61 and codon 146 mutations in colorectal cancer: cohort study and literature review. Mol Cancer. 2014;13(1):135. doi:10.1186/1476-4598-13-135.
- Ogino S, Kawasaki T, Brahmandam M, Yan L, Cantor M, Namgyal C, Mino-Kenudson M, Lauwers GY, Loda M, Fuchs CS. Sensitive sequencing method for KRAS mutation detection by Pyrosequencing. J Mol Diagn. 2005;7(3):413–421. doi:10.1016/S1525-1578(10)60571-5.
- Chan AT, Ogino S, Fuchs CS. Aspirin and the risk of colorectal cancer in relation to the expression of COX-2. N Engl J Med. 2007;356(21):2131–2142. doi:10.1056/NEJMoa067208.
- Fujiyoshi K, Bruford EA, Mroz P, Sims CL, O'Leary TJ, Lo AWI, Chen N, Patel NR, et al. Opinion: standardizing gene product nomenclature-a call to action. Proc Natl Acad Sci U S A. 2021;118(3):e2025207118.
- Masugi Y, Nishihara R, Yang J, Mima K, Da Silva A, Shi Y, Inamura K, Cao Y, Song M, Nowak JA, et al. Tumour CD274 (PD-L1) expression and T cells in colorectal cancer. Gut. 2017;66(8):1463–1473. doi:10.1136/gutjnl-2016-311421.
- Lotan TL, Gurel B, Sutcliffe S, Esopi D, Liu W, Xu J, Hicks JL, Park BH, Humphreys E, Partin AW, et al. PTEN protein loss by immunostaining: analytic validation and prognostic indicator for a high risk surgical cohort of prostate cancer patients. Clin Cancer Res. 2011;17(20):6563–6573. doi:10.1158/1078-0432.CCR-11-1244.
- Benjamin DJ, Berger JO, Johannesson M, Nosek BA, Wagenmakers EJ, Berk R, Bollen KA, Brembs B, Brown L, Camerer C, et al. Redefine statistical significance. Nat Hum Behav. 2018;2:6–10.
- Haruki K, Kosumi K, Hamada T, Twombly TS, Väyrynen JP, Kim SA, Masugi Y, Qian ZR, Mima K, Baba Y, et al. Association of autophagy status with amount of fusobacterium nucleatum in colorectal cancer. J Pathol. 2020;250(4):397–408. doi:10.1002/path.5381.
- Liu L, Nevo D, Nishihara R, Cao Y, Song M, Twombly TS, Chan AT, Giovannucci EL, VanderWeele TJ, Wang M, et al. Utility of inverse probability weighting in molecular pathological epidemiology. Eur J Epidemiol. 2018;33(4):381–392. doi:10.1007/s10654-017-0346-8.
- Inaguma S, Lasota J, Wang Z, Felisiak-Golabek A, Ikeda H, Miettinen M. Clinicopathologic profile, immunophenotype, and genotype of CD274 (PD-L1)-positive colorectal carcinomas. Mod Pathol. 2017;30(2):278–285. doi:10.1038/modpathol.2016.185.
- Mayer IA, Arteaga CL. The PI3K/AKT pathway as a target for cancer treatment. Annu Rev Med. 2016;67(1):11–28. doi:10.1146/annurev-med-062913-051343.
- Chida K, Kawazoe A, Kawazu M, Suzuki T, Nakamura Y, Nakatsura T, Kuwata T, Ueno T, Kuboki Y, Kotani D, et al. A low tumor mutational burden and PTEN mutations are predictors of a negative response to PD-1 blockade in MSI-H/dMMR gastrointestinal tumors. Clin Cancer Res. 2021;27(13):3714–3724. doi:10.1158/1078-0432.CCR-21-0401.
- O’Donnell JS, Massi D, Teng MWL, Mandala M. PI3K-AKT-mTOR inhibition in cancer immunotherapy, redux. Semin Cancer Biol. 2018;48:91–103. doi:10.1016/j.semcancer.2017.04.015.
- Ni JM, Ni AP. Landscape of PD-1/PD-L1 regulation and targeted immunotherapy. Chin Med Sci J. 2018;33:174–182.
- Balan M, Mier y Teran E, Waaga-Gasser AM, Gasser M, Choueiri TK, Freeman G, Pal S. Novel roles of c-Met in the survival of renal cancer cells through the regulation of HO-1 and PD-L1 expression. J Biol Chem. 2015;290(13):8110–8120. doi:10.1074/jbc.M114.612689.
- Peng W, Chen JQ, Liu C, Malu S, Creasy C, Tetzlaff MT, Xu C, McKenzie JA, Zhang C, Liang X. Loss of PTEN promotes resistance to T cell-mediated immunotherapy. Cancer Discov. 2016;6(2):202–216. doi:10.1158/2159-8290.CD-15-0283.
- Llosa NJ, Cruise M, Tam A, Wicks EC, Hechenbleikner EM, Taube JM, Blosser RL, Fan H, Wang H, Luber BS, et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2015;5(1):43–51. doi:10.1158/2159-8290.CD-14-0863.
- Bates SE, Longo DL. Epigenetic Therapies for Cancer. N Engl J Med. 2020;383(7):650–663. doi:10.1056/NEJMra1805035.
- Akimoto N, Zhao M, Ugai T, Zhong R, Lau MC, Fujiyoshi K, Kishikawa J, Haruki K, Arima K, Twombly TS, et al. Tumor Long Interspersed Nucleotide Element-1 (LINE-1) hypomethylation in relation to age of colorectal cancer diagnosis and prognosis. Cancers (Basel). 2021;13:(9):2016.
- Swets M, Zaalberg A, Boot A, van Wezel T, Frouws MA, Bastiaannet E, Gelderblom H, van de Velde CJ, Kuppen PJ. Tumor LINE-1 methylation level in association with survival of patients with stage II colon cancer. Int J Mol Sci. 2016;18(1):36.
- Vedeld HM, Merok M, Jeanmougin M, Danielsen SA, Honne H, Presthus GK, Svindland A, Sjo OH, Hektoen M, Eknaes M, et al. CpG island methylator phenotype identifies high risk patients among microsatellite stable BRAF mutated colorectal cancers. Int J Cancer. 2017;141(5):967–976. doi:10.1002/ijc.30796.
- Levine AJ, Phipps AI, Baron JA, Buchanan DD, Ahnen DJ, Cohen SA, Lindor NM, Newcomb PA, Rosty C, Haile RW, et al. Clinicopathologic risk factor distributions for MLH1 promoter region methylation in CIMP-Positive tumors. Cancer Epidemiol Biomarkers Prev. 2016;25(1):68–75. doi:10.1158/1055-9965.EPI-15-0935.
- Inamura K. Colorectal cancers: an update on their molecular pathology. Cancers (Basel). 2018;10(1):26.
- Advani SM, Advani P, DeSantis SM, Brown D, VonVille HM, Lam M, Loree JM, Mehrvarz Sarshekeh A, Bressler J, Lopez DS, et al. Clinical, pathological, and molecular characteristics of CpG island methylator phenotype in colorectal cancer: a systematic review and meta-analysis. Transl Oncol. 2018;11(5):1188–1201. doi:10.1016/j.tranon.2018.07.008.
- Tsai JH, Jeng YM, Yuan CT, Lin Y-L, Cheng M-L, Liau J-Y. Traditional serrated pathway-associated colorectal carcinoma: morphologic reappraisal of serrated morphology, tumor budding, and identification of frequent PTEN alterations. Am J Surg Pathol. 2019;43(8):1042–1051. doi:10.1097/PAS.0000000000001274.
- Akimoto N, Ugai T, Zhong R, Hamada T, Fujiyoshi K, Giannakis M, Wu K, Cao Y, Ng K, Ogino S. Rising incidence of early-onset colorectal cancer — a call to action. Nat Rev Clin Oncol. 2021;18(4):230–243. doi:10.1038/s41571-020-00445-1.
- Ogino S, Nowak JA, Hamada T, Milner DA, Nishihara R. Insights into pathogenic interactions among environment, host, and tumor at the crossroads of molecular pathology and epidemiology. Annu Rev Pathol. 2019;14(1):83–103. doi:10.1146/annurev-pathmechdis-012418-012818.
- Haruki K, Kosumi K, Li P, Arima K, Väyrynen JP, Lau MC, Twombly TS, Hamada T, Glickman JN, Fujiyoshi K, et al. An integrated analysis of lymphocytic reaction, tumour molecular characteristics and patient survival in colorectal cancer. Br J Cancer. 2020;122(9):1367–1377. doi:10.1038/s41416-020-0780-3.
