ABSTRACT
Anti-PD1/PD-L1-directed immune checkpoint inhibitors are game changers in advanced non-small-cell lung cancer, but biomarkers are lacking. The aim of our study was to find clinically relevant biomarkers of the efficacy of ICI in non-squamous NSCLC. We conducted a retrospective study of patients receiving ICI for advanced non squamous NSCLC in two cohorts. For a subset of patients, RNAseq data were generated on tumor biopsy taken before ICI. The primary end point was progression-free survival under ICI. Secondary end point was overall survival from ICI initiation. In the cohort, we studied 231 patients. Clinico-pathological characteristics included KRAS mutant status (n = 88), TTF1-positive expression (n = 136), LIPI (Lung Immune Prognostic Index) score of 0 (n = 116). In our cohort, lack of TTF1 expression, LIPI score >0, line of treatment >1, and liver metastases were associated with poorer PFS. TTF1 and PD-L1 status could be used to stratify survival and improve the AUC for prediction of prognosis in comparison with the PD-L1 gold standard. Using an external cohort of 154 patients, we confirmed the independent prognostic role of TTF1. TTF1 expression and PD-L1 can be used to stratify risk and predict PFS and OS in patients treated with ICI for NS-NSCLC.
Introduction
Non Small Cell Lung Cancer (NSCLC) is the most frequent thoracic cancer, and incidence reached 2 million new cases in 2019.Citation1 Non-squamous NSCLC (NS-NSCLC) is the most common histological subtype.Citation2 NS-NSCLC are characterized by various molecular alterations. The most common of these genetic alterations in lung adenocarcinoma are epidermal growth factor receptor (EGFR) and KRAS activating mutations. EGFR insertions and deletions are found in roughly 15% of NS-NSCLC,Citation3 while KRAS mutation incidence reaches 30% in patients with NS-NSCLC in Western countries.Citation4,Citation5 The majority (95%) of KRAS mutations in non NS-NSCLC occur in codons 12 (>80%) and 13.Citation3 The G12 C point mutation accounts for more than one-third of all cases.Citation3,Citation4 In the area of cytotoxic chemotherapies, KRAS mutation was thought to be associated with poorer clinical outcomes overall in advanced-stage NSCLC.Citation6–8 However, the negative impact of KRAS mutation on outcome has not been unequivocally confirmed in all studies.Citation9 Additional driver mutations in lung adenocarcinoma occur with a frequency of <1–4%, including ALK gene rearrangements, ROS1 translocations, HER2 mutations, BRAF mutations, and RET translocations.Citation10
In contrast to other activating genomic alterations found in NS-NSCLC, the development of targeted approaches for patients with KRAS-mutant NSCLC only very recently led to target therapies which could be used as standard treatment.Citation11 Thus, these patients are classically treated with cytotoxic chemotherapies and checkpoint inhibitors used either concomitantly or sequentially.Citation12 With the rapid clinical development of anti-programmed death 1 (PD-1/PD-L1) immune checkpoint inhibitors (ICI), patients with advanced NS-NSCLC have come to be treated with chemotherapy followed by anti PD-1 monoclonal antibodies (mAb) in recent years. Moreover, ICI are now becoming standard in the first-line treatment of advanced NS-NSLC as monotherapy for patients with high expression of PD-L1,Citation13,Citation14 and in association with chemotherapy for all-comers with NS-NSCLC.Citation15
Predicting the response to ICI remains an unmet need. Although a large number of biological studies have tested complex biomarkers,Citation16 the only approved biomarker remains PD-L1 immunohistochemistry. Nevertheless, despite the substantial correlation between PD-L1 expression and response or clinical efficacy on ICI, PD-L1 remains a controversial biomarker of immunotherapy response with several limits.Citation17,Citation18 Moreover, few studies have tested clinical biomarkers that are easy to implement in clinical routine practice. The inflammation process has been proposed as a mechanism of immunoresistance in cancer patients, and peripheral inflammatory status has been shown to be associated with worse outcomes in various cancer types.Citation19–21 Many blood parameters, such as elevated white blood cells, neutrophil count, platelet count, and high levels of lactate dehydrogenase (LDH) were found to be associated with poor outcomes in several cancer types.Citation21–24 In the context of ICI treatment, a recent large retrospective cohort of NSCLC patients underlined that a score called LIPI (Lung Immune Prognostic Index), based on a combination of the derived neutrophil/lymphocyte (leukocytes minus neutrophils) ratio (dNLR) and LDH levels, was associated with resistance to immunotherapy.Citation25
The role of KRAS as a biomarker of response to ICI remains an open question. In clinical trials comparing ICI with chemotherapy in second-line treatment, a benefit was suggested in patients with KRAS-mutant NS-NSCLC on the basis of an unplanned subgroup analysis.Citation26 KRAS mutations are frequently associated with high tumor mutational burden and smoking habits, two classical factors associated with better response to checkpoint inhibitors, and previous reports underlined that KRAS status was associated with response to immunotherapy.Citation27 Moreover, other recent data indicate that KRAS-mutant NS-NSCLC patients display heterogenous immune profilesCitation28 and, consequently, various levels of sensitivity to immunotherapy. No clinical trial with ICI for NS-NSCLC has used KRAS-mutant status as a stratification factor to address this question.Citation29
In this study, we aimed to determine the prognostic role of classically used routine markers such as KRAS status and LIPI score, in patients with NS-NSCLC treated with ICI. We also evaluate, in addition to the classically described biomarkers, the role of Thyroid Transcription Factor-1 (TTF1) expression as predictor of ICI efficacy.
Materials and methods
Study population
Four hundred and five patients with metastatic NSCLC receiving treatment with anti-programmed death 1 (PD-1/PD-L1) checkpoint inhibitors in the Georges François Leclerc Cancer Center between 2014 and 2020 were selected. Patients receiving immunotherapy treatment in adjuvant conditions were excluded. Patients with squamous carcinoma, small cell neuroendocrine carcinoma, unknown KRAS status or an EGFR alteration were excluded (Supplementary Figure 1).
A total of 231 patients were finally included and composed cohort 1. Only patients from whom informed consent was obtained and recorded in the medical chart were included in this retrospective study.
We also used a second cohort (called cohort 2) of 154 patients with advanced NS-NSCLC and treated with anti-PD1 ICI in the University Hospital of Caen (France).
This study was approved by the CNIL (French national commission for data privacy) and the local ethics committee, and was performed in accordance with the Helsinki Declaration and European legislation. For RNAseq analysis, this study falls within the scope of the biological collection authorization registered under the name AC-2014-2260.
Molecular analysis
Mutations in KRAS, EGFR, BRAF, PIK3CA and HER2 were investigated for all patients by either allelic discrimination or fragment analysis (Supplementary Table 1).
ALK receptor tyrosine kinase and ROS1 rearrangement were detected using an immunohistochemical procedure, and positive cases were controlled by fluorescent in situ hybridization.
TTF1 is a tissue-specific transcription factor which is often use by pathologist to determine the origin (thyroid, lung, or diencephalon) of tumors or to differentiate lung adenocarcinoma from squamous cell carcinoma. TTF1 staining was performed using Ventana automates with the 8G7 G3/1 clone (Zymed, France), which is the most specific antibody to identify lung adenocarcinoma.Citation30,Citation31 A focal positivity for TTF1 result was considered a positive reaction indicating pulmonary adenocarcinoma in the proper clinical context.
The pathologists were A.L.L.P, F.B. and V.D; they were blinded to the treatment outcome.
PD-L1 expression analysis
PD-L1 protein expression in tumor cells was assessed using immunohistochemistry with a ready-to-use PDL1 commercial kit with QR1 or 22C3 antibodies, PD-L1 positivity was defined as >1% of cells in tumor.
LIPI score calculation
Blood cell counts and LDH levels at baseline before ICI treatment (within 30 d prior to the first treatment) were obtained from the electronic medical records. Demographic, clinical, pathological and molecular data were also collected.
The LIPI (Lung Immune Prognostic Index) was developed on the basis of dNLR (leukocytes/ (leukocytes – neutrophils) >3) or LDH > 230 U/L (considered as the upper limit of normal in our center).Citation32 We considered two distinct groups: negative if neither of these two conditions was met, and positive if one or both conditions were met.
Statistical analysis
The evaluation of tumor response was performed after four to six immunotherapy injections. Progression-free-survival (PFS) was calculated from the date of first immunotherapy administration until disease progression or death from any cause, and was evaluated at 6 months. Patients who were alive with no progression at 6 months were censored. Overall survival (OS) was calculated from the date of first immunotherapy administration until death from any cause, and was censored at 1 y. Patient and disease characteristics were compared between KRAS mutated and WT patients using the Chi-2 or Fisher’s exact test for qualitative variables, or the Wilcoxon test for continuous variables, as appropriate.
Survival analysis was performed using the survival R library. The prognostic value of the different variables was tested using univariate and multivariate Cox regression models for OS and PFS. Survival probabilities were estimated using the Kaplan–Meier method and survival curves were compared using the log-rank test. P-values less than or equal to 0.05 were considered statistically significant. Composite scores were computed to test the combined predictive value of a combination of variables of interest. To this end, we separated patients into four groups, classified by a score ranging from 0 to 3, where scores were computed as the sum of each the variables of interest. The predictive values of these scores were estimated on the discovery cohort (cohort 1) and on the external cohort (cohort 2).
Statistical analyses were performed using the R software (http://www.R-project.org/) and graphs were drawn using GraphPad Prism version 7.0.3.
Results
Patients’ clinical characteristics of the discovery cohort (cohort 1)
In total, 231 patients treated with ICI (in first-line or more) for non-squamous NSCLC with available KRAS status were retained for analysis. Eighty-eight patients (38%) had KRAS mutation, and 143 (62%) had a wild-type KRAS phenotype. The main characteristics of the population are reported in , and molecular abnormalities in Supplementary Table 2.
Table 1. Summary of clinical and genomic characteristics
No significant statistical difference was found between the clinical characteristics of KRAS mutated and WT KRAS carriers, except for a trend toward a higher presence of PD-L1 positive tumor in the KRAS mutated group, as classically reported. The G12C point mutation accounted for approximately half of KRAS-mutation carriers (41 patients, 47%), G12V and G12D being other frequently found variants. Details are reported in Supplementary Table 3.
In the whole cohort, median dNLR was 2.23 (Interquartile range (IQR) = 1.54) and was >3 in 61 patients. Median LDH was 226 (IQR = 106) and was above the Upper Limit of Normal (ULN = 230) in 92 patients. One hundred and sixty-six (50.2%) patients presented a LIPI score ≥1. Patients with high LIPI presented more liver metastasis (Fisher’s exact test: p = .03). PD-L1 status was positive in 129 patients (56%), negative in 65 (28%) and not available in 37 (16%) patients. Patients with positive status PD-L1 presented less liver metastasis (Fisher’s exact test: p = .02).
One hundred and thirty-six (58.8%) patients presented a TTF1 positive tumor. No significant statistical difference was found between the clinical characteristics of TTF1 positive and TTF1 negative groups (Supplementary Table 4).
Clinical and genomic biomarkers of PFS and OS
In this study, the median follow-up was 2.9 months for PFS. In the overall cohort, using a univariate Cox model, presence of liver and bones metastasis, absence of PD-L1 and absence of TTF1 expression, and treatment in second line or more were significantly associated with poor PFS () and Supplementary Figure 2(a,b)). Surprisingly, WHO performance status >0 at initiation of immunotherapy was not associated with PFS. Using a multivariate model, we observed that treatment in second line or more, presence of liver metastasis and absence of TTF1 expression were independently associated with poor PFS ()).
Figure 1. Univariate and multivariate Cox models for progression-free and overall survival

For OS, by Cox univariate analysis, LDH > ULN, dNLR >3, LIPI score ≥1, absence of TTF1 expression, line of therapy >1, WHO performance status >0, absence of PD-L1, KRAS mutated status, presence of liver, cerebral and bone metastasis were associated with significantly poorer overall survival () and Supplementary Figure 2(c,d)). Using a multivariate model, we observed that treatment absence of PD-L1 or TTF1, presence of liver or bone metastasis and presence of KRAS mutation were independently associated with poor OS ()).
OS was significantly poorer in KRAS mutation carriers (HR = 1.62 [1.07; 2.45], p-value = 0.02); median OS was 10 months (IQR = 29.1) in KRAS mutated patients and 15.1 months (IQR = 20.1) in WT KRAS patients. No difference in prognosis was observed between KRAS G12C mutations and other mutations (median OS was 9.33 (IQR = 29.5) months in KRAS G12C mutations and 13.9 (IQR = 52.8) months in other KRAS mutations; HR = 0.71 [0.39; 1.32], p-value = 0.28).
Since LIPI and TTF1 were found to be associated (Fisher test p-value = 0.04), we generated two separate multivariate models, one with the LIPI score and one with TTF1 status. By multivariate Cox regression analysis in the model with the LIPI score, we found that WHO performance status >0, line of therapy >1, LIPI score ≥1 and liver metastasis were independent prognostic factors for PFS. Multivariate Cox analysis with TTF1 status showed that KRAS mutated status, absence of PD-L1, absence of TTF1 expression, and presence of liver or bone metastasis were associated with poorer OS ()).
By subgroup analysis, in the KRAS mutant group, LIPI score ≥1 was associated with poor PFS (HR = 2.39 [1.31; 4.33] p-value = 0.004) and poor OS (HR = 3.24 [1.58; 6.7], p-value = 0.001). In the KRAS WT group, the LIPI score ≥1 was not associated with poor PFS (HR = 0.76 [0.47; 1.23], p-value = 0.26) but remain associated poor OS (HR = 1.87 [0.97; 3.6] p-value = 0.06) (). Similarly, only in the KRAS mutant group, we observed that tumors harboring KRAS mutation and TTF1 negative expression were associated with poor PFS (HR = 2.82 [1.65; 4.83], p-value<1.10−3) and OS (HR = 3.83 [2; 7.29], p-value<1.10−3) ().
Figure 2. Multivariate Cox models with KRAS status for progression-free and overall survival
PD-L1 expression (>1% in cancer cells) in RAS WT carriers was none significantly associated with longer PFS and significantly with OS (respectively HR = 0.64 [0.4; 1.05], p-value = 0.08 and HR = 0.36 [0.19; 0.69], p-value = 0.002). In contrast, in RAS mutated patients, PD-L1 expression was not significantly associated with better outcome (HR = 0.63 [0.35; 1.14], p-value = 0.13 for PFS, and HR = 0.55 [0.29; 1.11], p-value = 0.1 for OS) (). Note that subgroup analysis underlined that TTF1 might be a better predictive factor for patients treated with PD-1 mAb (results not shown). However due to the low number of patients confirmatory studies are needed.
Together, these data underline that TTF1 and LIPI are prognostic factors and that KRAS mutated status is a factor of poor prognosis only in patients with LIPI score ≥1 or TTF1 negative tumors.
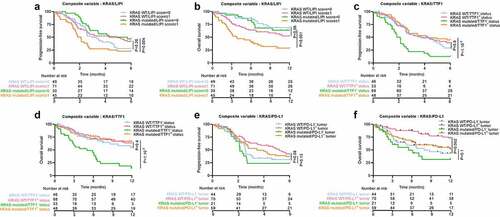
Using LIPI and KRAS status as a prognostic score to predict outcome
In view of the relevance of LIPI and KRAS as biomarkers, we hypothesized that combining these two variables with the classical PD-L1 biomarker would improve assessment of prognosis. To this end, we separated patients into four groups, classified by a score ranging from 0 to 3, where scores were computed as the sum of each the three variables: variables were scored 1 in case of KRAS mutated status, LIPI score ≥1 and absence of PD-L1 positive tumor cells. Despite the good predictive power for OS and PFS for models with LIPI/KRAS (Area Under the Curve (AUC) = 0.53 for PFS at 6 months and AUC = 0.65 for OS at 1 y), LIPI/PD-L1 (respectively AUC = 0.57 and AUC = 0.67) and KRAS/PD-L1 (respectively AUC = 0.59 and AUC = 0.69) ( for PFS and for OS), the KRAS/LIPI/PD-L1 score was a slightly better biomarker in particular for OS (respectively likelihood ratio test p-value = 0.002, 0.004 and 0.003 for OS and p = .02, 0.13 and 0.45 for PFS). AUC of the model combining LIPI score, KRAS status and PD-L1 expression was estimated at 0.59 for 6-month PFS ()) and 0.72 for 1-y OS ()). All composite biomarkers which include the gold standard PD-L1 biomarker improved the predictive value for OS compared to the gold standard PD-L1 biomarker (AUC = 0.61, likelihood ratio test p-value = 0.008 with KRAS/PD-L1, 0.004 with LIPI/PD-L1 and <1.10−3 with LIPI/KRAS/PD-L1) but results were not significant for PFS (AUC = 0.55, likelihood ratio test p-value = 0.15 with KRAS/PD-L1, 0.55 with LIPI/PD-L1 and 0.27 with LIPI/KRAS/PD-L1) () and ) and )).
Figure 3. Association between progression-free survival, and LIPI, KRAS, and PD-L1 status
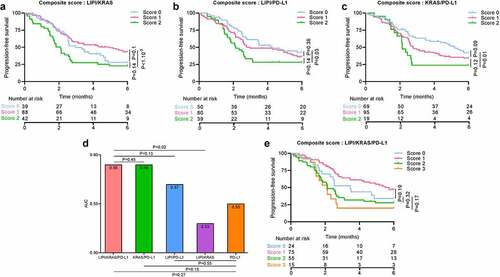
Figure 4. Association between overall survival, and LIPI, KRAS, and PD-L1 status
Taking patients with a score = 0 as a reference, patients with scores of 2 and 3 had worse OS (respectively HR = 3.95 [1.66; 9.42], p-value = 0.002; HR = 5.6 [2.1; 15.3], p-value < 1.10−3). No difference was observed compared to patients with a score = 1. For PFS, no significant differences were found between patients with score 0 and those with higher scores () and )).
Using TTF1 and KRAS to predict outcome in NS-NSCLC
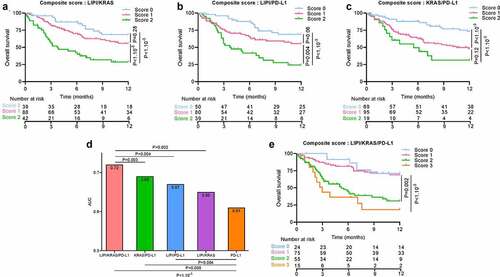
We investigated whether TTF1 could be also used together with KRAS as a valuable biomarker associated with outcome in this category of patients. As above, we separated patients in 4 groups, classified by a score ranging from 0 to 3, whereby one point was attributed for occurrence of KRAS mutated status, absence of TTF1 expression, and absence of PD-L1 positive tumor cells.
All pairs of biomarkers had good predictive power for PFS and OS: TTF1/KRAS (AUC = 0.61 for PFS and AUC = 0.65 for OS), TTF1/PD-L1 (respectively AUC = 0.62 and AUC = 0.69) and KRAS/PD-L1 (respectively AUC = 0.57 and AUC = 0.68) ( for PFS and for OS). Nevertheless, the PD-L1, TTF1, KRAS score outperformed the capacity of the paired-biomarker scores (AUC = 0.62 for PFS and AUC = 0.73 for OS, likelihood ratio test p-value for PFS respectively: 0.02, 0.08 and 0.01, for OS: <1.10−3, 0.006 and 0.005). All composite biomarkers including PD-L1 improved compared to the gold standard PD-L1 biomarker prediction for OS (AUC = 0.61, likelihood ratio test p-value = 0.05 with KRAS/PD-L1, 0.003 with TTF1/PD-L1 and <1.10−3 with TTF1/KRAS/PD-L1) and PFS (AUC = 0.55, likelihood ratio test p-value = 0.05 with KRAS/PD-L1, 0.01 with TTF1/PD-L1 and 0.001 with TTF1/KRAS/PD-L1) () and ) and )).
Figure 5. Association between progression-free survival and TTF1, KRAS, and PD-L1 status
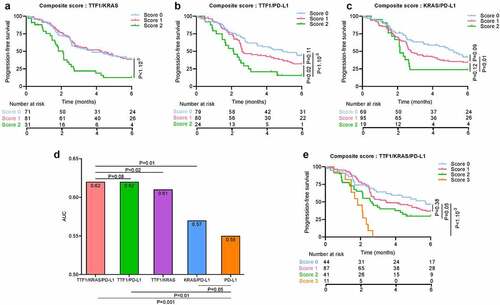
Figure 6. Association between overall survival and TTF1, KRAS, and PD-L1 status
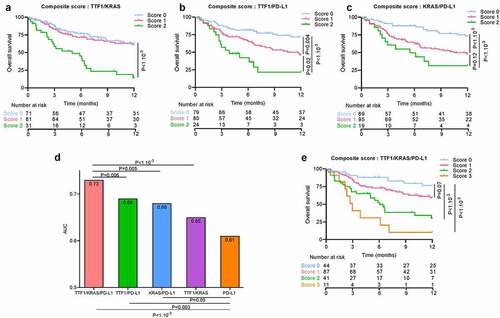
Taking patients with a score of 0 as the reference, patients with a score of 2 or 3 had worse PFS (respectively HR = 1.76 [1; 3.09], p-value = 0.05, HR = 5.01 [2.37; 10.61], p-value <b1.10−3) and worse OS (respectively HR = 4.41 [2.04; 9.5], p-value <b1.10−3, HR = 8.3 [3.3; 21.12], p-value <b1.10−3). No significant difference was observed for patients with a score of 1 () and )).
Transcriptomic and histological features related to LIPI, TTF1, and KRAS status
To link this clinical observation with a transcriptomic molecular pattern, we performed RNAseq analysis in a subset of 48 patients included in this cohort, namely 26 (54.2%) wild-type patients, 22 (45.8%) mutated tumors, 20 (41.7%) patients with a LIPI score of 0, 28 (58.3%) with LIPI score ≥1. Twenty-nine (60.4%) patients had TTF1 positive tumors and 19 (39.6%) patients had TTF1 negative tumors. Results are presented as Supplementary data.
External evaluation of the predictive role of TTF1 and KRAS to predict outcome in NS-NSCLC
To externally evaluate our results, we tested the prognostic role of TTF1 in a cohort of NS-NSCLC treated by ICI in Caen hospital (n = 154). The clinical characteristic of this cohort 2 were similar to our cohort, but with a higher percentage of TTF1 positive tumors (Supplementary Table 5).
In cohort 2, no significant difference was found for PFS or OS according KRAS status (). As in our cohort, we observed a non-significant trend toward poor PFS (HR = 1.6 [0.96; 2.68], p-value = 0.07) and significantly poorer OS (HR = 2.23 [1.3; 3.83], p-value = 0.004) in patients with TTF1 negative tumors (). The multivariate model for OS showed that TTF1 negative tumors, male sex and line of therapy >1 were associated with poor prognosis ()). As in our cohort, we showed that the TTF1/PD-L1 score outperformed the gold standard PD-L1 biomarker for OS (AUC = 0.59 for TTF1/PD-L1 and 0.55 for PD-L1 and likelihood ratio test p-value = 0.006) and was equivalent for PFS (AUC = 0.58 for both and likelihood ratio test p-value = 0.06) ().
Figure 7. Evaluation of the predictive role of TTF1 and KRAS in the external cohort
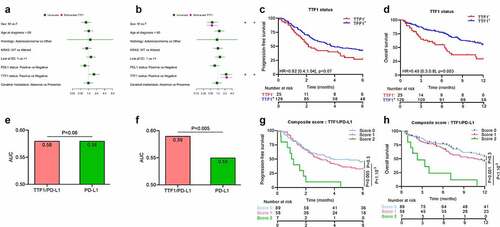
We separated patients into 3 groups classified by a score ranging from 0 to 2 points, attributing one point for absence of TTF1 expression, and absence of PD-L1 positive tumor cells. Taking patients with scores of 0 or 1 as a reference, patients with a score = 2 had worse PFS (respectively HR = 3.98 [1.8; 8.85], p-value < 1.10−3, HR = 3.23 [1.43; 7.25], p-value = 0.005) and a worse OS (respectively HR = 5.34 [2.34; 12.16], p-value < 1.10−3, HR = 4.05 [1.8; 9.35], p-value = 0.001) ().
Discussion
This retrospective study including two different cohorts brings to light some important clinico-pathological data that may help to better stratify patients treated with ICI. The most salient observation is that TTF1 expression is a predictive factor for better response to ICI. Secondly, we observed that in the first cohort of patients with NS-NSCLC treated with an ICI, KRAS mutated status is independently associated with poor prognosis only in TTF1-negative tumors. However, these findings could not be validated in the second cohort, likely due to a lack of statistical power. Finally, adding TTF1 assessment to the standard PD-L1 biomarker improves the prediction of prognosis in NS-NSCLC.
Conflicting data have previously been observed when analyzing interactions between KRAS status and response to ICI. For example, in the Checkmate 057 study,Citation26 a potential advantage for immunotherapy in KRAS mutant NSCLC was observed, whereas in a larger retrospective study, Jeanson et al did not observe any significant relation between KRAS status and objective response rate (ORR), PFS or OS in patients treated with ICI.Citation29 A recently published meta-analysis that included five randomized clinical trials testing the antitumor effect of anti PD-1/PD-L1 mAb for the treatment of NSCLC in second-line treatment, highlighted a greater benefit of ICI in KRAS-mutant non-squamous NSCLC.Citation33 However, KRAS mutation status was not independently related to survival. The discrepancies between these results may stem from patient selection, with squamous lung cancer included in some cases, or EGFR-positive tumors in other cases. The recent ImmunoTarget registry underlined that the response of KRAS carriers to ICI was better than that of patients with other driver alterations, especially for EGFR and ALK altered tumors.Citation34 To avoid such bias, in our study, we only included non-squamous adenocarcinoma and excluded EGFR mutation cancers, known to be resistant to ICI and mainly treated with target therapies.Citation35,Citation36 This patient selection seems logical, since it represents the panel of patients classically treated with ICI.
PD-L1 is a classical predictive marker of response to ICI and is used in most clinical trials involving NSCLC.Citation37–42 However, the predictive role of PD-L1 status is discordant in some trials that have demonstrated benefits for ICI therapy, regardless of the PD-L1 status.Citation43 In a previous study, a trend was reported toward better ORR and longer PFS in KRAS-mutant NSCLC with PD-L1–positive versus PD-L1–negative tumors, with increased benefit observed at a higher rate of PD-L1–positive tumor cells.Citation29 As in previous studies,Citation38,Citation39 PD-L1 positive tumors tend to be more frequent in the KRAS mutated group in our cohort. However, our study shows contrasting results in terms of prognosis. We observed that PD-L1 > 1% expression in cancer cells was associated with better PFS and OS in the whole cohort, but subgroup analysis showed that this association drove by the difference observed in WT KRAS tumors. Taken together with the report of poor OS in KRAS-mutated tumors, these data underline the specific intrinsically poor prognosis in this subgroup of patients, which may explain why they yield less benefit from immunotherapy.
A lung immune prognostic index (LIPI) based on a dNLR ratio >3 and LDH above the ULN was developed by Mezquita et al.,Citation25 and characterizes three prognostic groups (good, 0 factors; intermediate, 1 factor; poor, 2 factors). In a large cohort of NSCLC treated in second line or more with ICI, they demonstrated the predictive role of this marker and that systemic inflammatory status was closely correlated with worse prognosis in lung cancer. Similarly additional reports have shown the prognostic and predictive role of NLR or LDH level in either lung cancer or melanoma treated with ICI.Citation32,Citation44,Citation45
However, the interaction between LIPI and KRAS status has never been addressed. We show here that LIPI determines PFS and OS only in patients with KRAS-mutated tumors, thus suggesting a particular behavior of KRAS-mutated tumors as regards their capacity to react against the inflammatory process. A possible explanation is provided by the RNAseq data from our study, which showed that patients with high LIPI had a decrease in myeloid dendritic cells, classically reported to be related to better response to immunotherapy.Citation46 When we studied the two variables of the LIPI score, we noted that the difference in terms of prognosis between WT KRAS type and mutated tumors was only dependent on the dNLR variable. Similarly, a low level of myeloid dendritic cells is only dependent on dNLR status, and not on LDH status. Additional studies are warranted to elucidate how LDH levels affect immune response.Citation47–49
Interestingly, we made the surprising observation that LIPI score is linked to a specific transcriptomic feature of cancer cells, with the signature of exocrine bronchiolar cells. Previous studies showed that basal cells (BCs) are considered the candidate “cell of origin” of lung squamous cell carcinoma.Citation50 In contrast, the cellular origin of lung adenocarcinoma is less clear.Citation51 Centrally located adenocarcinomas are thought to arise from the surface or glandular epithelium of the bronchi.Citation52 By contrast, an exocrine bronchiolar cell (Clara cell) has been observed in peripheral adenocarcinoma.Citation2,Citation53 These data suggest that cancer differentiation type may in part explain local and systemic immune contexture.
The relation between TTF1 expression and response to ICI is surprising. To date, no study has addressed this question. Previous studies in patients untreated with ICI report in most cases that TTF1 expression was associated with better outcome in either early stage or metastatic disease,Citation54–57 but the prognostic role of TTF1 in the context of ICI remains unknown. Interestingly, the subgroup analysis of our cohort suggests that the prognostic role of TTF1 is mainly observed in KRAS mutant tumors, but this analysis requires confirmation in another external cohort. Transcriptomic data support a particular biological feature of TTF1-positive tumors with an exocrine bronchiolar phenotype, and presence of an inflammatory response, which is classically associated with better efficacy of ICI.
Limitations of this study include the retrospective nature of data, with no determination of sample size, and the presence of missing clinical and pathological data albeit only for a low proportion of patients. In contrast to many retrospective reports, the PD-L1 status was known for most patients (83%). TTF1 status is heterogeneous between both cohorts. These differences may stem from the use of different mAbs to detect TTF1. However, despite this heterogeneity in the method, TTF1 remained of prognostic value in both groups, supporting the robustness of the marker. Another possible bias relates to the pooling of patients treated with different ICI mAb at different lines of treatment and to the pooling of patients with immunotherapy alone with patients with a combination of immunotherapy and chemotherapy. Molecular biology testing and PD-L1 was performed homogeneously in our biology department, so these data strengthen the results by limiting the analytic heterogeneity. We chose to include patients with BRAF mutation, because at the time of the study, these patients were treated as WT and RAS mutated NSCLC, and had similar response rates to ICI, but we acknowledge that this could represent a potential source of bias. Moreover, presence of LKB1 loss is reported to be associated with a lack of efficacy of immunotherapy in KRAS lung adenocarcinoma.Citation58 This is why, in an exploratory data set, we analyzed LKB1 expression on 100 patients, 50 with TTF1+ and 50 with TTF1-. In this group, 15 patients had a loss of LKB1 expression: 7 were TTF1+ and 8 TTF1- (data not shown). In this small cohort, LKB1 was not associated with outcome, but probably due to the small number of patients.
Lastly, subgroup analysis, especially for the different types of mutation, included only small numbers of patients, and therefore, RNAseq data should be interpreted with caution.
To conclude, our data reveal that TTF1 expression could be used to stratify and predict PFS and OS in patients treated with ICI for NS-NSCLC. The assessment of TTF1 and PDL1 could be combined to improve prognostic prediction in these patients, in association or not with LIPI score which had already show to be useful for identifying patients unlikely to benefit from immunotherapy. Nevertheless, this approach warrants exploration in future prospective clinical trials.
List of abbreviations
Non Small Cell Lung Cancer (NSCLC)
Non squamous NSCLC (NS-NSCLC)
Epidermal growth factor receptor (EGFR)
Anti-programmed death (Ligand) 1 (PD-1/PD-L1)
Immune checkpoint inhibitors (ICI)
Lactate dehydrogenase (LDH)
Thyroid Transcription Factor-1 (TTF1)
Derived neutrophils/lymphocyte (leukocytes minus neutrophils) ratio (dNLR)
LIPI (Lung Immune Prognostic Index)
Progression-free-survival (PFS)
Overall survival (OS)
Interquartile range (IQR)
Upper Limit of Normal (ULN)
WHO (World health organization)
HR (Hazard ratio)
Interleukine 2 (IL2)
Basal cells (BCs)
Authors’contributions
L.G., J.L., A-L.L., F.B., Y.O., V.D., C.T. and F.G. contributed to the design and implementation of the research, to the analysis of the results and to the writing of the manuscript.
Ethics approval and consent to participate
The study was approved by the CNIL (French national commission for data privacy) and the local ethics committee, and was performed in accordance with the Helsinki Declaration and European legislation. Autorisation registered under the name AC-2014-2260.
Consent for publication
Only patients from whom informed consent was obtained and recorded in the medical chart were included in this retrospective study.
Supplemental Material
Download ()Acknowledgments
We wish to thank Caen hospital and Anne-Laure Lepage for the external cohort. We wish to thank Fiona Ecarnot, PhD (EA3920, University of Franche-Comté, Besançon) for English correction and helpful comments. We thank Romain Boidot and Sandy Chevrier for RNAseq data generation.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Availability of data and material
The datasets analyzed during the current study are available from the corresponding author on reasonable request.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
Additional information
Funding
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA. Cancer J. Clin. 2019;69(1):7–12. doi:10.3322/caac.21551.
- Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger KR, Yatabe Y, et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol Off Publ Int Assoc Study Lung Cancer. févr 2011;6(2):244‑85.
- Dogan S, Shen R, Ang DC, Johnson ML, D’Angelo SP, Paik PK, Brzostowski EB, Riely GJ, Kris MG, Zakowski MF, et al. Molecular Epidemiology of EGFR and KRAS mutations in 3,026 lung adenocarcinomas: higher susceptibility of women to smoking-related KRAS-mutant cancers. Clin Cancer Res Off J Am Assoc Cancer Res. 2012;18(22):6169–6177. doi:10.1158/1078-0432.CCR-11-3265.
- El Osta B, Behera M, Kim S, Berry LD, Sica G, Pillai RN, et al. Characteristics and Outcomes of Patients With Metastatic KRAS-Mutant Lung Adenocarcinomas: The Lung Cancer Mutation Consortium Experience. J Thorac Oncol Off Publ Int Assoc Study Lung Cancer. mai 2019;14(5):876–89.
- Barlesi F, Mazieres J, Merlio J-P, Debieuvre D, Mosser J, Lena H, et al. Routine molecular profiling of patients with advanced non-small-cell lung cancer: results of a 1-year nationwide programme of the French Cooperative Thoracic Intergroup (IFCT). Lancet Lond Engl. 2 avr 2016;387(10026):1415‑26.
- Mascaux C, Iannino N, Martin B, Paesmans M, Berghmans T, Dusart M, Haller A, Lothaire P, Meert A-P, Noel S, et al. The role of RAS oncogene in survival of patients with lung cancer: a systematic review of the literature with meta-analysis. Br J Cancer. 2005;92(1):131–139. doi:10.1038/sj.bjc.6602258.
- Johnson ML, Sima CS, Chaft J, Paik PK, Pao W, Kris MG, Ladanyi M, Riely GJ. Association of KRAS and EGFR mutations with survival in patients with advanced lung adenocarcinomas. Cancer. 2013;119(2):356–362. doi:10.1002/cncr.27730.
- Jancík S, Drábek J, Radzioch D, Hajdúch M. Clinical relevance of KRAS in human cancers. J Biomed Biotechnol. 2010;2010:150960. doi:10.1155/2010/150960.
- Macerelli M, Caramella C, Faivre L, Besse B, Planchard D, Polo V, et al. Does KRAS mutational status predict chemoresistance in advanced non-small cell lung cancer (NSCLC)? Lung Cancer Amst Neth. mars 2014;83(3):383‑8.
- Barta JA, Powell CA, Wisnivesky JP. Global Epidemiology of Lung Cancer. Ann Glob Health. 22 janv 2019;85(1):8.
- Tomasini P, Walia P, Labbe C, Jao K, Leighl NB. Targeting the KRAS pathway in non-small cell lung cancer. The Oncologist. 2016;21(12):1450–1460. doi:10.1634/theoncologist.2015-0084.
- Planchard D, Popat S, Kerr K, Novello S, Smit EF, Faivre-Finn C, et al. Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol Off J Eur Soc Med Oncol. 1 oct 2018;29\(Suppl 4):iv192‑237.
- Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, Patnaik A, Aggarwal C, Gubens M, Horn L, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372(21):2018–2028. doi:10.1056/NEJMoa1501824.
- Reck M, Rodríguez-Abreu D, Robinson AG, Hui R, Csőszi T, Fülöp A, Gottfried M, Peled N, Tafreshi A, Cuffe S, et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med. 2016;375(19):1823–1833. doi:10.1056/NEJMoa1606774.
- Gandhi L, Rodríguez-Abreu D, Gadgeel S, Esteban E, Felip E, De Angelis F, Domine M, Clingan P, Hochmair MJ, Powell SF, et al. Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. N Engl J Med. 2018;378(22):2078–2092. doi:10.1056/NEJMoa1801005.
- Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer. 2019;19:133–150.
- Doroshow DB, Bhalla S, Beasley MB, Sholl LM, Kerr KM, Gnjatic S, Wistuba II, Rimm DL, Tsao MS, Hirsch FR, et al. PD-L1 as a biomarker of response to immune-checkpoint inhibitors. Nat Rev Clin Oncol. 2021;18(6):345–362. doi:10.1038/s41571-021-00473-5.
- Takada K, Toyokawa G, Shoji F, Okamoto T, Maehara Y. The significance of the PD-L1 expression in non-small-cell lung cancer: trenchant double swords as predictive and prognostic markers. Clin Lung Cancer. 2018;19(2):120–129. doi:10.1016/j.cllc.2017.10.014.
- Zhu L, Zhu L, Li X, Shen Y, Cao Y, Fang X, Chen J. A new prognostic score based on the systemic inflammatory response in patients with inoperable non-small-cell lung cancer. OncoTargets Ther. 2016;9:4879–4886. doi:10.2147/OTT.S107279.
- Laird BJA, Fallon M, Hjermstad MJ, Tuck S, Kaasa S, Klepstad P, McMillan DC. Quality of life in patients with advanced cancer: differential association with performance status and systemic inflammatory response. J Clin Oncol Off J Am Soc Clin Oncol. 2016;34(23):2769–2775. doi:10.1200/JCO.2015.65.7742.
- McMillan DC. The systemic inflammation-based Glasgow Prognostic Score: a decade of experience in patients with cancer. Cancer Treat Rev. 2013;39(5):534–540. doi:10.1016/j.ctrv.2012.08.003.
- Petrelli F, Cabiddu M, Coinu A, Borgonovo K, Ghilardi M, Lonati V, Barni S. Prognostic role of lactate dehydrogenase in solid tumors: a systematic review and meta-analysis of 76 studies. Acta Oncol. Stockh. Swed. 2015;54(7):961–970. doi:10.3109/0284186X.2015.1043026.
- Templeton AJ, Ace O, McNamara MG, Al-Mubarak M, Vera-Badillo FE, Hermanns T, Šeruga B, Ocaña A, Tannock IF, Amir E, et al. Prognostic role of platelet to lymphocyte ratio in solid tumors: a systematic review and meta-analysis. Cancer Epidemiol. Biomark. Prev. Publ. Am. Assoc Cancer Res Cosponsored Am Soc Prev Oncol. 2014;23(7):1204–1212. doi:10.1158/1055-9965.EPI-14-0146.
- Paramanathan A, Saxena A, Morris DL. A systematic review and meta-analysis on the impact of pre-operative neutrophil lymphocyte ratio on long term outcomes after curative intent resection of solid tumours. Surg Oncol. 2014;23(1):31–39. doi:10.1016/j.suronc.2013.12.001.
- Mezquita L, Auclin E, Ferrara R, Charrier M, Remon J, Planchard D, Ponce S, Ares LP, Leroy L, Audigier-Valette C, et al. Association of the lung immune prognostic index with immune checkpoint inhibitor outcomes in patients with advanced non-small cell lung cancer. JAMA Oncol. 2018;4(3):351–357. doi:10.1001/jamaoncol.2017.4771.
- Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, Chow LQ, Vokes EE, Felip E, Holgado E, et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl J Med. 2015;373(17):1627–1639. doi:10.1056/NEJMoa1507643.
- Sun L, Hsu M, Cohen RB, Langer CJ, Mamtani R, Aggarwal C. Association between KRAS variant status and outcomes with first-line immune checkpoint inhibitor–based therapy in patients with advanced non–small-cell lung cancer. JAMA Oncol. 2021;7(6):937. doi:10.1001/jamaoncol.2021.0546.
- Skoulidis F, Byers LA, Diao L, Papadimitrakopoulou VA, Tong P, Izzo J, Behrens C, Kadara H, Parra ER, Canales JR, et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov. 2015;5(8):860–877. doi:10.1158/2159-8290.CD-14-1236.
- Jeanson A, Tomasini P, Souquet-Bressand M, Brandone N, Boucekine M, Grangeon M, et al. Efficacy of Immune Checkpoint Inhibitors in KRAS-Mutant Non-Small Cell Lung Cancer (NSCLC). J Thorac Oncol Off Publ Int Assoc Study Lung Cancer. juin 2019;14(6):1095‑101.
- Yatabe Y, Dacic S, Borczuk AC, Warth A, Russell PA, Lantuejoul S, et al. Best Practices Recommendations for Diagnostic Immunohistochemistry in Lung Cancer. J Thorac Oncol Off Publ Int Assoc Study Lung Cancer. mars 2019;14(3):377‑407.
- Kaufmann O, Dietel M. Thyroid transcription factor-1 is the superior immunohistochemical marker for pulmonary adenocarcinomas and large cell carcinomas compared to surfactant proteins A and B. Histopathology. 2000;36(1):8–16. doi:10.1046/j.1365-2559.2000.00801.x.
- Ferrucci PF, Ascierto PA, Pigozzo J, Del Vecchio M, Maio M, Antonini Cappellini GC, Guidoboni M, Queirolo P, Savoia P, Mandalà M, et al. Baseline neutrophils and derived neutrophil-to-lymphocyte ratio: prognostic relevance in metastatic melanoma patients receiving ipilimumab. Ann Oncol Off J Eur Soc Med Oncol. 2016;27(4):732–738. doi:10.1093/annonc/mdw016.
- Rothschild SI. KRAS and immune checkpoint inhibitors-serendipity raising expectations. J Thorac Oncol Off Publ Int Assoc Study Lung Cancer. 2019;14:951–954.
- Mazieres J, Drilon A, Lusque A, Mhanna L, Cortot AB, Mezquita L, Thai AA, Mascaux C, Couraud S, Veillon R, et al. Immune checkpoint inhibitors for patients with advanced lung cancer and oncogenic driver alterations: results from the IMMUNOTARGET registry. Ann Oncol Off J Eur Soc Med Oncol. 2019;30(8):1321–1328. doi:10.1093/annonc/mdz167.
- Yamada T, Hirai S, Katayama Y, Yoshimura A, Shiotsu S, Watanabe S, Kikuchi T, Hirose K, Kubota Y, Chihara Y, et al. Retrospective efficacy analysis of immune checkpoint inhibitors in patients with EGFR-mutated non-small cell lung cancer. Cancer Med. 2019;8(4):1521–1529. doi:10.1002/cam4.2037.
- Gainor JF, Shaw AT, Sequist LV, Fu X, Azzoli CG, Piotrowska Z, Huynh TG, Zhao L, Fulton L, Schultz KR, et al. EGFR mutations and ALK rearrangements are associated with low response rates to PD-1 pathway blockade in non–small cell lung cancer: a retrospective analysis. Clin Cancer Res Off J Am Assoc Cancer Res. 2016;22(18):4585–4593. doi:10.1158/1078-0432.CCR-15-3101.
- Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, et al.. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–128. doi:10.1126/science.aaa1348.
- Falk AT, Yazbeck N, Guibert N, Chamorey E, Paquet A, Ribeyre L, Bence C, Zahaf K, Leroy S, Marquette C-H, et al. Effect of mutant variants of the KRAS gene on PD-L1 expression and on the immune microenvironment and association with clinical outcome in lung adenocarcinoma patients. Lung Cancer Amst Neth. 2018;121:70–75. doi:10.1016/j.lungcan.2018.05.009.
- Calles A, Liao X, Sholl LM, Rodig SJ, Freeman GJ, Butaney M, et al. Expression of PD-1 and Its Ligands, PD-L1 and PD-L2, in Smokers and Never Smokers with KRAS-Mutant Lung Cancer. J Thorac Oncol Off Publ Int Assoc Study Lung Cancer. déc 2015;10(12):1726‑35.
- D’Incecco A, Andreozzi M, Ludovini V, Rossi E, Capodanno A, Landi L, Tibaldi C, Minuti G, Salvini J, Coppi E, et al. PD-1 and PD-L1 expression in molecularly selected non-small-cell lung cancer patients. Br J Cancer. 2015;112(1):95–102. doi:10.1038/bjc.2014.555.
- Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJM, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515(7528):568–571. doi:10.1038/nature13954.
- Scheel AH, Ansén S, Schultheis AM, Scheffler M, Fischer RN, Michels S, Hellmich M, George J, Zander T, Brockmann M, et al. PD-L1 expression in non-small cell lung cancer: correlations with genetic alterations. Oncoimmunology. 2016;5(5):e1131379. doi:10.1080/2162402X.2015.1131379.
- Rittmeyer A, Barlesi F, Waterkamp D, Park K, Ciardiello F, von Pawel J, et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet Lond Engl. 21 janv 2017;389(10066):255‑65.
- Weide B, Martens A, Hassel JC, Berking C, Postow MA, Bisschop K, et al. Baseline Biomarkers for Outcome of Melanoma Patients Treated with Pembrolizumab. Clin Cancer Res Off J Am Assoc Cancer Res. 15 nov 2016;22(22):5487‑96.
- Bagley SJ, Kothari S, Aggarwal C, Bauml JM, Alley EW, Evans TL, Kosteva JA, Ciunci CA, Gabriel PE, Thompson JC, et al. Pretreatment neutrophil-to-lymphocyte ratio as a marker of outcomes in nivolumab-treated patients with advanced non-small-cell lung cancer. Lung Cancer Amst Neth. 2017;106:1–7. doi:10.1016/j.lungcan.2017.01.013.
- Mayoux M, Roller A, Pulko V, Sammicheli S, Chen S, Sum E, et al. Dendritic cells dictate responses to PD-L1 blockade cancer immunotherapy. Sci Transl Med. 11 mars 2020;12(534).
- Diem S, Kasenda B, Spain L, Martin-Liberal J, Marconcini R, Gore M, Larkin J. Serum lactate dehydrogenase as an early marker for outcome in patients treated with anti-PD-1 therapy in metastatic melanoma. Br J Cancer. 2016;114(3):256–261. doi:10.1038/bjc.2015.467.
- Fiala O, Pesek M, Finek J, Topolcan O, Racek J, Svaton M, et al. Change in Serum Lactate Dehydrogenase Is Associated with Outcome of Patients with Advanced-stage NSCLC Treated with Erlotinib. Anticancer Res. mai 2016;36(5):2459‑65.
- Inomata M, et al.Elevated levels of plasma lactate dehydrogenase is an unfavorable prognostic factor in patients with epidermal growth factor receptor mutation-positive non-small cell lung cancer, receiving treatment with gefitinib or erlotinib. Mol Clin Oncol. 2016;4(5):774–778. doi:10.3892/mco.2016.779
- Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, Randell SH, Hogan BLM. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci U S A. 2009;106(31):12771–12775. doi:10.1073/pnas.0906850106.
- Wistuba II, Gazdar AF. Lung cancer preneoplasia. Annu Rev Pathol. 2006;1(1):331–348. doi:10.1146/annurev.pathol.1.110304.100103.
- Travis WD, Brambilla E, Burke AP, Marx A, Nicholson AG. Introduction to the 2015 world health organization classification of tumors of the lung, pleura, thymus, and heart. J Thorac Oncol. 2015;10(9):1240–1242. doi:10.1097/JTO.0000000000000663.
- Yatabe Y. EGFR mutations and the terminal respiratory unit. Cancer Metastasis Rev. 2010;29(1):23–36. doi:10.1007/s10555-010-9205-8.
- Da Cruz V, Yvorel V, Casteillo F, Tissot C, Luchez A, Bayle-Bleuez S, Fournel P, Tiffet O, Péoc’h M, Forest F, et al.. Histopathological subtyping is a prognostic factor in stage IV lung adenocarcinoma. Lung Cancer Amst Neth. 2020;147:77–82. doi:10.1016/j.lungcan.2020.07.010.
- Qian H, Xu T, Cai X, Ji T, Guo H. Prognostic value of TTF-1 expression in patients with non-small cell lung cancer: a meta-analysis. Clin Chim Acta Int J Clin Chem. 2015;451:208–214. doi:10.1016/j.cca.2015.01.023.
- Kim JH, Kim HS, Kim BJ, Han B, Choi DR, Kwon JH. Prognostic Impact of TTF-1 Expression in Non-Squamous Non-Small-Cell Lung Cancer: A Meta-Analysis. J Cancer. 2018;9(22):4279‑86.
- Schilsky JB, Ni A, Ahn L, Datta S, Travis WD, Kris MG, et al. Prognostic impact of TTF-1 expression in patients with stage IV lung adenocarcinomas. Lung Cancer Amst Neth. juin 2017;108:205‑11.
- Skoulidis F, Goldberg ME, Greenawalt DM, Hellmann MD, Awad MM, Gainor JF, et al. STK11/LKB1 Mutations and PD-1 Inhibitor Resistance in KRAS-Mutant Lung Adenocarcinoma. Cancer Discov. juill 2018;8(7):822‑35.
