ABSTRACT
Immune checkpoint inhibitors (ICIs) have improved the survival of patients with non-small cell lung cancer (NSCLC) by reinvigorating tumor-specific T cell responses. However, the specificity of such T cells and the human leukocyte antigen (HLA)-associated epitopes recognized, remain elusive. In this study, we identified NSCLC T cell epitopes of recently described NSCLC-associated antigens, termed keratinocyte differentiation antigens. Epitopes of these antigens were presented by HLA-A 03:01 and HLA-C 04:01 and were associated with responses to ICI therapy. Patients with CD8+ T cell responses to these epitopes had improved overall and progression-free survival. T cells specific for such epitopes could eliminate HLA class I-matched NSCLC cells ex vivo and were enriched in patient lung tumors. The identification of novel lung cancer HLA-associated epitopes that correlate with improved ICI-dependent treatment outcomes suggests that keratinocyte-specific proteins are important tumor-associated antigens in NSCLC. These findings improve our understanding of the mechanisms of ICI therapy and may help support the development of vaccination strategies to improve ICI-based treatment of these tumors.
Introduction
The use of immune checkpoint inhibitors (ICIs) has revolutionized therapy for patients with non-small cell lung cancer (NSCLC). Compared to standard chemotherapy, overall survival (OS) and progression-free survival (PFS) have significantly increased with ICIs.Citation1–4 However, very few NSCLC antigens and specific epitopes have been identified.Citation5–7 The identification of novel NSCLC antigens and related epitopes can improve the efficacy of immunotherapy, for example by combining ICIs with anti-cancer vaccination or with adoptive cell transfer of T cells expressing synthetic T cell receptors (TCRs). In addition, the validation of new tumor targets may help explain the mechanisms of ICI-induced immune-related adverse events (irAEs), which frequently occur in patients treated with ICIs.Citation8 In a previous study, we identified nine novel NSCLC antigens that likely mediate ICI-induced autoimmune skin toxicity, and possibly anti-tumor responses.Citation6 Here, we aimed to determine specific HLA-associated epitopes of the identified tumor antigens as recognized by CD8+ T cells identified in human NSCLC tissues and associate these with survival of ICI-treated patients with NSCLC.
Results
Screening of predicted lung tumor CD8 epitopes
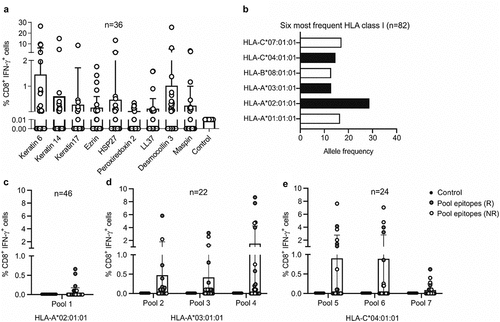
We aimed to further elaborate on the role of the recently described NSCLC-associated antigens in ICI therapy.Citation6 These tumor antigens were identified using a novel approach of tissue profiling and bioinformatics, as recently described,Citation6 and had to fulfill the following properties:Citation1 evidence of self-immunogenicity,Citation2 high expression in NSCLC, andCitation3 skin or lung specificity. The antigens were more highly expressed in lung squamous cell carcinoma compared to adenocarcinoma (Supplementary Fig. S1). As these antigens are also expressed in skin keratinocytes, we named this group of tumor-associated antigens (TAAs) keratinocyte differentiation antigens (KDAs). This is an analogy to the extensively studied melanocyte differentiation antigens in melanoma. We found that these antigens link the appearance of skin irAEs with responses to ICIs.Citation6 Here, we used these antigens in T cell stimulation assays by testing overlapping 15-mer peptide pools. These assays were carried out with PBMCs from patients with NSCLC under treatment with an ICI, to determine whether the antigens could elicit CD8+ T cell activation. Indeed, all of the previously described candidate antigens were able to stimulate IFN-γ secretion in CD8+ T cells. Six of nine antigens tested lead to a significantly higher frequency of CD8+ IFN-γ+ T cells compared to the medium-only negative control (). While healthy donors also showed CD8+ T cell responses to the antigens, the frequencies were significantly lower compared to NSCLC patients (Supplementary Fig. S2a-b). In addition, we found that the antigens could also elicit CD4+ T cell activation and that the frequencies were again significantly higher in NSCLC patients compared to healthy donors (Supplementary Fig. S2c-e). When comparing the CD8+ IFN-γ+ T cell frequencies before and during therapy in NSCLC patients, we found that the differences were not significant, showing that patients harbored these antigen-specific T cells before the start of therapy (Supplementary Fig. S3). The fact that patients harbor tumor-specific T cells before the start of treatment is not unusual, and has been previously exploited for the expansion and transfer of tumor-infiltrating lymphocytes in melanoma patients.Citation9 We next aimed to identify specific CD8 epitopes of the keratinocyte differentiation antigens. Using next generation sequencing we therefore carried out human leukocyte antigen class I (HLA-I) typing (HLA-A, HLA-B and HLA-C) of our NSCLC cohort consisting of 82 patients treated with ICI therapy (nivolumab, pembrolizumab or atezolizumab) (Supplementary Table S1). Peptide epitopes were predicted for HLA-A 02:01, HLA-A 03:01 and HLA-C 04:01 which represent the most frequent HLA class I alleles in our cohort. This was initially predicted on the basis of analyses of the first patients but after typing the entire cohort, they remained among the six most frequent HLA class I alleles expressed (, Supplementary Table S2). The HLA presented peptide epitopes for the keratinocyte differentiation antigens (keratin 6, keratin 14, keratin 17, ezrin, HSP27, peroxiredoxin 2, LL37, desmocollin 3 and maspin) were predicted using the IEDB epitope prediction resource, and peptides were selected based on their percentile rank and IC50 HLA-binding scores as previously describedCitation10 (Supplementary Fig. S4). For each antigen, five nonamer and five decamer peptides were selected. A list of all peptides can be found in Supplementary Table S3. T cell stimulation assays were initially carried out with pools of the predicted epitopes using cryopreserved PBMCs from HLA-A 02:01, HLA-A 03:01 or HLA-C 04:01 positive NSCLC patients, isolated while being treated with an ICI. This screening process identified the patients that had CD8+ IFN-γ+ T cell responses to the epitope pools. Of note, several patients with CD8+ IFN-γ+ T cell responses to the epitope pools were responders to ICI therapy; more specifically, of patients harboring T cell responses to the pools, 66.6% were responders to treatment ().
Figure 1. Screening of predicted CD8 epitopes in ICI-treated patients with NSCLC. (a) NSCLC antigens were predicted using a novel approach of tissue profiling and bioinformaticsCitation6 and used to stimulate PBMC cultures of ICI-treated patients with NSCLC (n = 36). The frequency of CD8+ IFN-γ+ T cells among total CD8+ T cells was detected as a measure of T cell activation for each stimulation. These frequencies were compared to the background frequency of CD8+ IFN-γ+ cells in medium-only negative control cultures (Kruskal Wallis test; keratin 6, p = .70; keratin 14, p = .02; keratin 17, p = .002; ezrin, p = .05; HSP27, p = .01; peroxiredoxin 2, p = .0004; LL37, p = .01; desmocollin 3, p = .72; maspin, p = .01). (b) HLA class I typing of the patient cohort was carried out to identify the most frequent HLA class I alleles. The alleles shown in black bars were selected for further analysis. (c-e) An initial screen was carried out by pooling the predicted CD8 epitopes into seven different peptide pools. Pool 1 contained epitopes predicted for HLA-A 02:01 (c), pools 2–4 contained epitopes for the nine antigens predicted for HLA-A 03:01 (d) and pools 5–7 contained epitopes predicted for HLA-C 04:01 (e). Responders (r) are shown in red and non-responders (NR) in white. Pool 1 contained epitopes from all predicted antigens; pools 2 and 5 contained epitopes from keratin 6, keratin 17 and keratin 14; pools 3 and 6 contained epitopes from LL37, ezrin and HSP27; pools 4 and 7 contained epitopes from desmocollin 3, maspin and peroxiredoxin 2. The frequency of CD8+ IFN-γ+ T cells among total CD8+ T cells was detected as a measure of T cell activation for each stimulation. These frequencies were compared to the background frequency of CD8+ IFN-γ+ cells in medium-only negative control cultures
CD8 epitopes of keratinocyte differentiation antigens are associated with improved survival in ICI-treated NSCLC patients
The PBMCs of patients that showed an IFN-γ+ T cell response to the epitope pools were then stimulated with the single peptides, which lead to the identification of several HLA-A 03:01 and HLA-C 04:01 specific CD8 epitopes. Surprisingly, no epitopes for HLA-A 02:01 were identified. Interestingly, seven of the identified peptide epitopes induced a CD8+ IFN-γ+ T cell response in at least two patients. Four peptides were shared among HLA-A 03:01-positive patients () and three were shared among HLA-C 04:01-positive patients (). A summary of the shared and non-shared epitopes identified is presented in Supplementary Table S4. Epitopes were identified for all keratinocyte differentiation antigens with the exception of keratin 17. Representative flow cytometry plots of the identified epitopes are shown in Supplementary Fig. S5. When comparing survival between patients harboring T cells specific for the identified epitopes, and patients where we failed to identify such T cells, we observed better overall survival (OS) (HR = 0.37 [95% CI, 0.20–0.68]; log-rank p-value =0.01) and progression-free survival (PFS) (HR = 0.37 [95% CI, 0.20–0.68]; log-rank p-value = 0.01) for patients with CD8+ IFN-γ+ T cell responses to the predicted epitope(s) ().
Figure 2. ICI-treated patients with NSCLC harboring CD8+ T cell responses to predicted CD8 epitopes have improved survival. (a) Identified CD8 epitopes for the predicted antigens for HLA-A 03:01 that are shared among patients (i.e. two or more patients showed a CD8+ T cell response to a specific epitope). (b) Identified CD8 epitopes for the predicted antigens for HLA-C 04:01 that are shared among patients (as in A). For (a-b) PBMCs from patients with NSCLC were stimulated with the single epitopes of the predicted tumor antigens. T cell activation upon stimulation was detected by measuring ex vivo IFN-γ production by CD8+ T cells. The background frequency of CD8+ IFN-γ+ cells in medium-only negative control cultures was subtracted for all stimulations. IMIT refers to the patient ID. (c-d) Patients positive for HLA-A 03:01 and HLA-C 04:01 with CD8+ T cell responses to the predicted epitopes (n = 17) had significantly better OS (HR = 0.37 [95% CI, 0.20–0.68], log-rank p-value = 0.01) (c) and PFS (HR = 0.37 [95% CI, 0.20–0.68], log-rank p-value = 0.01) (d) compared to patients with the same HLAs that did not have a CD8+ T cell response to the epitopes (n = 71)
![Figure 2. ICI-treated patients with NSCLC harboring CD8+ T cell responses to predicted CD8 epitopes have improved survival. (a) Identified CD8 epitopes for the predicted antigens for HLA-A 03:01 that are shared among patients (i.e. two or more patients showed a CD8+ T cell response to a specific epitope). (b) Identified CD8 epitopes for the predicted antigens for HLA-C 04:01 that are shared among patients (as in A). For (a-b) PBMCs from patients with NSCLC were stimulated with the single epitopes of the predicted tumor antigens. T cell activation upon stimulation was detected by measuring ex vivo IFN-γ production by CD8+ T cells. The background frequency of CD8+ IFN-γ+ cells in medium-only negative control cultures was subtracted for all stimulations. IMIT refers to the patient ID. (c-d) Patients positive for HLA-A 03:01 and HLA-C 04:01 with CD8+ T cell responses to the predicted epitopes (n = 17) had significantly better OS (HR = 0.37 [95% CI, 0.20–0.68], log-rank p-value = 0.01) (c) and PFS (HR = 0.37 [95% CI, 0.20–0.68], log-rank p-value = 0.01) (d) compared to patients with the same HLAs that did not have a CD8+ T cell response to the epitopes (n = 71)](/cms/asset/038e4bfe-e27b-4850-88de-305f03353df4/koni_a_2006893_f0002_oc.jpg)
Keratinocyte differentiation antigen-specific T cells are enriched in patient lung tumors and mediate killing of lung tumor cells
We next determined whether peptides of the nine keratinocyte differentiation antigens are naturally presented via HLA molecules on human lung tumor cells. We therefore performed mass spectrometry-based immunopeptidome analyses of 16 primary human NSCLC tumor samples. Indeed, we identified HLA class I naturally presented peptides from seven of the keratinocyte differentiation antigens (desmocollin 3, ezrin, HSP27, keratin 6, keratin 14, keratin 17 and peroxiredoxin 2) on patient lung tumors (, Supplementary Table S5). In addition, we found naturally presented HLA class II ligands from all nine antigens on lung tumors (Supplementary Fig. S6, Supplementary Table S6). Naturally presented HLA ligands from keratin 6, keratin 14 and keratin 17 could not be assigned to a single keratin protein sequence due to their high sequence similarity and were therefore grouped together as keratins. Mapping of the tumor ligandome by mass spectrometry confirms that the identified peptides initiating CD8+ T cells responses in the earlier assays are truly presented peptides on the tumor HLA.
Figure 3. Predicted peptide antigens are naturally presented on HLA class I of human NSCLC tissue, and epitope-specific T cells are found in ICI-treated lung tumors. (a) Frequency of donors in which HLA class I ligands derived from the predicted NSCLC antigens in NSCLC tissue (n = 16) were detected by mass spectrometry. *HLA ligands from keratin 6, keratin 14 and keratin 17 could not be assigned to a single keratin protein sequence due to their high sequence similarity and were therefore grouped together as keratins. (b) PBMC cultures from patients with NSCLC (n = 3) were stimulated with single predicted CD8 epitopes and co-cultured overnight with an immortalized HLA class I-matched NSCLC cell line pulsed with the same peptides. The following day the frequency of NSCLC cell death was analyzed using the Apotracker green/propidium iodide apoptosis detection kit. (c) PBMC cultures from patients with NSCLC were stimulated with single predicted CD8 epitopes and CD8+ IFN-γ+ T cells were sorted after the stimulations and used for TCR sequencing. IMIT refers to the patient ID. The epitope-specific TCRs were compared with the TCRs found in human NSCLCs (primary lung tumors or metastases) treated with ICI therapy (n = 5). A fuzzy string matching analysis using a maximal edit distance of one was carried out to identify highly similar TCRs in sorted T cells and NSCLCs. Edit distance is defined as the Levenshtein distance, which is the number of removals, insertions, or substitutions of a character needed to transform one string into the other
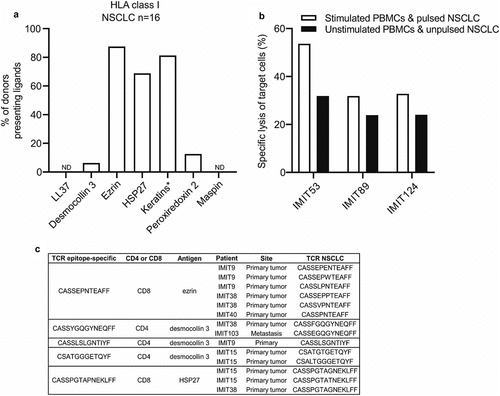
Next, we wanted to determine whether the identified epitopes could mediate the killing of immortalized HLA class I-matched NSCLC cells. We therefore used the predicted epitopes to stimulate the PBMCs of patients with NSCLC and pulse the NSCLC cells, and subsequently co-cultured the PBMCs and NSCLC cells overnight. After this overnight incubation, we determined the specific lysis of the target cells and found that T cells stimulated with the identified epitopes could indeed mediate ex vivo killing of NSCLC cells pulsed with the same epitope (). Lastly, to determine whether epitope-specific T cells could be found in patient lung tumors, we stimulated PBMCs isolated from patients with NSCLC with the identified epitopes and sorted the CD8+ IFN-γ+ T cells followed by TCR sequencing of patient lung tumors and sorted T cells. We found that tumors of patients with NSCLC harbor T cells with TCRs that are almost identical to the antigen-specific TCRs – TCRs differed by only one amino acid within the TCRβ CDR3 region. It has been shown that TCRs that differ by only one amino acid in this region usually cluster together and have the same specificity (based on so-called global similarity).Citation11 These epitope-specific T cells therefore localize within tumors and likely mediate anti-tumor effects in NSCLC (). To further validate these findings, we interrogated a publicly available TCR database and found that NSCLC patients harbor identical KDA epitope-specific T cells at significantly higher frequencies compared to melanoma and breast cancer patients in their tumors (Supplementary Fig. S7a), showing that these TCRs are enriched in NSCLCs. In addition, we found that the frequencies of these clones were significantly higher in blood of NSCLC patients compared to the blood of melanoma patients, and higher compared to the blood of breast cancer patients, although the difference was not significant (Supplementary Fig. S7b). Lastly, we found that these clones are enriched in NSCLCs compared to blood of NSCLC patients and adjacent lung tissue (Supplementary Fig. S7c).
Discussion
The identification of tumor antigens is crucial for a better understanding of how T cells recognize and kill tumor cells. To date, only a few NSCLC antigens have been identified.Citation12–14 In this study we elaborate on the findings of our previous articleCitation6 where we identified NSCLC antigens and proposed that these antigens mediated autoimmune skin toxicity and probably also anti-tumor responses in ICI-treated patients with NSCLC. Here we elucidate the exact CD8 epitopes that may explain these effects in HLA-A 03:01- and HLA-C 04:01-positive patients. Of note, specific HLA alleles have been previously associated with ICI-induced irAEs such as skin toxicity and colitis.Citation15 We also show that T cells specific for these epitopes are able to kill NSCLC cells ex vivo and are enriched in patient lung tumors.
A major question in tumor immunotherapy is why only a subset of patients treated with ICIs develop such autoimmune toxicities, despite containing the antigen-specific T cells. This phenomenon is also observed in melanoma patients, where many patients harbor melanocyte differentiation antigen-specific T cells with only a fraction of these patients developing vitiligo. A fundamental next step is to understand the conditions required for these T cells to become activated in organs other than the tumor and whether the underlying triggers can be avoided to reduce autoimmune toxicities. This would allow further exploration of these self-antigens to further improve immunotherapy, such as exploiting them for anti-cancer vaccination or generating T cells with synthetic TCRs targeting the relevant antigens. Earlier observations suggest that self-antigens are able to induce strong anti-tumor responses, as seen with melanocyte differentiation antigens in melanoma.Citation16 It is then important to identify which self-antigens can elicit such responses, and which antigens do not cause strong autoimmune side effects, as these can be treatment limiting. Still, under conditions of limited and acceptable autoimmune responses, strong anti-cancer immune responses will result in the best treatment outcomes. Our definition of tissue-specific antigens presented by HLAs in NSCLC patients improves our understanding of lung tumor immunology and may support future studies aimed at the development of more specific cancer vaccination or TCR transfer combination therapies.
Materials and methods
Clinical sample collection
A prospective cohort study of patients with NSCLC treated with an ICI was conducted in four different centers in Switzerland (Kantonsspital St Gallen, Spital Grabs, Spital Wil, and Spital Flawil) from July 1, 2016, to September 30, 2019. Patient follow-up time was at least one year. The study protocol was approved by the Ethics Committee of Eastern Switzerland (Ethikkommission Ostschweiz, EKOS 16/079) and was conducted in accordance with the Declaration of Helsinki guidelines. All patients gave written, informed consent. Peripheral blood mononuclear cells (PBMCs) were collected before the start of therapy and at each cycle of therapy. Where possible, paraffin-embedded tumor tissue from remaining tumor resection and diagnostic biopsies was used for TCR sequencing analyses.
Patients were divided into responders and non-responders, according to the following criteria: responders were defined as experiencing partial remission (PR) or stable disease (SD) with progression-free survival (PFS) of over 200 days; non-responders were defined as experiencing progressive disease (PD) or SD with PFS of less than 200 days.
HLA immunoaffinity data from snap-frozen human lung tumor samples was provided by the University of Tübingen, Germany.
Isolation of PBMCs
Patient blood was collected into EDTA-containing tubes and PBMCs were isolated following standard Ficoll-Paque separation. After isolation PBMCs were cryopreserved at −150°C.
HLA sequencing
HLA class I sequencing was performed by Histogenetics in 6-digit resolution using cryopreserved PBMCs.
CD8 epitope prediction and synthesis
CD8 T cell epitopes for the nine previously published shared antigens between NSCLC and skin were predicted for HLA-A 02:01, HLA-A 03:01 and HLA-C 04:01. MHC class I binding prediction was performed with IEDB Analysis Resource based on MHC class I binding affinity. The five nonamers and the five decamers with the highest immunogenicity score were selected for peptide synthesis (percentile rank < 1.5%, IC50 < 160 nm). The peptides were synthesized by GenScript and had >85% purity. Lyophilized peptides were dissolved in DMSO, diluted in RPMI, aliquoted and stored at −20°C.
T cell stimulation assays and flow cytometry
Cryopreserved PBMCs of patients with HLA-A 03:01, HLA-A 02:01 and HLA-C 04:01 were plated at a concentration of 1 × 106 cells per well in low-adherence 24-well plates (Sarstedt, ref. 83.3922.500) and resuspended in 2 mL RPMI per well containing 8% human serum (Biowest, ref. S4190-100), 1% penicillin-streptomycin, 1% L-glutamine, 1% non-essential amino acids, 1% sodium pyruvate, 0.1 mg/mL kanamycin and 0.1% β-mercaptoethanol. Cells were kept at 37°C. The following day, cells were stimulated with the predicted epitopes individually at a final concentration of 2ug/mL/peptide. After 48 hours, 1 mL medium was replaced with fresh RPMI (as above) containing IL-2 (final concentration of IL-2 was 150 U/mL). Cells were kept in culture for ten days and 1 mL medium was replaced with fresh IL-2-containing medium every second day. On day ten, cells were washed and transferred to 96-well plates and re-stimulated with each peptide individually for six hours in the absence of IL-2 at a final concentration of 2ug/mL/peptide. Brefeldin A was added at the same time of stimulation at a final concentration of 10ug/mL. After six hours cells were washed and stained for viability, CD3, CD4, CD8, CD45RA and CD14. Cells were then fixed and permeabilized and stained for IFN-γ and TNF. Samples were acquired using the BD LSRFortessa flow cytometer and data was analyzed using the FlowJo software. IFN-γ and TNF production were analyzed to identify antigen-specific T cells. For three patients, the experiment was repeated and CD8+ IFN-γ+ T cells were sorted using the BD FACSMelody flow cytometer.
HLA immunoaffinity purification
HLA-I and -II molecules were isolated from snap-frozen tissue by standard immunoaffinity chromatographyCitation17 using the mABs W6/32, Tü39, and L243.
Mass spectrometric data acquisition
HLA ligand extracts were analyzed as previously described.Citation18,Citation19 Peptides were separated by nanoflow high-performance liquid chromatography. Eluted peptides were analyzed on an on-line coupled Orbitrap Fusion Lumos or an LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific, San Jose, CA).
Mass spectrometric data processing
Data processing was performed as previously describedCitation17 with the Proteome Discoverer software (v1.4, Thermo Fisher Scientific, San Jose, CA) to integrate the search results of the SEQUEST HT search engine (University of Washington)Citation20 with the human proteome (Swiss-Prot database). The false discovery rate (FDR; estimated by the Percolator algorithm 2.04)Citation21 was limited to 5% for HLA class I and 1% for HLA class II. HLA class I annotation was performed using SYFPEITHI 1.0Citation22 and NetMHCpan 4.0.Citation23
Analysis of target cell lysis
PBMCs were counted and plated at a concentration of 1 × 106 cells per well in low-adherence 24-well plates in 2 mL RPMI containing 8% human serum, 1% penicillin-streptomycin, 1% L-glutamine, 1% non-essential amino acids, 1% sodium pyruvate, 0.1 mg/mL kanamycin and 0.1% β-mercaptoethanol. After overnight incubation at 37°C single peptides were added to the wells at a final concentration of 2ug/mL/peptide. After 48 hours, 1 mL medium was replaced with fresh RPMI (as above) containing IL-2 (final concentration of IL-2 was 150 U/mL). Cells were kept in culture for ten days and 1 mL medium was replaced with fresh IL-2-containing medium every second day. On day ten, cells were collected, washed and transferred to 96-well plates before re-stimulation with peptide(s) for 6 hours in the absence of IL-2 at a final concentration of 2ug/mL/peptide. Simultaneously, target cells (EPLC-272 H, HLA-A 03:01 or NCI-H2030, HLA-C 04:01) were stained with a cell dye (detected in the APC channel) and half of the target cells were then also pulsed with the same peptide(s) for two hours at a final concentration of 2ug/mL/peptide. PBMCs and target cells were then co-cultured overnight in round-bottom 96-well plates in RPMI at an effector-to-target ratio of 10:1 or 30:1. The following day an apoptosis assay was carried out using the Apotracker green/PI kit to determine target cell death. Cells were acquired using the BD LSRFortessa flow cytometer and data was analyzed using the FlowJo software.
TCR sequencing
DNA was extracted from human NSCLC samples (formalin-fixed paraffin-embedded samples or fresh biopsy samples) and from sorted PBMCs. DNA was used for T cell receptor beta (TCRβ) sequencing using the ImmunoSEQ “Survey” resolution from Adaptive Biotechnologies. In brief, the ImmunoSEQ assay utilizes a biased-controlled multiplex PCR-based assay coupled with high-throughput sequencing and sophisticated bioinformatics to deliver highly accurate data sets. The data provided from the ImmunoSEQ platform was then analyzed using the Adaptive Biotechnologies Analyzer platform.
A fuzzy string matching analysis using a maximal edit distance of 1 was carried out to identify highly similar TCRs between sorted T cells and NSCLCs. Edit distance is defined as the Levenshtein distance, which is the number of removals, insertions, or substitutions of a character needed to transform one string into the other.
Analysis of specific TCRs using publicly available TCR database
Projects for which TCR sequences of blood, tumor and non-tumor samples of patients with NSCLC,Citation24,Citation25 melanomaCitation26,Citation27 and breast cancerCitation28,Citation29 were made available online were chosen by using the TCR data base TCRdb (http://bioinfo.life.hust.edu.cn/TCRdb/#/). The TCR sequences generated during these projects were reposited on the ImmunoSeq Analyzer platform, which were used to conduct the search for specific clonotypes. By employing the so-called CMV search tool, clones with matching sequences were identified in each sample and their total productive frequency was retrieved, which was then compared between the different compartments and tumor types.
Statistical analysis
Kaplan-Meier plots were generated for overall survival (OS) and progression-free survival (PFS) using GraphPad Prism. The association between the T cell response to the epitopes and OS or PFS was examined using the log-rank test. The hazard ratios and 95% confidence intervals were calculated using GraphPad Prism.
Student’s t-test and One-way ANOVA were also performed using GraphPad Prism.
Authors’ Contributions
Lukas Flatz had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Experimental design: Berner, Niederer, Flatz
Data acquisition: Berner, Niederer, Luimstra, Pop, AK Jochum, Purde, Bomze, Bauer, Freudenmann, Marcu, Wolfschmitt, Gross, Haen, Dubbelaar, Lenz, W Jochum, Rammensee, Walz, Neefjes
Data interpretation and analysis: Berner, Niederer, Luimstra, Bauer, Bomze, AK Jochum, Walz, Neefjes, Flatz
Sample and patient data collection and processing: Berner, Niederer, Pop, AK Jochum, Purde, Hasan Ali, Abdou, Appenzeller, Cicin-Sain, Jörger, Früh, Driessen, W Jochum, Cozzio
Manuscript writing: Berner, Flatz
Critical revision of the manuscript: Berner, Flatz
Clinical information: Niederer, Hasan Ali, Appenzeller, Cicin-Sain, Jörgen, Früh, W Jochum, Flatz
Technical or material support: Abdou, Baumgaertner, Speiser, Ludewig
Funding and study supervision: Flatz
Supplemental Material
Download ()Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website
Additional information
Funding
References
- Ribas A, Puzanov I, Dummer R, Schadendorf D, Hamid O, Robert C, Hodi FS, Schachter J, Pavlick AC, Lewis KD, et al. Pembrolizumab versus investigator-choice chemotherapy for ipilimumab-refractory melanoma (KEYNOTE-002): a randomised, controlled, phase 2 trial. Lancet Oncol. 2015;16(8):908–8. doi:10.1016/S1470-2045(15)00083-2.
- Robert C, Long GV, Brady B, Dutriaux C, Maio M, Mortier L, Hassel JC, Rutkowski P, McNeil C, Kalinka-Warzocha E, et al. Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med. 2015;372(4):320–330. doi:10.1056/NEJMoa1412082.
- Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, Chow LQ, Vokes EE, Felip E, Holgado E, et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl J Med. 2015;373(17):1627–1639. doi:10.1056/NEJMoa1507643.
- Carbone DP, Reck M, Paz-Ares L, Creelan B, Horn L, Steins M, Felip E, van Den Heuvel MM, Ciuleanu T-E, Badin F, et al. First-Line Nivolumab in stage IV or recurrent non-small-cell lung cancer. N Engl J Med. 2017;376(25):2415–2426. doi:10.1056/NEJMoa1613493.
- Djureinovic D, Hallstrom BM, Horie M, Mattsson JSM, La Fleur L, Fagerberg L, Brunnström H, Lindskog C, Madjar K, Rahnenführer J, et al. Profiling cancer testis antigens in non-small-cell lung cancer. JCI Insight. 2016;1(10):e86837. doi:10.1172/jci.insight.86837.
- Berner F, Bomze D, Diem S, Ali OH, Fassler M, Ring S, et al. Association of checkpoint inhibitor-induced toxic effects with shared cancer and tissue antigens in non-small cell lung cancer. JAMA Oncol. 2019;5(7):94–98. doi:10.1001/jamaoncol.2019.0402.
- Thomas A, Hassan R. Immunotherapies for non-small-cell lung cancer and mesothelioma. Lancet Oncol. 2012;13(7):e301–e10. doi:10.1016/S1470-2045(12)70126-2.
- Martins F, Sofiya L, Sykiotis GP, Lamine F, Maillard M, Fraga M, Shabafrouz K, Ribi C, Cairoli A, Guex-Crosier Y, et al. Adverse effects of immune-checkpoint inhibitors: epidemiology, management and surveillance. Nat Rev Clin Oncol. 2019;16(9):563–580. doi:10.1038/s41571-019-0218-0.
- Wang RF, Robbins PF, Kawakami Y, Kang XQ, Rosenberg SA. Identification of a gene encoding a melanoma tumor antigen recognized by HLA-A31-restricted tumor-infiltrating lymphocytes. J Exp Med. 1995;181(2):799–804. doi:10.1084/jem.181.2.799.
- Fleri W, Paul S, Dhanda SK, Mahajan S, Xu X, Peters B, Sette A. The immune epitope database and analysis resource in epitope discovery and synthetic vaccine design. Front Immunol. 2017;8:278. doi:10.3389/fimmu.2017.00278.
- Glanville J, Huang H, Nau A, Hatton O, Wagar LE, Rubelt F, Ji X, Han A, Krams SM, Pettus C, et al. Identifying specificity groups in the T cell receptor repertoire. Nature. 2017;547(7661):94–98. doi:10.1038/nature22976.
- Mirandola L, Figueroa JA, Phan TT, Grizzi F, Kim M, Layeequr Rahman R, Jenkins MR, Cobos E, Jumper C, Alalawi R, et al. Novel antigens in non-small cell lung cancer: SP17, AKAP4, and PTTG1 are potential immunotherapeutic targets. Oncotarget. 2015;6(5):2812–2826. doi:10.18632/oncotarget.2802.
- Yasumoto K, Hanagiri T, Takenoyama M. Lung cancer-associated tumor antigens and the present status of immunotherapy against non-small-cell lung cancer. Gen Thorac Cardiovasc Surg. 2009;57(9):449–457. doi:10.1007/s11748-008-0433-6.
- Wei X, Lai Y, Li J, Qin L, Xu Y, Zhao R, et al. PSCA and MUC1 in non-small-cell lung cancer as targets of chimeric antigen receptor T cells. Oncoimmunology. 2017;6(3):e1284722. doi:10.1080/2162402X.2017.1284722.
- Hasan Ali O, Berner F, Bomze D, Fassler M, Diem S, Cozzio A, Jörger M, Früh M, Driessen C, Lenz TL, et al. Human leukocyte antigen variation is associated with adverse events of checkpoint inhibitors. Eur J Cancer. 2019;107:8–14. doi:10.1016/j.ejca.2018.11.009.
- Dudley ME, Rosenberg SA. Adoptive-cell-transfer therapy for the treatment of patients with cancer. Nat Rev Cancer. 2003;3(9):666–675. doi:10.1038/nrc1167.
- Nelde A, Kowalewski DJ, Stevanovic S. Purification and identification of naturally presented MHC Class I and II ligands. Methods Mol Biol. 2019;1988:123–136.
- Kowalewski DJ, Schuster H, Backert L, Berlin C, Kahn S, Kanz L, et al. HLA ligandome analysis identifies the underlying specificities of spontaneous antileukemia immune responses in chronic lymphocytic leukemia (CLL). Proc Natl Acad Sci U S A. 2014;112(2):E166–75. doi:10.1073/pnas.1416389112.
- Marcu A, Bichmann L, Kuchenbecker L, Kowalewski DJ, Freudenmann LK, Backert L, et al. The HLA ligand Atlas. A Resource of Natural HLA Ligands Presented on Benign Tissues. 2020.
- Milner E, Gutter-Kapon L, Bassani-Strenberg M, Barnea E, Beer I, Admon A. The effect of proteasome inhibition on the generation of the human leukocyte antigen (HLA) peptidome. Mol Cell Proteomics. 2013;12(7):1853–1864. doi:10.1074/mcp.M112.026013.
- Käll L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods. 2007;4(11):923–925. doi:10.1038/nmeth1113.
- Schuler MM, Nastke MD, Stevanovic S. SYFPEITHI: database for searching and T-cell epitope prediction. Methods Mol Biol. 2007;409:75–93.
- Jurtz V, Paul S, Andreatta M, Marcatili P, Peters B, Nielsen M. NetMHCpan-4.0: improved Peptide-MHC Class I interaction predictions integrating eluted ligand and peptide binding affinity data. J Immunol. 2017;199(9):3360–3368. doi:10.4049/jimmunol.1700893.
- Reuben A, Zhang J, Chiou SH, Gittelman RM, Li J, Lee WC, Fujimoto J, Behrens C, Liu X, Wang F, et al. Comprehensive T cell repertoire characterization of non-small cell lung cancer. Nat Commun. 2020;11(1):603. doi:10.1038/s41467-019-14273-0.
- Kargl J, Busch SE, Yang GH, Kim KH, Hanke ML, Metz HE, Hubbard JJ, Lee SM, Madtes DK, McIntosh MW, et al. Neutrophils dominate the immune cell composition in non-small cell lung cancer. Nat Commun. 2017;8(1):14381. doi:10.1038/ncomms14381.
- Valpione S, Galvani E, Tweedy J, Mundra PA, Banyard A, Middlehurst P, Barry J, Mills S, Salih Z, Weightman J, et al. Immune-awakening revealed by peripheral T cell dynamics after one cycle of immunotherapy. Nat Cancer. 2020;1(2):210–221. doi:10.1038/s43018-019-0022-x.
- Pruessmann W, Rytlewski J, Wilmott J, Mihm MC Jr., Attrill GH, Dyring-Andersen B, Fields P, Zhan Q, Colebatch AJ, Ferguson PM, et al. Molecular analysis of primary melanoma T cells identifies patients at risk for metastatic recurrence. Nat Cancer. 2020;1(2):197–209. doi:10.1038/s43018-019-0019-5.
- Beausang JF, Wheeler AJ, Chan NH, Hanft VR, Dirbas FM, Jeffrey SS, Quake SR. T cell receptor sequencing of early-stage breast cancer tumors identifies altered clonal structure of the T cell repertoire. Proc Natl Acad Sci U S A. 2017;114(48):E10409–E17. doi:10.1073/pnas.1713863114.
- Page DB, Yuan J, Redmond D, Wen YH, Durack JC, Emerson R, Solomon S, Dong Z, Wong P, Comstock C, et al. Deep sequencing of T-cell receptor DNA as a biomarker of clonally expanded TILs in breast cancer after immunotherapy. Cancer Immunol Res. 2016;4(10):835–844. doi:10.1158/2326-6066.CIR-16-0013.
