ABSTRACT
Lung tumor-infiltrating neutrophils are known to support growth and dissemination of cancer cells and to suppress T cell responses. However, the precise impact of tissue neutrophils on programming and differentiation of anticancer CD8 T cells in vivo remains poorly understood. Here, we identified cancer cell-autonomous secretion of CXCL5 as sufficient to drive infiltration of mature, protumorigenic neutrophils in a mouse model of non-small cell lung cancer (NSCLC). Consistently, CXCL5 transcripts correlate with neutrophil density and poor prognosis in a large human lung adenocarcinoma compendium. CXCL5 genetic deletion, unlike antibody-mediated depletion, completely and selectively prevented neutrophils accumulation in lung tissues. Depletion of tumor-infiltrating neutrophils promoted expansion of tumor-specific CD8 T cells, differentiation into effector cells and acquisition of cytolytic functions. Transfer of effector CD8 T cells into neutrophil-rich tumors, inhibited IFN-ϒ production, indicating active suppression of effector functions. Importantly, blocking neutrophils infiltration in the lung, overcame resistance to checkpoint blockade. Hence, this study demonstrates that neutrophils curb acquisition of cytolytic functions in lung tumor tissues and suggests targeting of CXCL5 as a strategy to restore anti-tumoral T cell functions.
Introduction
Tumor-infiltrating neutrophils (neu) have been associated with worst prognosis in the majority of human cancers and experimental models, with the exception of some selected tumor types.Citation1 Multiple direct protumorigenic functions related to cancer cell proliferation,Citation2,Citation3 metastasisCitation4–7 and angiogenesisCitation8,Citation9 have been described. In addition, the capacity of neutrophils to suppress proliferation of T cells ex vivo has been largely documented in humans and experimental mouse models.Citation10–14 Lung tumors are characterized by increased neutrophil counts in periphery and in tumor tissues and neu accumulation correlates to decreased CD8 T cells and reduced cytotoxicity.Citation15–19 Moreover, accumulating evidences indicate that neutrophils in lung cancer determine resistance to checkpoint blockade and chemotherapy.Citation20–24 Sparse evidences documented changes in the T cell compartment upon neutrophils depletion or drugs blocking recruitment.Citation4,Citation13,Citation24,Citation25 Single-cell sequencing and mass-cytometry recently resolved the diversity and complexity of neu subsets in lung tumor bearing hosts, unveiling multiple neu subsets with graded protumorigenic potential.Citation26,Citation27 In parallel, characterization of dysfunctional T cell states within lung tumor tissue provides novel markers to assess the interplay between the myeloid suppressive immune infiltrate and the anticancer CD8 response.Citation28,Citation29 What causal relationship connects subsets of tumor-infiltrating neutrophils to dysfunctional endogenous tumor-specific CD8 T cell responses in vivo in lung tissues remains poorly defined.
Infiltration by neu is often caused by oncogene-driven overexpression of chemokines.Citation30 Overexpression of CXCL5 that binds CXCR2 drives neutrophils infiltration in hepatocellular carcinoma (HCC), intrahepatic cholangiocarcinoma (ICC),Citation31,Citation32 melanomaCitation33,Citation34 bladder cancer,Citation35 prostate cancerCitation31 and breast cancer.Citation36,Citation37 In lung tissues, seminal studies described CXCL5 as critical to recruit neutrophils during inflammation and infection.Citation38–40 In lung cancer, CXCL5 was shown to induce cancer cell proliferation, migration,Citation41,Citation42 autophagyCitation43 and epithelial mesenchymal transitionCitation44 and it is associated with poor prognosis.Citation45,Citation46 CXCL5 is also expressed in experimental models of lung cancer,Citation47,Citation48 however the direct link to neutrophils recruitment and interference with anti-tumoral T cell responses has not been demonstrated.
The orthotopic KrasG12D/WT Tp53 mouse adenocarcinoma model (KP) recapitulates the neutrophils structure found in human lung tumors and it is emerging as a potent preclinical tool to identify immune-cancer networks in human NSCLC.Citation29,Citation49,Citation50 KP tumors contain a population of mature aged protumorigenic neutrophils expressing the lectin SiglecF,Citation26,Citation51 whose precise mechanism of accumulation and interference with anticancer CD8 responses has not been assessed.
Here, we generated an immunogenic variant of the KP model to explore the interplay between neutrophils and anticancer CD8 responses. By RNAseq, we identified CXCL5 as the dominant cancer-cell autonomous upregulated chemokine. Gene editing to delete CXCL5 expression in cancer cells resulted in tumors that selectively lack neutrophils without gross changes in the remaining immune infiltrate. CXCL5-null KP clones permitted clonal expansion and activation of effector CD8 T cells functions in lung tissues, and neutrophil-depleted tumors became sensitive to treatment with checkpoint inhibitors.
Results
Parental KP and immunogenic KP lung tumors are dominated by SiglecFhigh neu
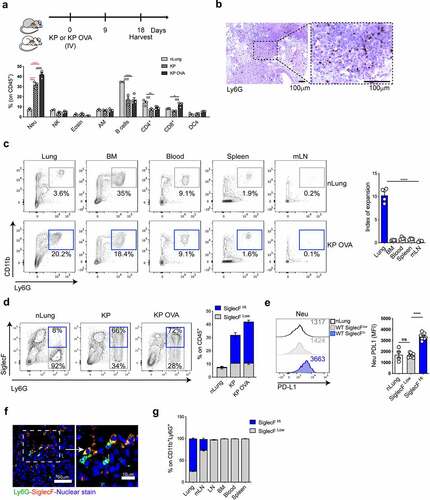
To study the interplay between neu and anticancer T cell responses in lung tumor tissue, we established an orthotopic lung adenocarcinoma model (KP cell line),Citation27,Citation52 engineered to express the OVA neoantigen (KP-OVA). As previously shown, cancer-specific CD8 T cell responses are established in early tumors but progressively decline due to defective antigen presentation by type 1 DCsCitation53 and accumulation of dysfunctional T cells.Citation28,Citation29 Flow cytometry of the immune infiltrate showed a three- to fourfold expansion of CD11b+/Ly6G+ in both parental and immunogenic KP-OVA tumors (). KP and KP-OVA showed a slight decrease in CD4 and B cells and KP-OVA, as expected, showed a higher frequency of CD8 T cells ( and S1A). Staining of lung sections by anti-Ly6G antibodies confirmed that neu are deeply infiltrated in tumor nodules (). Neu expansion was not present in the blood, spleen or tumor draining mediastinal lymph node (mLN), suggesting that orthotopic KP-OVA lung tumors, unlike subcutaneous colon or other lung tumor models,Citation54 accumulate neu only locally (). We next analyzed expression of the lectin SiglecF on neu accumulating in orthotopic lung tumors. Up to 70% of CD11b+/Ly6G+ cells in both KP and KP-OVA tumors expressed SiglecF (SiglecFhigh neu), whereas the majority of neu in normal lungs were SiglecF low (SiglecFlow neu) (). In agreement with previous data, we observed increased PD-L1 expression on SiglecFhigh neu.Citation50 Tissue fluorescence confirmed that SiglechFhigh neu are found within nodules (). A small fraction of SiglecFhigh neu was found as well in the mLN but not in the non-draining LNs, blood, bone marrow or spleen, in line with previous studies indicating that this subset differentiates specifically in tumor tissuesCitation50 ().
Figure 1. Neutrophils density in KP and KP OVA orthotopic lung tumors. (a–f) Mice were challenged intravenously with KP, KP-OVA cells or PBS (nLung) as depicted in the scheme. The immune composition was evaluated by flow cytometry in normal lungs (nLung) and established lung tumors, 18 d upon inoculation. (a) Relative abundance of each subset expressed as a fraction of live CD45+ cells (neu = neutrophils, Eosin = eosinophils, AM = alveolar macrophages). Data represent mean ± SEM of two to four mice each group. Significance was determined by two-way ANOVA with *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ****p ≤ 0.0001. (b) Representative 10× image of paraffine-embedded KP-OVA tumor tissue labelled with Ly6G antibody (brown) and the corresponding 40× magnification. Scale bars (100 μm) are showed. (c) Representative dot plot of neu in lungs (gated on live CD45+,Lin−), BM (bone marrow), blood, spleen and mLN (mediastinal lymph node) of normal and KP OVA tumor bearing mice. The expansion index represents the ratio between neu in KP-OVA and nLung. Data represent mean ± SEM of four mice each group. (d) Representative dot plots showing SiglecF expression on lung neu (gated on CD45+, Lin−, Ly6G/CD11b+ cells). Numbers in quadrant show percentages of SiglecFhigh/low among CD11b+Ly6G+ cells. Bars show quantification of SiglecFhigh/low neu frequencies on CD45+ total lung cells. Data are mean ± SEM of three to five mice/group. (e) PD-L1 expression on lung-infiltrating neu. Representative histogram of PD-L1 expression on neu from nLung or KP-OVA WT tumors. PD-L1 MFI (median fluorescence intensity) are indicated in the histogram and plotted on the right as mean ± SEM of two experiments with two to four mice each group. Significance was determined by one-way ANOVA with ns p > 0.05, ****p≤ 0.0001. (f) Representative cryo-sections showing Ly6G (green) and SiglecF (red) in tumor nodules and the corresponding magnification. Scale bars (50 or 10 μm) are indicated. (G) Percentage of SiglecFhigh/low neu within CD11b+Ly6G+ cells in the indicated organs isolated from KP OVA tumor bearing mice (mLN = mediastinal lymph node, LN = inguinal lymph node, BM = bone marrow). Data represent the mean ± SEM of three to six mice each group.
CXCL5 is highly expressed by mouse lung tumors and correlates to neutrophil infiltrate in human cancers
We tested a classical protocol for neutrophil depletion by administration of anti-Ly6G antibodies into KP-OVA bearing mice.Citation4,Citation55 Despite efficient depletion in the blood, a large fraction of neu persisted in lung tumor tissues as previously reported,Citation56 precluding further analysis (Fig S2A-D). We thus turned into gene expression data to identify mechanisms of neu accumulation in KP tumors. We identified three chemokines (Cxcl5, Cxcl16 and Ccl21a) in common between the top 10% highly expressed chemokine genes in isolated KP cells and KP-OVA tumor tissues (). Of these, Cxcl5 that is directly implicated in neu recruitment in various cancer tissues and in inflamed lungs was chosen for subsequent analysis. A 20-fold induction of Cxcl5 transcripts in tumor lung tissues was also confirmed using a gene expression array (Fig S3A). To isolate the source of Cxcl5 we analyzed tumor tissues upon fractionation into CD45 negative and positive cells and in isolated KP cells ex vivo. We found high expression of Cxcl5 primarily in CD45 negative cells in cancer tissues and in KP cells (). We conclude that murine KP lung tumors express high levels of Cxcl5 by a cancer-cell autonomous mechanism.
Figure 2. CXCL5 is highly expressed in mice tumors and human LUAD. (a) Venn diagram showing chemokines and chemokine receptors genes that were common between nLung tissues, KP lung tissues and the KP cell line. Top 10% quantile was used as a cut-off for highly expressed genes in each sample. nLung data are from normal C57Bl6/J lung of the Expression Atlas; KP-OVA lung and KP cell line data are from in-house RNA-seq. (b) Cxcl5 transcripts in CD45+ or CD45− cells isolated from nLung or KP OVA tumor bearing lungs were quantified by RT-PCR. Graph shows mean ± SEM of three replicates, significance was determined by one-way ANOVA with ***p ≤ 0.001; ****p ≤ 0.0001. (c) Cxcl5 transcripts in the KP-OVA cell line and in nLung. Mean ± SEM are from five to eight independent RNA extractions, significance was determined by unpaired t-test with **p ≤ 0.01. (d) Patients from the compendium in , were stratified based on low, medium or high neu content based on CIBERSORT and correlated to the level of expression of CXCL5. (e) Correlation between a human neu gene signature (hNeutrophils) and expression of CXCL5 within the Compendium in . (f) Prognostic value of CXCL5 for overall survival of human cancer patients in the compendium (n = 989), comparing top and bottom quartiles.
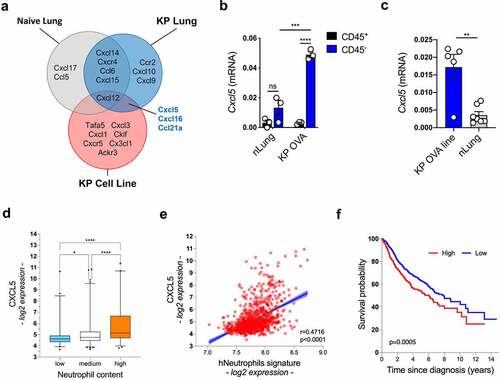
To explore the correlation between CXCL5 expression and neutrophils recruitment in human lung tumors, we next compiled a large compendium of lung adenocarcinoma (LUAD) (), that integrates multiple gene expression datasets. Patients were stratified using CIBERSORT Citation57 to create three categories corresponding to low, medium and high neu density. Interestingly, we observed a high significant correlation between CXCL5 expression and neu density (). Moreover, expression of a gene signature corresponding to tumor-infiltrating neuCitation27 highly correlates to CXCL5 expression (). In line with previous reports,Citation46 we confirmed that expression of CXCL5 is associated with worst prognosis also in this large compendium (). Together, these data indicate that in humans, as in mice, enrichment of neu in lung cancer tissues is associated with overexpression of CXCL5.
Table 1. Datasets composing the lung cancer compendium
CXCL5 deletion inhibits accumulation of tumor associated SiglecFhigh neu
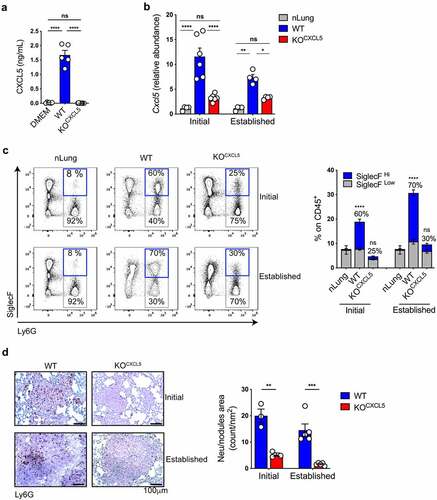
We next tested the possibility to interfere with neu recruitment by targeting CXCL5 expression in KP cells by CRISPR/CAS9 genome editing (). WT and KOCXCL5 KP-OVA clones were selected by ELISA and validated for OVA expression and growth rate ( and S3B-D). Lung tissues of animals challenged with parental KP-OVA cells showed an early peak of Cxcl5 expression that persisted in late tumors. In contrast, lungs challenged with the CRISPR-targeted clone had only a modest increase in Cxcl5, both at early and late time points (). These data demonstrate efficient gene deletion and confirm chemokine production primarily by cancer cells and not by stromal cells conditioned by tumor environmental factors. To test whether deletion of CXCL5 was sufficient to blunt neu arrival we analyzed the immune infiltrate at two consecutive time points. Tumors formed by the parental KP-OVA line showed the expected accumulation of SiglecFhigh neu already at initial stages, which slightly intensified at later stages (). In sharp contrast, tumors induced by CXCL5 deficient cells were devoid of neu at early stages and showed only a modest increase at later stages. Of the few infiltrating neu in KOCXCL5 tumors, a minor fraction expressed SiglecF (). Immunohistochemistry confirmed lack of Ly6G+ cells in nodules of KOCXCL5 challenged mice, both at initial stages and in more advanced tumors (). Lack of CXCL5 secretion by KP-OVA cells had no impact on the frequency of circulating and splenic neu whereas it resulted in higher neu content within the bone marrow, likely reflecting reduced mobilization (S3E-G). We conclude that CXCL5 is a non-redundant mechanism driving neu accumulation in KP lungs. To ascertain that lack of neu recruitment in KOCXCL5 tumors is causally linked to chemokine expression, we reintroduced CXCL5 expression by lentiviral transduction, creating KOCXCL5(lenti-CXCL5) or control KOCXCL5(lenti-vec) (Fig S4A). Reconstituted KP-OVA cells were validated for CXCL5 chemokine production in vitro and expression in lung tissues (Fig S4B,C). Importantly, recruitment of neu was restored in the TME of KOCXCL5(lenti-CXCL5) tumors, but not in control KOCXCL5(lenti-vec), indicating that chemokine expression alone is sufficient to induce neu influx (Fig S4D).
Figure 3. CXCL5 deletion blunts neu recruitment in immunogenic KP lung. (a) Secretion of CXCL5 by parental and gene deleted KP-OVA clones was assessed by ELISA. Data represent the mean ± SEM of five to nine independent measurements. Significance was determined by a one-way ANOVA with ****p ≤ 0.0001. (b–d) Mice were inoculated with WT or KOCXCL5 KP-OVA clones. Lung tissues were harvested after 9 (initial) or 18 (established) d for analysis. (b) Relative abundance of Cxcl5 transcripts in total lung were evaluated by qRT-PCR. Data are expressed as fold induction over nLung. Data represent the mean ± SEM of four to six independent RNA extraction. Significance was determined by two-way ANOVA with *p ≤ 0.05, **p ≤ 0.01, ****p ≤ 0.0001. (c) Representative dot plots showing infiltrating neu and SiglecF expression (gated on CD45+CD11b+ cells). Numbers in quadrants refer to percentages of SiglecF+ on CD11b+/Ly6G+ cells. Bars show the frequencies (%) of neu on live CD45+ lung cells. The fractions of SiglecFHigh and SiglecFLow within neu is indicated. Data represent the mean ± SEM of two independent experiment, three to four mice each group. Significance was determined by a two-way ANOVA with ****p ≤ 0.0001. (d) Representative IHC tissue sections of initial or established nodules labelled with Ly6G antibody (brown dots). Scale bars (100 μm) are indicated. Graph on the right represents quantification of Ly6G+ on nodule area (count/nm2). Data are mean ± SEM of three to five mice each group. Significance was determined by a two-way ANOVA with **p ≤ 0.01; ***p ≤ 0.001.
Neutrophil density inversely correlates with CD8 T cells in tumor nodules
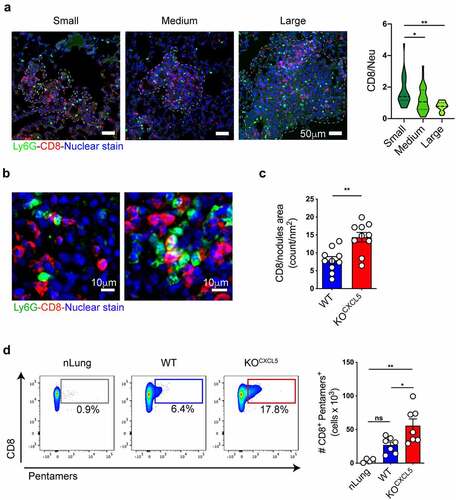
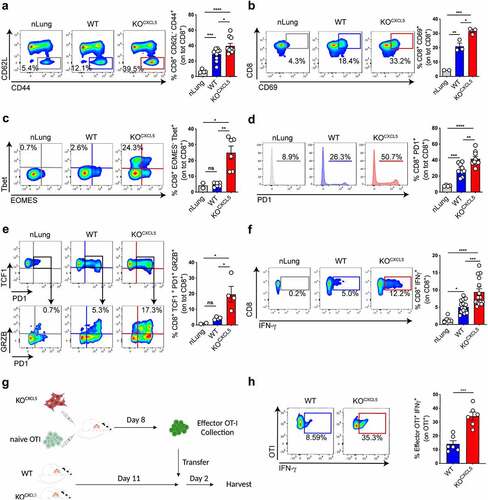
Given the efficient and selective depletion of infiltrating neu observed in KOCXCL5 tumors, we moved to examine the consequences on tumor-specific CD8 T cell responses. Previous studies showed exclusion of CD8 T cells by infiltrating neu in a genetic model of colorectal cancer.Citation25 To explore the relative distribution of the two cell types in lung tumors, we classified nodules into three categories corresponding to small (area <0.009 mm2), medium (area between 0.0091 mm2 and 0.02 mm2) and large (area >0.021 mm2). We found that CD8 T cells outnumbered neu in small nodules and were equal to neu in medium nodules, whereas the proportion was inverted in large nodules that contained more neu than CD8 T cells (), suggesting T cell exclusion. Higher magnification images revealed several neu-CD8 synapses and some areas where CD8 T cells were surrounded by multiple neu, indicative of tight intercellular interactions (). Moreover, the density of CD8 T cell was significantly higher in KOCXCL5 tumor nodules that lack neu (). In line with this observation, flow cytometry to measure the fraction of tumor specific, endogenous, CD8 by pentamer staining showed a threefold expansion in neu depleted tumors ().
Figure 4. Neu depleted lung tumors promote infiltration and expansion of cancer-specific CD8 T cells. (a) Nodules were classified into 3 categories based on the area (small<0.009 mm2, 0.0091 mm2 < medium<0.02 mm2, large>0.021 mm2). Scale bars (50 μm) are indicated. On the right, quantification of the relative abundance of CD8 and neu within nodules of different sizes. Data are expressed as ratio of CD8/neu (mean ± SEM of 90 nodules from two independent experiments with 2 mice each). Significance was determined by one-way ANOVA with *p ≤ 0.05, **p ≤ 0.01. (b) High magnification examples of representative lung sections from WT KP OVA tumors co-labelled with antibodies to CD8 (red) and Ly6G (green) and counterstained with Hoechst to visualize nuclei. (c) Quantification of infiltrating CD8 (CD8 count/nm2) in WT or KOCXCL5 KP OVA nodules. Data are from lung tumor sections from two independent experiments with five mice each group (mean ±SEM). Significance was determined by an unpaired t-test with **p ≤ 0.01. Scale bars (100 μm) are indicated. (d) Representative dot-plot showing OVA class-I pentamers labelling on lung CD8+ T cells. Absolute number (left) of CD8+pentamer+ cells are plotted. Data are mean ± SEM of two independent experiments with 3/4 mice/group. Significance was determined by one-way ANOVA with *p ≤ 0.05, **p ≤ 0.01.
Neutrophils inhibit markers of effector T cell differentiation and cytolytic functions
To further explore the CD8 compartment in growing lung tumors, animals were challenged with WT or KOCXCL5 KP-OVA cells and tissues were harvested at the onset of the T cell responses. The fraction of CD8 T cells carrying activation (CD69) and effector memory differentiation (CD44high/CD62Llow) markers were significantly higher in the lung of KOCXCL5 tumors as compared to WT (. Deeper analysis of intracellular transcription factors showed a significantly higher fraction of cells committed toward the effector fate (T-bet+/Eomes−), in neu depleted tumors (). In both WT and KOCXCL5 tumors, CD8 T cells upregulated the exhaustion markers PD-1, indicating exposure to cognate antigen, which was more pronounced in KOCXCL5 tumors (). We next investigated expression of TCF-1 transcription, which defines a state of not terminally exhausted cells still retaining proliferative and effector functions. The majority of CD8 T cells in the lung of tumor-bearing mice expressed TCF-1, in agreement with recent data.Citation29 Among PD1+/TCF-1+ CD8, the fraction of cells producing granzyme was significantly higher in CXCL5KO tumors, indicating maintenance of effector functions and reduced exhaustion (). To further understand whether endogenous CD8 T cells displaying effector functions were tumor specific, we restimulated lung cell suspensions ex vivo with OVA class-I peptide. Notably, tumor tissue resident CD8 T cells secreted twice as much IFN-γ in neu depleted tumors than in control tumors (). Finally, to establish if neu actively suppresses effector functions in CD8 T cells, we generated tumor-specific effector CD8 T cells in KP-OVA neu-depleted tumors. Effector cells were then transferred into mice carrying WT or KOCXCL5 tumors and the production of IFN-ϒ by transferred CD8 T cells in the lung was evaluated 2 d later. IFN-ϒ production was strongly decreased upon transfer in hosts carrying WT tumors, indicating that effector functions are inhibited in the presence of tissue-infiltrating neu (.
Figure 5. Lung-infiltrating neu restrict differentiation into effector CD8 T cells. (a–e) Mice were challenged with WT or KOCXCL5 KP-OVA cells and endogenous T cell responses in the lungs were analysed by flow cytometry. (a) Representative plots showing CD62L and CD44 expression on lung CD8 T cells and corresponding quantification. (b) Representative plots and quantification of CD69 in lung CD8 T cells.(cC) Expression of T-bet and EOMES in lung CD8 T cells.)d) Representative histograms and quantification of PD-1 expression on total lung CD8 T cells. (e) Example and quantification of the percent of granzyme and PD-1 expression levels on TCF1-expressing lung CD8 T cells. (f) Single-cell lung suspensions were restimulated ex-vivo with OVA class-I peptide and intracellular staining was performed to determine IFN-γ expression levels. Representative dot-plots and quantifications are shown. Data (a–f) represent mean ± SEM of two to eight mice/group. Significance was determined by one-way ANOVA with ns p > 0.05, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001. (g) Scheme of OVA-specific CD8 effector adoptive transfer. CD45.2 mice were inoculated with KP-OVA KOCXCL5 cancer cells and adoptively transferred with OT-I. Effector OT-I cells were sorted from the mLN after 8 d and re-transferred into CD45.1 recipient carrying WT or KOCXCL5 tumors. Two days later lung tissues were harvested to perform intracellular staining for IFN-γ as described in (f). (h) Dot plot and quantification of effector OT-I on total lung OT-I T cells. Data represent mean ± SEM of four to six mice each group. Significance was determined by unpaired t-test with *p ≤ 0.05, ***p ≤ 0.001.
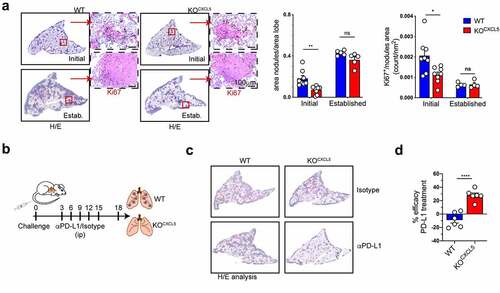
Lung tissue-infiltrating neu limit the capacity of anti PD-L1 antibodies to reduce tumor growth
We next examined the impact of selective deletion of infiltrating neu on growth of KP-OVA lung tumors. The size of tumor nodules and the number of proliferating tumor cells was evaluated in initial and established tumors. At early stages, nodules were significantly reduced in KOCXCL5 tumors and the fraction of proliferating cancer cells was diminished (). However, tumor growth was similar between the two genotypes at later time points, indicating that tumor containment by cytotoxic CD8 T cells was not sufficient to control growth at later stages. By decreasing the cancer cells input, we found delayed tumor growth also at later time point in KOCXCL5 tumors (Fig S5). The efficacy of checkpoint blockade was recently shown to inversely correlate to mutations in Stk11, CXCL5 levels and neu recruitment in NSCLC patients.Citation20,Citation58 Experimental data further suggested that neu depletion can increase the benefit of chemotherapy by reverting T cell suppressionCitation24 and radiation therapy was shown to synergizes with rewiring of aged neu in lung tumors.Citation51 Moreover, a recent study demonstrated that NETs counteract checkpoint blockade by inducing CD8 T cells exhaustion.Citation59 To evaluate whether blocking neu recruitment could synergize with anti-PD-L1 treatment in our model, we inoculated mice with WT or KOCXCL5 KP-OVA followed by administration of antibodies to PD-L1 or isotype every 3 d (). In line with previous studies,Citation60 the treatment had no impact on the growth of WT KP tumors. In contrast, PD-L1 blockade effectively decreased the growth of KOCXCL5 tumors (). We concluded that CXCL5-driven accumulation of neu in KP lung tumors prevent acquisition of anticancer CD8 effector functions and limits the activity of checkpoint blockade.
Figure 6. Targeting CXCL5-mediate neutrophils recruitment increase the effectiveness of checkpoint blockade. (a) Mice were challenged with WT or KOCXCL5 KP-OVA cells and lungs were harvested after 9 or 18 d to score tumor burden (area nodules/area lobe) and cancer cell proliferation. Representative images of tumor nodules labelled by HE and Ki67 staining. Bars show quantification of tumor burden (area nodules/total lobe area) and quantification of the number of Ki67+ cells (expressed as number of cells/nodule area). Data are mean ± SEM of one to two experiments with four to five mice each group. Significance was determined by two-way ANOVA with ns p > 0.05, *p ≤ 0.05, **p ≤ 0.01. (b) Mice were challenged with WT or KOCXCL5 tumor cells and treated with aPD-L1 or isotype control at d 3, 6, 9, 12. Lung tissue were harvested at d 18 to evaluate tumor burden by HE. (c) Representative examples of lung sections by IHC. D) The therapeutic efficacy of anti PD-L1 antibodies was expressed as fold difference over isotype treated controls on tumor area calculated as in (a). Data represent the mean ± SEM of two experiments with three mice each group. Significance was determined by two-way ANOVA with ns p > 0.05, *p ≤ 0.05, ****p ≤ 0.0001.
Discussion
This study shows that blunting neu recruitment in lung cancer tissues allows expansion, effector state commitment and cytoxicity of tumor-specific CD8 T cells. These findings expand and complement previous studies by providing a more detailed analysis of neu-driven changes in anti-tumoral CD8 responses in vivo, in the original environment of the tumor. Recent advances unveiled the complexity of neu and CD8 T cells states in cancer tissues, revealing the importance of studying tumors in the native tissue of origin and the need to increase the markers used to define effector functions and exhaustion.Citation27,Citation29 The negative association of neu density with patient outcome in lung adenocarcinoma remains mostly correlative, thus we focused on a valid lung cancer experimental model to address the crosstalk between neu and tumor-specific CD8 T cell responses in tissues. In line with past data, transplantable KP lung tumors and, as demonstrated here, immunogenic KP-OVA, accumulated a large fraction of SiglecFhigh neu, a mature subset with protumorigenic functions.Citation26,Citation51 Since neu depletion by conventional antibodies to Ly6G failed in lung tissues, we sought to identify targetable pathways guiding their recruitment to the lung. The chemokine CXCL5 turned out to be the dominant CXCR2 ligand expressed in KP cancer cells, in line with previous data showing upregulation of chemokines downstream oncogenic Kras.Citation48 Despite other pathways such as G-CSFCitation51 and IL-17Citation61 may operate in neu recruitment to the lung, genetic deletion of CXCL5 in murine cancer cells was sufficient to completely inhibit accumulation of mature neu. SiglecFhigh neu accumulate as well in slowly progressing autochthonous KP tumors,Citation51 yet it remains to be explored whether CXCL5 acts as a dominant factor controlling recruitment also in this context. Of relevance, CXCL5 expression in human lung cancer patients is emerging as a biomarker for prognosis and response to therapyCitation45,Citation58,Citation62 and we found that its expression highly correlates to neu density and predicts survival across a large human compendium suggesting a similar non-redundant role in lung cancer patients. It still remains to be established, however, what is the major cellular source of the chemokine in the human context.
Tumors lacking a neu infiltrate, offered a well-controlled approach to compare the phenotype of CD8 T cells in tissues. Tumor devoid of neu showed a larger expansion of tumor-specific CD8 T cells and a significant upregulation of surface markers and transcription factors defining effector T cells, as compared to neu-infiltrated tumors. Based on recent evidences describing diverse levels of T cell dysfunction in lung tumors,Citation28,Citation63 we propose that neu contribute to induce a hyporesponsive state with reduced effector functions in CD8 T cells. Consistently, neu confer resistance to PD-L1 blockade, which is in line with the correlation between neu density and response to immunotherapy in human cohorts of NSCLC treated with ICB.Citation64 One possibility is that neu depletion lower the load of PD-L1 molecules reducing terminal exhaustion and allowing reactivation upon therapy. Alternatively, other inhibitory signals arising from mature tissue neu render T cells refractory to reactivation by checkpoint inhibitors. Previous studies proposed several mechanisms of T cell suppression by neutrophils including NOS and ROS, Arginase1, Fas-FasL induced apoptosis, PD-L1 expression and PGE2 production.Citation4,Citation11,Citation12,Citation65,Citation66 The suppressive mechanisms operating preferentially in vivo in different cancer tissues need to be further elucidated and are likely to be context dependent. The present study suggests that future analysis should be performed in vivo using multiple parameters to define the extent of T cell dysfunction, beyond assays of ex-vivo T cell suppression. This study also provides a proof of concept that neutralizing pathological overexpression of CXCL5 by antibodies may be viable and more specific than blocking chemokine receptors Citation24 to reinforce the efficacy of checkpoint inhibitors.
In conclusion our findings uncover that SiglechFhigh neu accumulating in lung tissues curb development of effector CD8 T cell functions and impede reactivation by checkpoint blockade, suggesting CXCL5 as an interesting axis to target in order to restore an immune responsive tumor environment.
Material and methods
Mice
C57BL/6 and OT-I (C57BL/6-Tg(TcraTcrb)1100Mjb/J) mice were purchased from Envigo or Jackson Laboratories, respectively. Animals were maintained in sterile isolators at the ICGEB animal Bio-experimentation facility (12 h/12 h light and dark cycle, 21°C ± 2°C).
Sample size was determined based upon prior knowledge of the intragroup variation of tumor challenges by our laboratory and published studiesCitation49,Citation50 and was sufficient to detect meaningful biological differences with good reproducibility. The study was approved by International Centre for Genetic Engineering and Biotechnology (ICGEB) board for animal welfare and authorized by the Italian Ministry of Health (approval number 1133/2020-PR, issued on 12/11/2020). Animal care and treatment were conducted with national and international laws and policies (European Economic Community Council Directive 86/609; OJL 358; December 12, 1987). All experiments were performed in accordance with the Federation of European Laboratory Animal Science Association (FELASA) guidelines
Cell lines
The KP cells (LG1233) were generated from lung tumors of C57BL/6 KP mice (K-rasLSLG12D/+;p53fl/fl mice) and kindly provided by Dr Tyler Jacks (Massachusetts Institute of Technology, Cambridge, USA).Citation67 KP OVA cells were generated by transducing with the lentiviral vector Pdual-liOVAha-PuroR as described in.Citation53 To generate KOCXCL5 cells, the KP OVA cells were transiently co-transfected with pSpCas9(BB)-(PX458) and pZac2.1-U6sgRNA-CMV-ZsGreen plasmids, carrying 5’-caccgCTGCCGCAGCATCTAGCTGA-3’ guide. The ZsGreen+ cells were sorted and single clones were tested for CXCL5 expression by ELISA (Abcam ab100719).
To rescue CXCL5 expression in KP OVA KOCXCL5 cells were transduced with a lentivirus carrying the expression vector pLVX-IRES-G418-CXCL5 or empty vector pLVX-IRES-G418 as control. After antibiotic selection, CXCL5 expression was tested by ELISA.
All cell lines were maintained in DMEM media (containing 1 g/L of glucose) supplemented with 10% fetal bovine serum (FBS, Euroclone) and Gentamicin (50 μg/mL, Gibco) and routinely tested for mycoplasma contamination. The growth rate in vitro was assessed by crystal violet assay.
Tissue preparation for flow cytometry
Lung tissues from control or tumor bearing mice were harvested after PBS lung circulatory perfusion, mechanically cut into small pieces and digested with 0.1% Collagenase type 2 (265 U/mL; Worthington) and DNase I (250 U/mL; Thermo Scientific) at 37°C for 30’. Cells were filtered using a 70-μm cell strainer (Corning), to obtain single-cell suspensions. Blood was collected through subclavian vein puncture, followed by red blood cell lysate. Spleen, lymph nodes (mediastinal and inguinal) were smashed and filtered with a 70 - and 40 -μm cell strainer, respectively. Bone marrow was extracted from femur and tibia, flush to obtain a single-cell suspension.
Flow cytometry
For cell staining, FcR binding sites were blocked by using αCD16/CD32 and viability of cells was assessed by staining with LIVE/DEAD dye). The antibodies used for the experiments are listed in Supplementary Table 1. To analyze Cxcl5 expression, CD45+ or – populations from healthy or tumor-bearing lungs were stained with CD45-A647 antibody and sorted. MHC-I-OVA pentamers (SIINFEKL/H-2Kb Pro5, Proimmune) were used to identify OVA specific CD8 T cells following manufacturer’s instruction. For intracellular detection of IFNγ, single-cell suspensions were stimulated with OVA peptide (SIINFEKL) 2 µM 37°C for 4 h in the presence of Golgi Stop (monensin, BD Biosciences). Upon extracellular staining, cells were fixed and permeabilized using Cytofix/Cytoperm solution (BD Biosciences) following manufacturer’s instructions.
To identify effector EOMES−Tbet+ CD8+ T cells, we performed a nuclear staining analyzed using Foxp3/transcription factor staining buffer set (Thermo Fisher) following manufacturer’s instructions.
To measure OVA expression, cells were fixed and permeabilized using Cytofix/Cytoperm solution (BD Biosciences) following manufacturer’s instructions, stained with rat-monoclonal αHA and then with αRAT-AF488. Where indicated, absolute cell count was analyzed by adding TrueCount Beads (BioLegend) to the samples following manufacturer’s instructions. Samples were acquired with FACS Celesta (BD Biosciences) and analyzed with FlowJo software (Tree Star, Inc.).
Flow cytometry experiments are based on objective measurements and blinding was not required.
Real-time PCR
RNA from total lungs or sorted cells was extracted using Trizol reagent (Thermo Fisher Scientific), according to manufacturer’s instruction. cDNA was synthesized using SuperscriptII (ThermoFisher) and quantitative real-time PCR (qRT-PCR) was performed using SsoFast EvaGreen Supermix (Biorad) with specific primers: Gapdh For (AGAAGGTGGTGAAGCAGGCAT) Rev (CGAAGGTGGAAGAGTGGGAGT), Cxcl5 For (GCT GCC CCT TCC TCA GTC AT) Rev (CAC CGT AGG GCA CTG TGG AC). Gene expression profiling of inflammatory cytokines and receptors of normal and KP-OVA tumor bearing lungs was performed by custom RT2 Profiler PCR Array (Qiagen, cat. no. 330221) following manufacturer’s instructions.
Immunohistochemistry
To assess tumor burden, lung tissues were harvested and fixed in formaldehyde 10% and paraffine embedded following standard procedure. Consecutive sections of 8 μm were dewaxed and rehydrated and stained with the H&E using (Bio-Optica, Milano Spa). The area of tumor nodules was quantified manually over consecutive sections and averaged (three sections/sample).
To identify neutrophils or proliferating cells within nodules, sections were treated with antigen-retrieval solution (Vector laboratories) for 20 min at 120°C. Slides were treated for 10 min in H2O2 and after blocking in 10% goat serum in 0.1% Tween 20 for 30 min and incubated overnight at 4°C with specific antibody diluted in PBS 0.1%Tween 20: anti-mouse Ly6G (1A8, BD Pharmingen, cat. no. 551459), or anti-mouse Ki67 (D3B5, Cell Signaling, cat. no. 12202s). Detection was performed using the ImmPRESS polymer detection system (Vector Laboratories), according to manufacturer’s Instructions. Automatic thresholding and measurements were performed using Ilastik or ImageJ software, respectively. Images were acquired by Leica microscope. For tumor burden, neutrophil and proliferating cells measurements slides were scored blindly by two independent operators.
Immunofluorescence staining
To identify SiglecFhigh neu, tumor tissues were intratracheally perfused with 1% paraformaldehyde (PFA), fixed in 4% PFA and embedded in a frozen tissue matrix following standard procedure. Sections of 5 μm were dried 10’ at RT, fixed with 4% PFA for 15’ and permeabilized for 15’ with PBS 0,5% Triton. After blocking in 5% mouse serum in PBS 1%BSA/0,1%NP-40 for 30’, slides were incubated overnight at 4°C with specific antibody diluted in PBS 1%BSA/0,1%NP-40: anti-mouse Ly6G-PE and anti-mouse SiglecF-BB515 (listed in Supplementary Table 1). Nucleus were labeled by Hoechst 15’ at RT. Images were acquired with LSM880 META reverse microscope.
To assess spatial distribution of neutrophils and CD8 T cells, tumor tissues were paraffine embedded. Sections of 8 μm were dewaxed, rehydrated, treated with antigen-retrieval solution, and incubated overnight at 4°C with rat anti-mouse CD8 (4SM15, Invitrogen) and rabbit anti-mouse Ly6G (E6Z1T, Cell Signaling) antibodies diluted in PBS 0.1% Tween 20 followed by α-rat 647 (Invitrogen) and α-rabbit AF488 (Invitrogen) nucleus were labeled by Hoechst.
Images were acquired with C1 Nikon reverse microscope. Automatic thresholding was performed by Ilastik. ImageJ software was used to quantify CD8 T cells and neu and to measure nodule’s area. Nodules having an area <0.09 mm2 were classified as small, the ones having an area ranging from 0.091 to 0.2 mm2 as medium and those with an area >0.2 mm2 as large nodules. Spatial distribution of neu and T cells was performed blindly on code-labeled slides.
In vivo tumor challenge and blocking studies
To establish the adenocarcinoma tumor models C57BL/6 WT female mice at 8–10 weeks of age, were intravenously injected with 7 × 104 KP cells or with 2 × 105 KP OVA variants (WT or KOCXCL5, KOCXCL5 (lenti-CXCL5) or KOCXCL5 (lenti-vec)). In one experiment (supplementary Figure 5) mice were challenged with 5 × 104 cells. Otherwise indicated, mice were sacrificed at initial (9 days) or at established stage of tumorigenesis (18 d). To deplete neutrophils in vivo, KP OVA bearing mice were intraperitoneally treated every 3 d, starting from the 6th to 12th d, with 200 µg of anti-Ly6G antibody (InVivo Plus, clone 1A8, Bio X Cell) or isotype control (Rat IgG2a isotype control, clone 2A3, Bio X Cell) and sacrificed at d 13.
In PD-L1 blockade experiments, mice were challenged with KP OVA WT or KOCXCL5 cells, intraperitoneally treated every 3 d with 200 µg of αPD-L1 (InVivoMab, clone 10 F.9G2, BioXcell) or isotype control (InVivoMab, rat IgG2b isotype control, clone LTF-2, BioXcell). Mice were sacrificed at d 18.
Adoptive transfer of effector OVA-specific CD8
2x106 naïve OTI T cells were intravenously injected into C57BL6/CD45.2 congenic mice followed by inoculation of 4 × 105 KP-OVA KOCXCL5 tumor cells. After 8 d, the mLNs were collected and effector CD44+ CD62l− CD8+ T cells were isolated by cell sorting and intravenously transferred into CD45.1 recipient (0.1 × 105/mouse) mice that had been challenged with WT or KOCXCLKP-OVA cells 10 d before. Lung tissues were harvested 2 d after adoptive transfer and processed for intracellular IFN-ϒ detection as described above.
RNA seq and data analysis
RNA from total lungs was extracted using RNeasy Micro Kit (Qiagen), according to manufacturer’s instruction. Libraries were prepared using TruSeq Stranded mRNA LT Sample Prep Kit and sequenced on an Illumina platfor. Raw sequences files’ quality was checked via FastQC (v 0.11.9, http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and was found to satisfy quality check. We have performed reads alignment to the mouse reference genome using Salmon alignment tool, used for transcript quantification, with the following parameters: read length of 100, Mus musculus GRCm39.103 (GTF file), Mus musculus GRCm39 DNA primary assembly fasta (Sequence file). Data was further elaborated performing a trimming with Salmon.
We have therefore obtained the generated gene counts, which was analyzed using R’s package DESeq2. Using vsd method we generated the normalized expression matrix which was investigated with Principal Component Analysis (PCA) together with a dimensionality reduction algorithm present in the DESeq2 package. Chemokines and chemokine receptors were ranked based on their normalized expression in KP lung and KP cell line and top 10% expressed genes were selected. The same ranking and cut-off were applied to TPM values of naïve lung (nLung), as retrieved from a C57Bl6/J mouse background from Expression Atlas.
Collection and processing of human lung cancer gene expression data
A lung cancer compendium has been created from seven major datasets comprising microarray data of lung cancer samples annotated with clinical outcome. All data were measured on Affymetrix arrays and have been downloaded from NCBI Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) GSE3141, GSE10245, GSE14814, GSE19188, GSE31210, GSE68465, and form the Ladanyi and Gerald Laboratories Lung Adenocarcinoma microarray repository (http://cbio.mskcc.org/public/lung_array_data/). Prior to analysis, we eliminated duplicate samples and renamed all original sets after the medical center where patients were recruited. This re-organization returned 1,136 unique samples from ten independent cohorts comprising 989 adenocarcinomas, 778 of which with complete clinical outcome information (). The type and content of clinical and pathological annotations of the compendium samples have been derived from the original cohorts. Since raw data (.CEL files) were available for all samples, the integration, normalization and summarization of gene expression signals has been obtained applying the procedure described in Rustighi et al.Citation68 Briefly, expression values were generated in R from intensity signals using a custom CDF obtained merging HG-U133A, HG-U133A2 and HG-U133 Plus2 original CDFs and transforming the original CEL files accordingly. Intensity values have been background-adjusted, normalized using quantile normalization, and gene expression levels calculated using median polish summarization (multi-array average procedure, RMA). Gene expression data for a total of 21,995 probe sets have been collapsed to 12,391 unique gene symbols using the hgu133a.db annotation package (version 3.2.3) and the aggregate function of R stats package. To remove possible batch effects due to the different cohorts, we applied the ComBat function of the sva Bioconductor package to the merged matrix. ComBat was used with default parameters and the batch variable imputed as a vector of dataset labels (see ). Clinical information among the various datasets has been standardized as described in Cordenonsi et al.Citation69 Average signature expression has been calculated as the average expression of all signature genes in sample subgroups.
Human neutrophil fraction analysis
Neutrophil cell fractions have been quantified using CIBERSORTCitation57 on the lung adenocarcinoma samples of the LUAD compendium. Briefly, the non-log linear expression matrix with the 989 cases was uploaded to the CIBERSORT R script (version 1.04) as a mixture file and CIBERSORT was run in absolute mode with the LM22 signature gene file, 100 permutations, and quantile normalization. In absolute mode, CIBERSORT scales relative cellular fractions into a score that reflects the absolute proportion of each cell type in a mixture. Although not expressed as a fraction, the absolute score can be directly compared both between- and within-samples.Citation70 Samples have been divided into three groups based on the upper (high neutrophil content) and lower (low neutrophil content) quartiles of the absolute scores for the neutrophil cell type. Samples that fell in between the quartile cut-offs have been termed the medium neutrophil content group.Citation71
Survival analysis
To evaluate the prognostic level of CXCL5 expression, we separated the samples into two groups based on CXCL5 standardized expression.Citation69 Tumor samples have been classified as ‘Low’ if CXCL5 standardized expression was negative and as ‘High’ if the CXCL5 standardized expression was positive. The overall survival probability in the two groups has been estimated using the Kaplan–Meier method and the Kaplan–Meier curves compared using the log-rank (Mantel–Cox) test. P values were calculated according to the standard normal asymptotic distribution. Survival analysis was performed in GraphPad Prism.
Statistic
Primary data were collected in Microsoft Excel, and statistical analysis was performed using GraphPad Prism 8 software. Values reported in figures are expressed as the standard error of the mean, unless otherwise indicated. For comparison between two or more groups with normally distributed datasets two-tailed Student’s T-test, multiple T-test, one-way ANOVA or two-way ANOVA were used as appropriate. For the comparison of matched groups, we used Wilcoxon test. The nonparametric Kruskal–Wallis test with Dunn’s multiple comparison was performed to compare three or more unmatched groups. p Values >0.05 were considered not significant, p values ≤0.05 were considered significant: *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001.
No exclusion criteria or data have been performed.
Contributors
FS and GMP performed experiments. FS and GMP analyzed data, prepared figures and contributed to manuscript writing. AC provided CXCL5 expression construct. SB and RA performed bioinformatic analysis. NC set up the tumor model and generated initial observations. FB ideated and supervised the study and wrote the manuscript.
Ethical approval
The study was approved by International Centre for Genetic Engineering and Biotechnology (ICGEB) board for animal welfare and authorized by the Italian Ministry of Health (approval number 1133/2020-PR, issued on 12/11/2020). Animal care and treatment were conducted with national and international laws and policies (European Economic Community Council Directive 86/609; OJL 358; December 12, 1987). All experiments were performed in accordance with the Federation of European Laboratory Animal Science Association (FELASA) guidelines
Supplemental Material
Download Zip (7.7 MB)Acknowledgments
We thank Luciano Morosi, ICGEB Trieste, for support on statistical analysis. We thank Prof. Annalisa del Prete, University of Brescia, for helpful discussions and advises on the KP mouse model. We thank Prof Mauro Giacca, ICGEB Trieste for providing the CXCL5 coding sequence.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website
Data availability statement
All data relevant to the study are included in the article or uploaded as supplementary information. The datasets used and analyzed during the current study are included in the article or uploaded as Supplementary Information. They are available from the corresponding author on reasonable request.
Additional information
Funding
References
- Ponzetta A, Carriero R, Carnevale S, Barbagallo M, Molgora M, Perucchini C, Magrini E, Gianni F, Kunderfranco P, Polentarutti N, et al. Neutrophils driving unconventional T cells mediate resistance against murine sarcomas and selected human tumors. Cell. 2019;178(2):346–60 e24. doi:10.1016/j.cell.2019.05.047.
- Houghton AM, Rzymkiewicz DM, Ji H, Gregory AD, Egea EE, Metz HE, Stolz DB, Land SR, Marconcini LA, Kliment CR, et al. Neutrophil elastase-mediated degradation of IRS-1 accelerates lung tumor growth. Nat Med. 2010;16(2):219–15. doi:10.1038/nm.2084.
- Butin-Israeli V, Bui TM, Wiesolek HL, Mascarenhas L, Lee JJ, Mehl LC, Knutson KR, Adam SA, Beyder A, Wiesmuller L, et al. Neutrophil-induced genomic instability impedes resolution of inflammation and wound healing. J Clin Invest. 2019;129(2):346–360.e24. doi:10.1172/JCI122085.
- Coffelt SB, Kersten K, Doornebal CW, Weiden J, Vrijland K, Hau CS, Verstegen NJM, Ciampricotti M, Hawinkels LJAC, Jonkers J, et al. IL-17-producing gammadelta T cells and neutrophils conspire to promote breast cancer metastasis. Nature. 2015;522(7556):345–348. doi:10.1038/nature14282.
- Wculek SK, Malanchi I. Neutrophils support lung colonization of metastasis-initiating breast cancer cells. Nature. 2015;528(7582):413–417. doi:10.1038/nature16140.
- Albrengues J, Shields MA, Ng D, Park CG, Ambrico A, Poindexter ME, Upadhyay P, Uyeminami DL, Pommier A, Küttner V, et al. Neutrophil extracellular traps produced during inflammation awaken dormant cancer cells in mice. Science. 2018;361(6409). doi:10.1126/science.aao4227.
- Wellenstein MD, Coffelt SB, Duits DEM, van Miltenburg MH, Slagter M, de Rink I, Henneman L, Kas SM, Prekovic S, Hau C-S, et al. Loss of p53 triggers WNT-dependent systemic inflammation to drive breast cancer metastasis. Nature. 2019;572(7770):538–542. doi:10.1038/s41586-019-1450-6.
- Nozawa H, Chiu C, Hanahan D. Infiltrating neutrophils mediate the initial angiogenic switch in a mouse model of multistage carcinogenesis. Proc Natl Acad Sci U S A. 2006;103(33):12493–12498. doi:10.1073/pnas.0601807103.
- Albini A, Bruno A, Noonan DM, Mortara L. Contribution to tumor angiogenesis from innate immune cells within the tumor microenvironment: implications for immunotherapy. Front Immunol. 2018;9:527. doi:10.3389/fimmu.2018.00527.
- Rodriguez PC, Ernstoff MS, Hernandez C, Atkins M, Zabaleta J, Sierra R, Ochoa AC. Arginase I-producing myeloid-derived suppressor cells in renal cell carcinoma are a subpopulation of activated granulocytes. Cancer Res. 2009;69(4):1553–1560. doi:10.1158/0008-5472.CAN-08-1921.
- Rice CM, Davies LC, Subleski JJ, Maio N, Gonzalez-Cotto M, Andrews C, Patel NL, Palmieri EM, Weiss JM, Lee J-M, et al. Tumour-elicited neutrophils engage mitochondrial metabolism to circumvent nutrient limitations and maintain immune suppression. Nat Commun. 2018;9(1):5099. doi:10.1038/s41467-018-07505-2.
- Veglia F, Tyurin VA, Blasi M, De Leo A, Kossenkov AV, Donthireddy L, To TKJ, Schug Z, Basu S, Wang F, et al. Fatty acid transport protein 2 reprograms neutrophils in cancer. Nature. 2019;569(7754):73–78. doi:10.1038/s41586-019-1118-2.
- Glodde N, Bald T, van den Boorn-konijnenberg D, Nakamura K, O’Donnell JS, Szczepanski S, Brandes M, Eickhoff S, Das I, Shridhar N, et al. Reactive neutrophil responses dependent on the receptor tyrosine kinase c-MET limit cancer immunotherapy. Immunity. 2017;47(4):789–802 e9. doi:10.1016/j.immuni.2017.09.012.
- Veglia F, Hashimoto A, Dweep H, Sanseviero E, De Leo A, Tcyganov E, Kossenkov A, Mulligan C, Nam B, Masters G, et al. Analysis of classical neutrophils and polymorphonuclear myeloid-derived suppressor cells in cancer patients and tumor-bearing mice. J Exp Med. 2021;218(4). doi:10.1084/jem.20201803.
- Templeton AJ, McNamara MG, Seruga B, Vera-Badillo FE, Aneja P, Ocana A, Leibowitz-Amit R, Sonpavde G, Knox JJ, Tran B, et al. Prognostic role of neutrophil-to-lymphocyte ratio in solid tumors: a systematic review and meta-analysis. J Natl Cancer Inst. 2014;106(6):dju124. doi:10.1093/jnci/dju124.
- Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, Nair VS, Xu Y, Khuong A, Hoang CD, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21(8):938–945. doi:10.1038/nm.3909.
- Kargl J, Busch SE, Yang GH, Kim KH, Hanke ML, Metz HE, Hubbard JJ, Lee SM, Madtes DK, McIntosh MW, et al. Neutrophils dominate the immune cell composition in non-small cell lung cancer. Nat Commun. 2017;8(1):14381. doi:10.1038/ncomms14381.
- Mitchell KG, Diao L, Karpinets T, Negrao MV, Tran HT, Parra ER, Corsini EM, Reuben A, Federico L, Bernatchez C, et al. Neutrophil expansion defines an immunoinhibitory peripheral and intratumoral inflammatory milieu in resected non-small cell lung cancer: a descriptive analysis of a prospectively immunoprofiled cohort. J Immunother Cancer. 2020;8(1):e000405. doi:10.1136/jitc-2019-000405.
- Shaul ME, Eyal O, Guglietta S, Aloni P, Zlotnik A, Forkosh E, Levy L, Weber LM, Levin Y, Pomerantz A, et al. Circulating neutrophil subsets in advanced lung cancer patients exhibit unique immune signature and relate to prognosis. FASEB J. 2020;34(3):4204–4218. doi:10.1096/fj.201902467R.
- Pore N, Wu S, Standifer N, Jure-Kunkel M, de Los Reyes M, Shrestha Y, Halpin R, Rothstein R, Mulgrew K, Blackmore S, et al. Resistance to durvalumab and durvalumab plus tremelimumab is associated with functional STK11 mutations in patients with non–small cell lung cancer and is reversed by STAT3 knockdown. Cancer Discov. 2021;11(11):2828–2845. doi:10.1158/2159-8290.CD-20-1543.
- Rapoport BL, Theron AJ, Vorobiof DA, Langenhoven L, Hall JM, Van Eeden RI, Smit T, Chan S-W, Botha MC, Raats JI, et al. Prognostic significance of the neutrophil/lymphocyte ratio in patients undergoing treatment with nivolumab for recurrent non-small-cell lung cancer. Lung Cancer Manag. 2020;9(3):LMT37. doi:10.2217/lmt-2020-0014.
- Russo A, Russano M, Franchina T, Migliorino MR, Aprile G, Mansueto G, Berruti A, Falcone A, Aieta M, Gelibter A, et al. Neutrophil-to-Lymphocyte Ratio (NLR), Platelet-to-Lymphocyte Ratio (PLR), and outcomes with nivolumab in pretreated Non-Small Cell Lung Cancer (NSCLC): a large retrospective multicenter study. Adv Ther. 2020;37(3):1145–1155. doi:10.1007/s12325-020-01229-w.
- Jiang T, Bai Y, Zhou F, Li W, Gao G, Su C, Ren S, Chen X, Zhou C. Clinical value of neutrophil-to-lymphocyte ratio in patients with non-small-cell lung cancer treated with PD-1/PD-L1 inhibitors. Lung Cancer. 2019;130:76–83. doi:10.1016/j.lungcan.2019.02.009.
- Cheng Y, Mo F, Li Q, Han X, Shi H, Chen S, Wei Y, Wei X. Targeting CXCR2 inhibits the progression of lung cancer and promotes therapeutic effect of cisplatin. Mol Cancer. 2021;20(1):62. doi:10.1186/s12943-021-01355-1.
- Germann M, Zangger N, Sauvain MO, Sempoux C, Bowler AD, Wirapati P, Kandalaft LE, Delorenzi M, Tejpar S, Coukos G, et al. Neutrophils suppress tumor-infiltrating T cells in colon cancer via matrix metalloproteinase-mediated activation of TGF β. EMBO Mol Med. 2020;12(1):e10681. doi:10.15252/emmm.201910681.
- Engblom C, Pfirschke C, Zilionis R, Da Silva Martins J, Bos SA, Courties G, Rickelt S, Severe N, Baryawno N, Faget J, et al. Osteoblasts remotely supply lung tumors with cancer-promoting SiglecF high neutrophils. Science. 2017;358(6367). doi:10.1126/science.aal5081.
- Zilionis R, Engblom C, Pfirschke C, Savova V, Zemmour D, Saatcioglu HD, Krishnan I, Maroni G, Meyerovitz CV, Kerwin CM, et al. Single-cell transcriptomics of human and mouse lung cancers reveals conserved myeloid populations across individuals and species. Immunity. 2019;50(5):1317–34 e10. doi:10.1016/j.immuni.2019.03.009.
- Schenkel JM, Herbst RH, Canner D, Li A, Hillman M, Shanahan SL, Gibbons G, Smith OC, Kim JY, Westcott P, et al. Conventional type I dendritic cells maintain a reservoir of proliferative tumor-antigen specific TCF-1(+) CD8(+) T cells in tumor-draining lymph nodes. Immunity. 2021;54(10):2338–53 e6. doi:10.1016/j.immuni.2021.08.026.
- Horton BL, Morgan DM, Momin N, Zagorulya M, Torres-Mejia E, Bhandarkar V, Wittrup KD, Love JC, Spranger S. Lack of CD8 + T cell effector differentiation during priming mediates checkpoint blockade resistance in non–small cell lung cancer. Sci Immunol. 2021;6(64):eabi8800. doi:10.1126/sciimmunol.abi8800.
- Li BH, Garstka MA, Li ZF. Chemokines and their receptors promoting the recruitment of myeloid-derived suppressor cells into the tumor. Mol Immunol. 2020;117:219–223. doi:10.1016/j.molimm.2019.11.014.
- Zhou SL, Dai Z, Zhou ZJ, Chen Q, Wang Z, Xiao YS, Hu Z-Q, Huang X-Y, Yang G-H, Shi Y-H, et al. CXCL5 contributes to tumor metastasis and recurrence of intrahepatic cholangiocarcinoma by recruiting infiltrative intratumoral neutrophils. Carcinogenesis. 2014;35(3):597–605. doi:10.1093/carcin/bgt397.
- Zhou SL, Dai Z, Zhou ZJ, Wang XY, Yang GH, Wang Z, Huang X-W, Fan J, Zhou J. Overexpression of CXCL5 mediates neutrophil infiltration and indicates poor prognosis for hepatocellular carcinoma. Hepatology. 2012;56(6):2242–2254. doi:10.1002/hep.25907.
- Soler-Cardona A, Forsthuber A, Lipp K, Ebersberger S, Heinz M, Schossleitner K, Buchberger E, Gröger M, Petzelbauer P, Hoeller C, et al. CXCL5 facilitates melanoma cell-neutrophil interaction and lymph node metastasis. J Invest Dermatol. 2018;138(7):1627–1635. doi:10.1016/j.jid.2018.01.035.
- Forsthuber A, Lipp K, Andersen L, Ebersberger S, Graña C, Ellmeier W, Petzelbauer P, Lichtenberger BM, Loewe R. CXCL5 as regulator of neutrophil function in cutaneous melanoma. J Invest Dermatol. 2019;139(1):186–194. doi:10.1016/j.jid.2018.07.006.
- Gao Y, Guan Z, Chen J, Xie H, Yang Z, Fan J, Wang X, Li L. CXCL5/CXCR2 axis promotes bladder cancer cell migration and invasion by activating PI3K/AKT-induced upregulation of MMP2/MMP9. Int J Oncol. 2015;47(2):690–700. doi:10.3892/ijo.2015.3041.
- Yang L, Huang J, Ren X, Gorska AE, Chytil A, Aakre M, Carbone DP, Matrisian L, Richmond A, Lin PC, et al. Abrogation of TGFβ signaling in mammary carcinomas recruits Gr-1+CD11b+ myeloid cells that promote metastasis. Cancer Cell. 2008;13(1):23–35. doi:10.1016/j.ccr.2007.12.004.
- Romero-Moreno R, Curtis KJ, Coughlin TR, Miranda-Vergara MC, Dutta S, Natarajan A, Facchine BA, Jackson KM, Nystrom L, Li J, et al. The CXCL5/CXCR2 axis is sufficient to promote breast cancer colonization during bone metastasis. Nat Commun. 2019;10(1):4404. doi:10.1038/s41467-019-12108-6.
- Walz A, Burgener R, Car B, Baggiolini M, Kunkel SL, Strieter RM. Structure and neutrophil-activating properties of a novel inflammatory peptide (ENA-78) with homology to interleukin 8. J Exp Med. 1991;174(6):1355–1362. doi:10.1084/jem.174.6.1355.
- Nouailles G, Dorhoi A, Koch M, Zerrahn J, Weiner J 3rd, Faé KC, Arrey F, Kuhlmann S, Bandermann S, Loewe D, et al. CXCL5-secreting pulmonary epithelial cells drive destructive neutrophilic inflammation in tuberculosis. J Clin Invest. 2014;124(3):186–194. doi:10.1172/JCI72030.
- Mei J, Liu Y, Dai N, Favara M, Greene T, Jeyaseelan S, Poncz M, Lee JS, Worthen GS. CXCL5 regulates chemokine scavenging and pulmonary host defense to bacterial infection. Immunity. 2010;33(1):106–117. doi:10.1016/j.immuni.2010.07.009.
- Saintigny P, Massarelli E, Lin S, Ahn Y-H, Chen Y, Goswami S, Erez B, O’Reilly MS, Liu D, Lee JJ, et al. CXCR2 expression in tumor cells is a poor prognostic factor and promotes invasion and metastasis in lung adenocarcinoma. Cancer Res. 2013;73(2):571–582. doi:10.1158/0008-5472.CAN-12-0263.
- Han N, Yuan X, Wu H, Xu H, Chu Q, Guo M, Yu S, Chen Y, Wu K. DACH1 inhibits lung adenocarcinoma invasion and tumor growth by repressing CXCL5 signaling. Oncotarget. 2015;6(8):5877–5888. doi:10.18632/oncotarget.3463.
- Wang J, Hu T, Wang Q, Chen R, Xie Y, Chang H, Cheng J. Repression of the AURKA-CXCL5 axis induces autophagic cell death and promotes radiosensitivity in non-small-cell lung cancer. Cancer Lett. 2021;509:89–104. doi:10.1016/j.canlet.2021.03.028.
- Zhou Y, Shurin GV, Zhong H, Bunimovich YL, Han B, Shurin MR. Schwann cells augment cell spreading and metastasis of lung cancer. Cancer Res. 2018;78(20):5927–5939. doi:10.1158/0008-5472.CAN-18-1702.
- Hu B, Fan H, Lv X, Chen S, Shao Z. Prognostic significance of CXCL5 expression in cancer patients: a meta-analysis. Cancer Cell Int. 2018;18(1):68. doi:10.1186/s12935-018-0562-7.
- Wu K, Yu S, Liu Q, Bai X, Zheng X, Wu K. The clinical significance of CXCL5 in non-small cell lung cancer. Onco Targets Ther. 2017;10:5561–5573. doi:10.2147/OTT.S148772.
- Mollaoglu G, Jones A, Wait SJ, Mukhopadhyay A, Jeong S, Arya R, Camolotto SA, Mosbruger TL, Stubben CJ, Conley CJ, et al. The lineage-defining transcription factors SOX2 and NKX2-1 determine lung cancer cell fate and shape the tumor immune microenvironment. Immunity. 2018;49(4):764–79.e9. doi:10.1016/j.immuni.2018.09.020.
- Koyama S, Akbay EA, Li YY, Aref AR, Skoulidis F, Herter-Sprie GS, Buczkowski KA, Liu Y, Awad MM, Denning WL, et al. STK11/LKB1 deficiency promotes neutrophil recruitment and proinflammatory cytokine production to suppress T-cell activity in the lung tumor microenvironment. Cancer Res. 2016;76(5):999–1008. doi:10.1158/0008-5472.CAN-15-1439.
- Maier B, Leader AM, Chen ST, Tung N, Chang C, LeBerichel J, Chudnovskiy A, Maskey S, Walker L, Finnigan JP, et al. A conserved dendritic-cell regulatory program limits antitumour immunity. Nature. 2020;580(7802):257–262. doi:10.1038/s41586-020-2134-y.
- Pfirschke C, Engblom C, Gungabeesoon J, Lin Y, Rickelt S, Zilionis R, Messemaker M, Siwicki M, Gerhard GM, Kohl A, et al. Tumor-promoting Ly-6G(+) SiglecF(high) cells are mature and long-lived Neutrophils. Cell Rep. 2020;32(12):108164. doi:10.1016/j.celrep.2020.108164.
- Ancey PB, Contat C, Boivin G, Sabatino S, Pascual J, Zangger N, Perentes JY, Peters S, Abel ED, Kirsch DG, et al. GLUT1 expression in tumor-associated neutrophils promotes lung cancer growth and resistance to radiotherapy. Cancer Res. 2021;81(9):2345–2357. doi:10.1158/0008-5472.CAN-20-2870.
- DuPage M, Cheung AF, Mazumdar C, Winslow MM, Bronson R, Schmidt LM, Crowley D, Chen J, Jacks T. Endogenous T cell responses to antigens expressed in lung adenocarcinomas delay malignant tumor progression. Cancer Cell. 2011;19(1):72–85. doi:10.1016/j.ccr.2010.11.011.
- Caronni N, Piperno GM, Simoncello F, Romano O, Vodret S, Yanagihashi Y, Dress R, Dutertre C-A, Bugatti M, Bourdeley P, et al. TIM4 expression by dendritic cells mediates uptake of tumor-associated antigens and anti-tumor responses. Nat Commun. 2021;12(1):2237. doi:10.1038/s41467-021-22535-z.
- Veglia F, Sanseviero E, Gabrilovich DI. Myeloid-derived suppressor cells in the era of increasing myeloid cell diversity. Nat Rev Immunol. 2021;21(8):485–498. doi:10.1038/s41577-020-00490-y.
- Zhu J, Powis de Tenbossche CG, Cane S, Colau D, van Baren N, Lurquin C, Schmitt-Verhulst A-M, Liljeström P, Uyttenhove C, Van den Eynde BJ, et al. Resistance to cancer immunotherapy mediated by apoptosis of tumor-infiltrating lymphocytes. Nat Commun. 2017;8(1):1404. doi:10.1038/s41467-017-00784-1.
- Boivin G, Faget J, Ancey PB, Gkasti A, Mussard J, Engblom C, Pfirschke C, Contat C, Pascual J, Vazquez J, et al. Durable and controlled depletion of neutrophils in mice. Nat Commun. 2020;11(1):2762. doi:10.1038/s41467-020-16596-9.
- Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi:10.1038/nmeth.3337.
- Deng J, Ma X, Ni Y, Li X, Xi W, Tian M, Zhang X, Xiang M, Deng W, Song C, et al. Identification of CXCL5 expression as a predictive biomarker associated with response and prognosis of immunotherapy in patients with non-small cell lung cancer. Cancer Med. 2022. doi:10.1002/cam4.4567.
- Kaltenmeier C, Yazdani HO, Morder K, Geller DA, Simmons RL, Tohme S. Neutrophil extracellular traps promote T cell exhaustion in the tumor microenvironment. Front Immunol. 2021;12:785222. doi:10.3389/fimmu.2021.785222.
- Maier BB, Hladik A, Lakovits K, Korosec A, Martins R, Kral JB, Mesteri I, Strobl B, Müller M, Kalinke U, et al. Type I interferon promotes alveolar epithelial type II cell survival during pulmonary Streptococcus pneumoniae infection and sterile lung injury in mice. Eur J Immunol. 2016;46(9):2175–2186. doi:10.1002/eji.201546201.
- Akbay EA, Koyama S, Liu Y, Dries R, Bufe LE, Silkes M, Alam MM, Magee DM, Jones R, Jinushi M, et al. Interleukin-17A promotes lung tumor progression through Neutrophil attraction to tumor sites and mediating resistance to PD-1 blockade. J Thorac Oncol. 2017;12(8):1268–1279. doi:10.1016/j.jtho.2017.04.017.
- Kowalczuk O, Burzykowski T, Niklinska WE, Kozlowski M, Chyczewski L, Niklinski J. CXCL5 as a potential novel prognostic factor in early stage non-small cell lung cancer: results of a study of expression levels of 23 genes. Tumour Biol. 2014;35(5):4619–4628. doi:10.1007/s13277-014-1605-x.
- Philip M, Schietinger A. CD8(+) T cell differentiation and dysfunction in cancer. Nat Rev Immunol. 2021. doi:10.1038/s41577-021-00574-3.
- Ren F, Zhao T, Liu B, Pan L. Neutrophil-lymphocyte ratio (NLR) predicted prognosis for advanced non-small-cell lung cancer (NSCLC) patients who received immune checkpoint blockade (ICB). Onco Targets Ther. 2019;12:4235–4244. doi:10.2147/OTT.S199176.
- Condamine T, Dominguez GA, Youn JI, Kossenkov AV, Mony S, Alicea-Torres K, Tcyganov E, Hashimoto A, Nefedova Y, Lin C, et al. Lectin-type oxidized LDL receptor-1 distinguishes population of human polymorphonuclear myeloid-derived suppressor cells in cancer patients. Sci Immunol. 2016;1(2). doi:10.1126/sciimmunol.aaf8943.
- Cheng Y, Li H, Deng Y, Tai Y, Zeng K, Zhang Y, Liu W, Zhang Q, Yang Y. Cancer-associated fibroblasts induce PDL1+ neutrophils through the IL6-STAT3 pathway that foster immune suppression in hepatocellular carcinoma. Cell Death Dis. 2018;9(4):422. doi:10.1038/s41419-018-0458-4.
- Dimitrova N, Gocheva V, Bhutkar A, Resnick R, Jong RM, Miller KM, Bendor J, Jacks T. Stromal expression of miR-143/145 promotes neoangiogenesis in lung cancer development. Cancer Discov. 2016;6(2):188–201. doi:10.1158/2159-8290.CD-15-0854.
- Rustighi A, Zannini A, Tiberi L, Sommaggio R, Piazza S, Sorrentino G, Nuzzo S, Tuscano A, Eterno V, Benvenuti F, et al. Prolyl-isomerase Pin1 controls normal and cancer stem cells of the breast. EMBO Mol Med. 2014;6(1):99–119. doi:10.1002/emmm.201302909.
- Cordenonsi M, Zanconato F, Azzolin L, Forcato M, Rosato A, Frasson C, Inui M, Montagner M, Parenti A, Poletti A, et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 2011;147(4):759–772. doi:10.1016/j.cell.2011.09.048.
- Sturm G, Finotello F, Petitprez F, Zhang JD, Baumbach J, Fridman WH, List M, Aneichyk T. Comprehensive evaluation of transcriptome-based cell-type quantification methods for immuno-oncology. Bioinformatics. 2019;35(14):i436–i45. doi:10.1093/bioinformatics/btz363.
- Craven KE, Gokmen-Polar Y, Badve SS. CIBERSORT analysis of TCGA and METABRIC identifies subgroups with better outcomes in triple negative breast cancer. Sci Rep. 2021;11(1):4691. doi:10.1038/s41598-021-83913-7.
