ABSTRACT
The human Epstein–Barr virus is associated with several human solid and hematopoietic malignancies. However, the underlying molecular mechanisms including virus-encoded microRNAs (miRs), which lead to the malignant transformation of infected cells and immune evasion of EBV-associated tumors, have not yet been characterized. The expression levels of numerous known EBV-specific miRs and their suitability as diagnostic and/or prognostic markers were determined in different human EBV-positive tissues followed by in silico analyses to identify putative EBV-miR-regulated target genes, thereby offering a suitable screening strategy to overcome the limited available data sets of EBV-miRs and their targeted gene networks. Analysis of microarray data sets from healthy human B cells and malignant-transformed EBV-positive B cells of patients with Burkitt’s lymphoma revealed statistically significant (p < 0.05) deregulated genes with known functions in oncogenic properties, immune escape and anti-tumoral immune responses. Alignments of in vivo and in silico data resulted in the prediction of putative candidate EBV-miRs and their target genes. Thus, a combinatorial approach of bioinformatics, transcriptomics and in situ expression analyses is a promising tool for the identification of EBV-miRs and their potential targets as well as their eligibility as markers for EBV detection in different EBV-associated human tissue.
Introduction
The human Epstein–Barr virus (EBV) is a double-stranded DNA virus belonging to the γ-subfamily of the human herpes viruses (HHV), which is known to exert a certain oncogenic potential.Citation1 Indeed, EBV infection is associated with several human malignancies including B cell Non-Hodgkin lymphoma (NHL), like Burkitt’s lymphoma (BL)Citation2 or NK/T-cell lymphoma,Citation3 classical Hodgkin’s lymphoma (cHL),Citation4 gastric carcinomaCitation5 and nasopharynx carcinoma (NPC).Citation6
The viral gene expression pattern differs between latent and lytic phase of the EBV infection, which could be further subgrouped based on their gene expression profile.Citation7 In detail, in the latency 0 phase present in infected and circulating memory B cells in vivo, only non-protein coding viral transcripts are produced. Latency 1 phase typically observed in BL exerts EBNA1 expression along with the non-coding viral transcripts, while the latency 2 phase characteristic for cHL and NPC exhibits an additional expression of LMP1 and LMP2. In contrast, in the latency 3 phase of B cell transformation, EBNA2, EBNA-LP, BHRF1, the three latent membrane proteins LMP1, 2A and 2B in addition to the non-coding transcripts can be detected.Citation7
The viral genome encodes for >80 genes and to date for 48 viral microRNAs (miRs), which are classified according to their genomic location (BamHI rightward reading frame 1 (BHRF1) and BamHI-A rightward transcript (BART)) into ebv-miR-BHRFs and ebv-miR-BARTs.Citation8–10 During the distinct EBV infection phases, a differential expression pattern of EBV-miRs was described.Citation7,Citation11 MiRs are small RNAs with approximately 22 nucleotides (nts) in length and regulate sequence-specific the post-transcriptional gene expression by binding preferentially, but not exclusively to the 3′ untranslated region (UTR) of mRNAs leading to translational inhibition and/or degradation of the targeted mRNA.Citation12,Citation13 Concerning canonical miR target/mRNA interactions featuring complementarity to the seed sequence, the second to eighth nt of the miR is critical for miR targeting, but recent studies also reported miR-target mRNA interactions without perfect seed pairing thereby possibly limiting the reliability of the in silico prediction tools.Citation14 Most of the EBV-miRs are involved in the development of EBV-driven malignancies and exert a variety of important regulatory functions in EBV-mediated tumorigenesis and cancer progression.Citation8,Citation15 Screening for EBV-miR targets is even more complex, since the miR target prediction tools available do not include the sequences of the EBV-miRs.
Due to the regulatory potential of tumor-relevant target genes, the expression of certain EBV-miRs has been shown to be of prognostic relevance in EBV-associated malignancies. For example, in gastric cancers, the ebv-miR-BART20-5p is linked to a poor patients’ survival,Citation16 while the ebv-miR-BART7 has been described as potential plasma marker in NPC patients.Citation17
The implementation of miRs as additional prognostic markers in formalin-fixed and paraffin-embedded (FFPE) tumor tissues of EBV-associated neoplasia is of growing importance due to their increased stability when compared to mRNAs or transcripts >80 nucleotides (nts) in length, which rapidly degrade in FFPE tissue specimens. Indeed, the quality and stability of miRs extracted from FFPE specimen was comparable to that of miRs derived from fresh tissues and was not affected by different fixation times.Citation18–20
Depending on the functions of the targeted mRNAs, miRs can exert tumor suppressive or even oncogenic functions.Citation21 Furthermore, the miR-dependent regulation of several (tumor) immunologic relevant target genes has even led to the definition of immune modulatory miRs involved in immune recognition and/or immune escape of tumors.Citation22 Furthermore, EBV-encoded molecules, like miRs (in particular miR-BART7-3p) and proteins (LMP1), are directly involved in the epithelial to mesenchymal transition (EMT), which in solid tumors might allow virus-infected cells to migrate into other tissues and locations in combination with a deregulated immune surveillance.Citation23–26
Recently, virus-encoded miRs have been shown to be involved in the various interactions between the EBV and the human host cells, thereby affecting different cell biological processes including immune surveillance, apoptosis induction, cell proliferation, tumor growth/suppression and cell migration.Citation27–32
The exact identification of the molecular targets and signaling pathways accompanying these processes is of general importance for the understanding of the EBV-driven malignant transformation and immune escape. Therefore, this study aims to identify deregulated relevant target genes with tumor biological importance, which reflect direct or indirect targets of EBV-encoded miRs thereby highlighting the combinatorial approach of in silico analyses with transcriptome analyses and in situ expression data.
Materials and methods
Cell culture, RNA extraction, cDNA synthesis and qPCR
The cell lines were obtained from the ATCC (www.atcc.org) and cultured in RPMI1640 medium (Gibco™, Fisher Scientific, Waltham, MA, USA) supplemented with 10% (V/V) fetal bovine serum (Anprotec, Germany), 1 x penicillin, streptomycin (Sigma-Aldrich, Taufkirchen, Germany), 1 x sodium pyruvate (Gibco™, Fisher Scientific, Germany), 1 x L-glutamine (Lonza, Basel, Switzerland) and 25 mM (final concentration) Hepes buffer (c-c-pro, Oberdorla, Germany) at 37°C and 5% CO2. RNAs of the EBV-positive cell lines were kindly provided by the Prof. Ofer Mandelboim laboratory.
Total RNA was extracted with TRIzol™ (Invitrogen™, Fisher Scientific, Waltham, MA, USA) and reverse transcribed by RevertAid First Strand cDNA Synthesis Kit (Fisher Scientific) according to Chen and co-authorsCitation33 and Jasinski-Bergner and co-authorsCitation12 followed by qPCR analyses as already described in detail.Citation12 For the determination of the relative miR expression values, the human endogenous hsa-miR-3960 was applied as housekeeping gene. All primers used and their conditions are listed in .
Table 1. List of the oligonucleotides used for the identification of EBV-miRs
Patient samples, EBER-CISH staining, immunohistochemistry and miR extraction from FFPE specimen
FFPE specimen from EBV-positive B cell lymphoma (n = 7), T cell lymphoma (n = 2), cHL (n = 2), NPCs (n = 12) and tonsils of patients with infectious mononucleosis (n = 13) as well as the respective EBV-negative controls (EBV-negative lymph nodes, n = 9; EBV-negative NPC adjacent mucosa tissues, n = 4; EBV-negative tonsils, n = 7) were provided as anonymised slides from the archive of the Institute of Pathology, Martin-Luther-University Halle-Wittenberg, Halle, Germany and conducted according to the principles expressed in the Declaration of Helsinki.
Scientific research on FFPE tissue samples from patients was approved by the Ethical Committee of the Medical Faculty of the Martin Luther University Halle-Wittenberg, Halle, Germany (2017–81).
The Epstein–Barr virus-encoded RNA chromogenic in situ hybridization (EBER-CISH) was performed on 4 µm thick FFPE tissue slides using the ZytoFast PLUS CISH Implementation Kit AP-Permanent Red (ZytoVision, Bremerhaven, Germany) according to the manufacturer’s instructions. Immunohistochemistry (IHC) was performed on a Bond III automated immunostainer (Leica Biosystems Nussloch GmbH, Wetzlar, Germany) using the Bond Polymer Refine Detection Kit (DS9800-CN). The primary antibody directed against LMP1 (clone CS.1–4, Agilent, Santa Clara, CA, USA) was applied as recommended by the manufacturer.
From each FFPE tissue block, three to four consecutive tissue slides (each 4 μm thick) were pooled for further miR extraction. Therefore, the MasterPure™ Complete DNA and RNA Purification Kit (Lucigen, Middleton, USA) was utilized according to the manufacturer’s protocol. The cDNA synthesis and qPCR analyses were performed as mentioned above.
In silico analyses of microarray data
The free online R2: Genomics Analysis and Visualization Platform (http://r2.amc.nl) was employed to determine the gene expression of various marker genes. Two data sets, both based on the same microarray chips (u133p2, Affymetrix Genechips, Santa Clara, CA, USA), were used: (i) healthy human B cells sorted for CD19Citation34 and (ii) the EBV-positive B cell line P493-6 derived from a human BL patientCitation35 cultured under hypoxic conditions, which contains a tetracycline-inducible myc transgene.Citation36
After selection of the data setCitation34,Citation35 at the R2 database, the option “view a gene” was selected. An Analysis was performed by submitting the respective gene symbols to obtain the exact expression levels in every single study sample.
Statistical evaluation and software
All statistically significant (two-tailed Mann-Whitney-U test) values were marked with * (p < 0.05), with ** (p < 0.005) and with *** (p < 0.0005). For statistical analyses and visualization Microsoft Excel 2016 (Microsoft Corporation, Redmond, WA, USA) and Prism GraphPad9 (GraphPad Software, San Diego, CA, USA) were applied.
Results
Identification of EBV-miR expression patterns in human EBV-positive tissue specimen of distinct origins and their role as diagnostic biomarkers
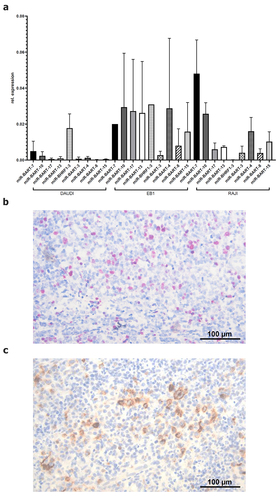
To identify host cell genes, which are targeted by EBV-encoded miRs expressed during latency, a selection of nine different EBV-miRs known to be expressed during latency was analyzed in three EBV-positive B cell lines (DAUDI, EB1, RAJI) in comparison to three EBV-negative B cell lines (BV173, CA46, DG75) by qPCR. The relative copy numbers were determined using the very abundantly expressed endogenous hsa-miR-3960 as housekeeping gene. While the EBV-encoded miRs were not detectable in the EBV-negative B cell lines (data not shown), all EBV-miRs were expressed in the EBV-positive cell lines. Some of the EBV-miRs exerted a heterogeneous expression within the three EBV-positive cell lines, while others were identified in all three EBV-positive cell lines with the highest expression of ebv-miR-BART7-3p and ebv-miR-BART10-3p (). To validate the postulated suitability of EBV-miRs as putative diagnostic markers for EBV detection in FFPE specimen as alternative and/or in addition to the EBER-CISH staining, the expression of the selected nine EBV-miRs was quantified by qPCR in a panel of EBV-positive human FFPE specimen. The EBV status of these FFPE specimen was proven by EBER-CISH staining and LMP1 expression determined by IHC as exemplarily shown in , respectively, and summarized in .
Table 2. Comparison of different EBV detection methods using EBV-positive malignant tissues
Figure 1. Expression and quantification of selected EBV-encoded factors in vitro and in vivo. (A) qPCR-based quantification of selected EBV-miRs in three EBV-positive cell lines. The relative copy numbers of the selected EBV-miRs were determined as described in Materials and Methods. The human endogenous hsa-miR-3960 served as housekeeping gene for calculation of the relative expression values, which are expressed as mean of three biological replicates. (B) representative EBER-CISH staining of an EBV-positive tonsil derived from a patient with infectious mononucleosis (sample ID: IM3). EBER-CISH staining was performed according to Materials and Methods. (C) representative anti-LMP1 immunohistochemistry staining of an EBV-positive tonsil derived from a patient with infectious mononucleosis (sample ID: IM3). LMP1 expression was determined by IHC using an anti-LMP1 antibody as described in Material and Methods.
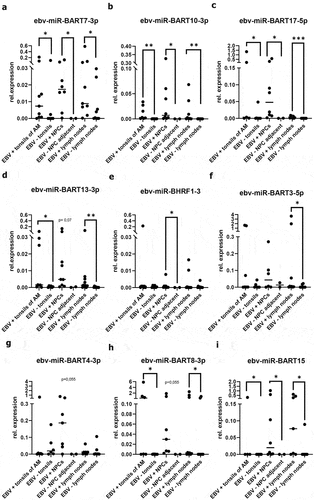
In addition, EBV-miRs were analyzed by qPCR in the same specimen from EBV-positive B cell lymphoma, T cell lymphoma, cHL, NPC and tonsils of patients with infectious mononucleosis as well as respective EBV-negative controls (for further information see Materials and Methods section). As shown in , EBV-miRs were detected in all EBV-positive samples.
In detail, the expression of ebv-miR-BART7-3p, ebv-miR-BART10-3p, ebv-miR-BART17-5p and ebv-miR-BART15 was statistically significant higher when compared to the respective EBV-negative controls (). With the exception of ebv-miR-BHRF 1–3, the other investigated EBV-miRs showed increased expression levels in at least one of the analyzed tissues. However, the expression levels of the different EBV-miRs varied within the analyzed EBV-positive specimen of distinct origin suggesting a tissue-specific expression. Interestingly, a statistically significant difference was found for ebv-miR-BART4-3p (p = 0.0344) and ebv-miR-BART17-5p (p = 0.0026) between EBV-positive NPC and EBV-positive lymphoma samples. Despite this heterogeneity, these results support the hypothesis of the eligibility of certain EBV-miRs as markers for EBV detection in addition to EBER-CISH.
Figure 2. Quantification of selected EBV-miRs in vivo in different EBV-positive and EBV-negative tissue samples. (A-I) qPCR-based quantification of selected EBV-miRs in EBV-positive tonsils of patients with IM, EBV-positive NPC tumors and EBV-positive lymph nodes of B cell lymphoma patients as well as respective EBV-negative controls was performed by determination of the relative copy numbers using the human endogenous hsa-miR-3960 as housekeeping gene as described in Materials and Methods. For calculation of statistically significance two-tailed Mann-Whitney-U test was performed.
Development of a strategy for identification of EBV-mediated deregulated, tumor-relevant genes
To investigate pathways, which might be altered during the process of EBV-induced malignant transformation, in silico analyses were performed. Since the standard in silico miR target prediction databases do not comprise EBV-miRs, including the candidate EBV-miRs, an alternative in silico screening strategy was established to identify potential human tumor relevant target genes, which are altered by EBV-encoded miRs expressed during latency.
Two microarray data sets available at the R2 microarray database (http://r2.amc.nl) were selected for comparison. The data set of Jima et al.Citation34 utilized HG-U133A gene chips (Affymetrix) for the analysis of flow cytometry sorted CD19-positive B cells of healthy human donors, while the study of Kim and co-authorsCitation35 applied the same gene chips and investigated EBV-positive BL.Citation37,Citation38
Implementing in silico analysis of these model systems, the molecular impact of EBV-driven malignant transformation affecting different tumor-relevant pathways was determined to get insights into the regulation of malignancy-related genes in the generation of EBV-associated B cell neoplasia, but perhaps also in general for EBV-linked diseases including NPCs.
Analysis of components of the innate immune system and immune surveillance
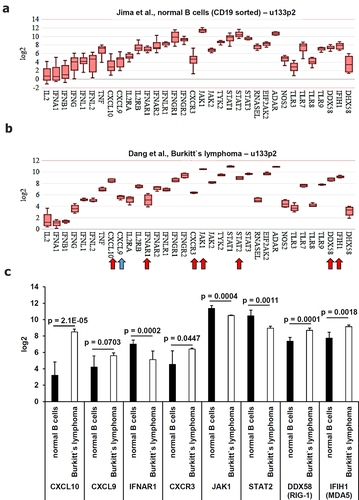
First, the expression of genes relevant for the functionality of the innate immune system known to exert anti-viral activities were compared. These include involving anti-viral cytokines/chemokines, their receptors, downstream signaling molecules and pattern recognition receptors with anti-viral functions like the toll-like receptors (TLR)3, TLR7, TLR8, and TLR9 and the intracellular pattern recognition factors RIG-1 (DDX58), MDA5 (IFIH1) and LGP2 (DHX58)Citation39–42 (). A statistically significant enhanced expression was detected for the anti-viral chemokine CXCL10 (p = 2.1E-05), an almost statistically significant gene expression was detected for CXCL9 (p = 0.0703) and for the receptor CXCR3 (p = 0.0447) in EBV-positive BL cells (). In contrast, a statistically significant downregulation in BL cells was found for some components of the type I interferon (IFN) signaling, such as IFNAR1 (p = 0.0002), JAK1 (p = 0.0004) and STAT2 (p = 0.0011), while RIG-1 (DDX58; p = 0.0001) and MDA5 (IFIH1; p = 0.0018) were downregulated in BL cells ().
Figure 3. Comparative analysis of the gene expression of anti-viral factors of the innate immune system derived from microarray data. Microarray data were analyzed as described in Material and Methods. The mean expression values were calculated (A) in healthy B cells and (B) in EBV-positive BL cells. Statistically significant deregulated genes were marked by a red arrow and almost statistically significant deregulated genes were marked by a blue arrow. These results were summarized in (C) including the calculated p values.
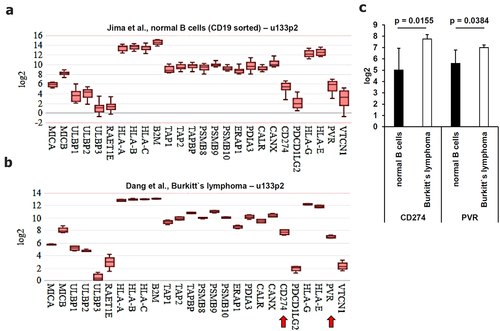
In addition, the expression of genes involved in the immune surveillance as well as immune evasion of tumors was analyzed (). These genes included the components of the antigen processing and presentation machinery (APM) and the NKG2D ligands MIC-A/B and UL16-binding proteins (ULBPs), which are frequently downregulated in tumors. It is noteworthy that the applied gene chips lack probes for ULBP5 and ULBP6. Furthermore, tumor-induced immune response-relevant molecules were determined. Interestingly, only two known immune modulatory genes, namely PD-L1 (CD274) and PVR (CD155 or NECL5), were statistically significantly upregulated (p = 0.0155 and p = 0.0384) in BL when compared to healthy B cells ().
Figure 4. Comparative analysis of the gene expression patterns of molecules involved in immune surveillance and immune modulation derived from microarray data. Microarray data were analyzed as described in Material and Methods. The mean expression values were calculated for (A-C) genes involved in immune surveillance and in immune modulation. Statistically significant deregulated genes were marked by a red arrow. These results were summarized in (C) including the calculated p values.
Analysis of marker genes with prognostic relevance
EBV-positive BL cells exerted a statistically significant induced expression of some proliferation marker genes, like MKI67, E2F1 and BUB1 with the strongest effect for the cell cycle regulators MCM2 (p = 0.0091), MCM3 (p = 0.0033) and MCM4 (p = 0.0052) ().
Figure 5. Comparative analysis of the gene expression patterns of proliferation markers of known oncogenes derived from microarray data. Microarray data were analyzed as described in Material and Methods. The mean expression values were calculated for (A-C) proliferation markers and for (D-F) oncogenes. Statistically significant deregulated genes were marked by a red arrow and almost statistically significant dysregulated genes were marked by a blue arrow. These results were summarized in (C) including the calculated p values.
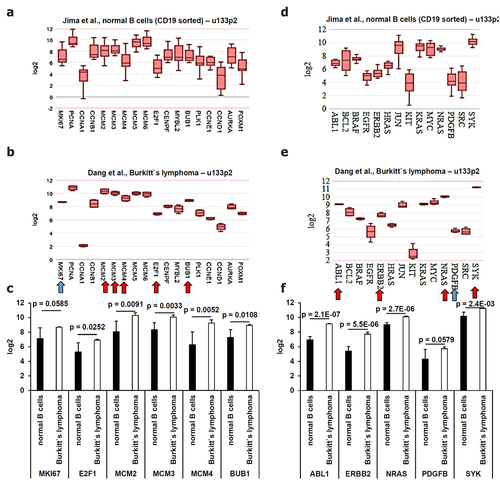
Since oncogenes are frequently involved in the regulation of cell proliferation and cell growth, the gene expression of 14 common oncogenes was analyzed in the EBV-positive BL cells (). As shown in , a statistically significant upregulation of ABL (p = 2.1E-07), ERBB2 (p = 5.5E-06), NRAS (p = 2.7E-06), SYK (p = 2.4E 03) and an almost statistically significant upregulation of PDGFB (0.0579) were found in BL cells when compared to healthy B cells ().
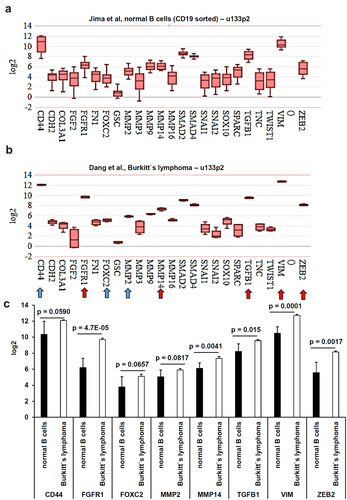
Based on the assumption that metastasis formation is a major cause of death from cancer,Citation43 markers of the epithelial–mesenchymal transition (EMT) and the mesenchymal–epithelial transition (MET) were analyzed. This is based on the fact that EBV infection is not only linked to BL or cHL,Citation37,Citation44 but also to solid tumor entities including inter alia NPCs and gastric cancers (GCs).Citation45–49 Since no data sets for solid EBV-associated tumor entities were available, the EMT and MET marker genes were also analyzed within the B cell and BL data sets, despite these results might only exert a surrogate for EBV-associated solid tumors.
It is known that at diagnosis some of the EBV-associated lymphoma are already at stage IV and infiltrated into the bone marrow, liver and lungs with differences within the lymphoma subtypes.Citation50 However, healthy B cells and the EBV-positive BL cells revealed a statistically significant downregulation of the two epithelial marker genes KLF4 (p = 0.0274) and KRT18 (p = 0.0338) (), while a statistically significant expression was found for FGFR1 (p = 4.7E-05), MMP14 (p = 0.0041), TGFB1 (p = 0.015), VIM (p = 0.0001) and ZEB2 (p = 0.0017) in healthy B cells () and a borderline statistically significant higher gene expression for CD44 (p = 0.059), FOXC2 (p = 0.0657) and MMP2 (p = 0.0817) in BL cells. This borderline statistical significance at the transcript level might be considered for putative protein analyses.
Figure 6. Comparative analysis of the gene expression from epithelial markers derived from microarray data. Microarray data were analyzed as described in Material and Methods. The mean expression values were calculated for (A-B) EMT markers. Statistically significant deregulated genes were marked by a red arrow. These results were summarized in (C) including the calculated p values.
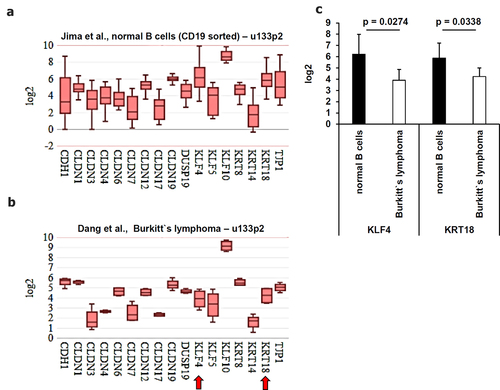
Figure 7. Comparative analysis of the gene expression from mesenchymal markers derived from microarray data. Microarray data were analyzed as described in Material and Methods. The mean expression values were calculated for (A-B) MET markers. Statistically significant dysregulated genes were marked by a red arrow and almost statistically significant deregulated genes were marked by a blue arrow. These results were summarized in (C) including the calculated p values.
Identification of EBV-regulated genes involved in the tumor metabolism
An altered metabolism in malignancies is a well-studied hallmark of cancer.Citation51 The increased glycolysis with a high catalysis and secretion of lactate in malignant tissues has been described as Warburg effectCitation52 contributing to an acidic tumor microenvironment (TME), thereby leading to a decreased activity of tumor-infiltrating immune effector cells and by acting as radical quencher enhancing the survival of tumor cells against radiation therapy and certain chemotherapies.Citation53 During EBV immortalization of B cells into LCLs, increased energetic demands for continued proliferation drive an upregulation of glycolysis and extracellular acidification.Citation54,Citation55 Therefore, the gene expression of enzymes involved in the glycolysis pathway leading to the synthesis of lactate were compared between healthy B cells and BL cells ().
Figure 8. Comparative analysis of the gene expression pattern for glycolysis-relevant enzymes focusing on to the Warburg effect from microarray data. Microarray data were analyzed as described in Material and Methods. The mean expression values were calculated (A) in healthy B cells and in EBV-positive BL cells. Statistically significant deregulated genes were marked by a red arrow and almost statistically significant dysregulated genes were marked by a blue arrow. These results were summarized in (B-C) including the calculated p values.
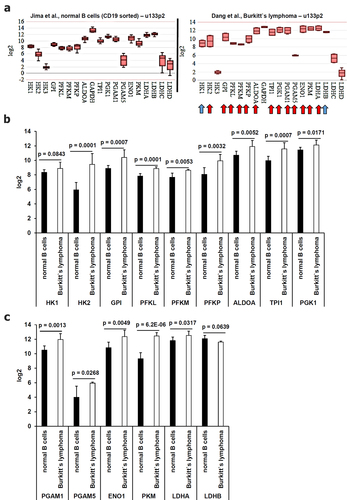
Indeed, 13 out of 19 investigated genes of glycolysis-relevant enzymes exerted a statistically significant increased expression and two genes showed an almost statistically significant gene expression in BL cells (). The strongest induction of glycolysis-relevant enzymes in BL cells was found for pyruvate kinase (p = 6.2E-06), triosephosphate isomerase (p = 0.0007), phosphofructokinase (p = 0.0001), glucose-6-phosphate isomerase (p = 0.0007) and hexokinase (p = 0.0001), which is accompanied by an increased proliferation of BL cells.
Chromosomal mapping of the deregulated tumor relevant target genes
In total, the expression of 161 cancer-related genes was analyzed between healthy B cells and malignant transformed EBV-positive BL cells. Compared to healthy B cells, statistically significant or almost significant altered gene expression patterns with either upregulated (n = 48) or downregulated (n = 5) genes by the EBV-driven malignant transformation of BL cells were found and summarized in including their genomic localization. The deregulated genes were randomly scattered over the human chromosomes, but most genes were located on chromosome 12 (n = 6), chromosome 9 (n = 5) and chromosome 10 (n = 5). These results might be of importance due to known genetic alterations on these chromosomes within tumor diseases in general, which will be addressed in more detail within the discussion section. None of the deregulated genes were located on the chromosomes 5, 7, 18 and Y.
Table 3. Identification of differentially expressed genes upon EBV-driven malignant transformation
Implementation of a combinatorial strategy for prediction of novel putative EBV-miR-regulated target genes
Due to the lack of in silico EBV-miR prediction databases, the RNA sequences including the 5’-untranslated region (UTR), the coding sequence (CDS) and the 3’-UTR of 5 down-regulated cancer-related host genes were analyzed for putative EBV-miR binding sites, involving all EBV-miR sequences, currently available within the mirbase online database (www.mirbase.org)Citation56,Citation57 by using the RNAhybrid prediction software (bibiserv.cebitec.uni-bielefeld.de/rnahybrid).Citation58,Citation59 This software allowed the calculation of the free energy of the EBV-miR mRNA target interaction (), but these in silico calculations have some limitations, since they do not consider any specifics of viral miRs, possibly containing putative viral specific RNA modifications with relevance for mRNA binding affinity and targeting.Citation60 Five EBV-miRs exerted strong interaction sites to even more than two of the five down-regulated tumor-relevant mRNA targets (IFNAR1, JAK1, STAT2, KLF4, KRT18). ebv-miR-BART4-5p and −13-5p could per se be involved in the down regulation of all five target genes, while the EBV-miRs ebv-miR-BART6-3p and ebv-miR-BART17-3p could be regulatory for four and ebv-miR-BART7-3p for three target genes (). The ebv-miR-BART7-3p was even among the most abundantly expressed EBV-miRs analyzed in the EBV-positive BL cell lines as well as in the EBV-positive FFPE specimen () suggesting a strong contribution of this EBV-miR in malignant transformation.
Table 4. In silico predicted free energies of putative EBV-miR-mRNA target interactions using RNA hybrid.Citation58,Citation59
In summary, this screening strategy allows to identify the most abundantly expressed EBV-miRs in the context of deregulated tumor biologic-/tumor immunologic-relevant target genes, which have to be subjected to additional molecular biological and functional studies to prove their predicted correlation and even the direct interactions.
Discussion
A putative host cell mRNA directly bound by viral encoded miRs should lead to a decrease of the respective protein and/or to the decay of the miR-bound mRNA. In addition, upregulated host cell gene expression could be caused by indirect effects of EBV-miRs, e.g. by direct binding and downregulation of suppressors/negative regulators of the respective gene. Based on this assumption, a downregulation of gene expression can be caused by miRs indirectly, e.g. by direct binding and downregulation of an inducer/positive regulator. In order to identify genes regulated by EBV-miRs, the expression of EBV-miRs was monitored in EBV-positive and EBV-negative tissues of distinct origin demonstrating even tissue-specific differences.
The highest expressed EBV-miRs were identified and were used as a prerequisite to reduce the number of putative EBV-miR candidates for follow up in silico analyses.
Next to the EBV-miRs, also the EBV-encoded lnc RNAs EBERs and BARTs are known to interfere with the gene expression of infected host cells. Recent studies report a shift towards an oncogenic phenotype by EBER1 and EBER2 overexpression in in vitro cell lines, but also in in vivo murine models,Citation61–63 which was due to an enhanced AKT signaling and an increased c-MYC (MYC) expression. In our study, an upregulated MYC expression was found in BL cells when compared to healthy B cells, which was not statistically significant (p = 0.57). The EBV-encoded protein LMP1 could also activate the PI3K-AKT signaling pathway thereby interfering with DNA repair mechanisms.Citation64 These observations strengthened the involvement of additional molecules in the process of virus-driven malignant transformation, including viral-encoded miRs. Recently, a growing number of target genes regulated by EBV-miRs have been identified, ranging exemplarily from immunological relevant targets, like IL-12 by ebv-miR-BART1, ebv-miR-BART2, and ebv-miR-BHRF1-2,Citation65 TAP2 by ebv-miR-BART17 and ebv-miR-BHRF1-3,Citation27 MICB by ebv-miR-BART2-5pCitation28 and IL-1 receptor 1 (IL1R1) by ebv-miR-BHRF1-2-5p,Citation66 CXCL11 by ebv-miR-BHRF1-3,Citation67 to the pro-apoptotic genes PTENCitation24 by ebv-miR-BART7-3p, PUMA by ebv-miR-BART20-5p and ebv-miR-BART8,Citation68 DAB2 by ebv-miR-BART1-3p,Citation69 DICE1 by ebv-miR-BART3Citation70 and TP53 by ebv-miR-BART-5-3p.Citation71
In the process of malignant transformation, an altered glucose metabolism known as Warburg effect is considered as an early event. It represents the metabolic basis for tumor cell proliferation and growth and also contributes to immune evasion due to the local acidic pH of the tumor microenvironment.Citation72 EBV infection has been shown to reorganize B cell metabolic pathways to promote immortalization as a precursor to the development of lymphomas. An increased aerobic glycolysis in EBV-infected B cells was associated with an upregulation of lactate transporters.Citation73 This is in line with a statistically significant higher expression in several key enzymes involved into the early steps of glycolysis in BL cells when compared to the healthy B cells. These findings might encourage further studies to investigate the impact of EBV-miRs towards metabolic alterations in the process of malignant transformation.
Next to this, other immune-relevant putative EBV target genes were identified, which mainly represent genes involved in the viral pattern recognition, in the recruitment of immune effector cells (chemokines) towards virus-infected host cells and in anti-viral interferon (IFN) signaling. These mechanisms might be beneficial for early or latent stages of EBV infection. Furthermore, immune modulatory molecules were detected to be statistically significant enhanced upon EBV infection, including PD-L1 and CD155, both involved in the inhibitory signaling to immune effector cells thereby possibly contributing to immune evasion of EBV-infected cells. This is in line with reports demonstrating an induction of PD-L1 upon EBV infection with an involvement of LMP1.Citation74,Citation75
Another hallmark of cancer is the activated invasiveness and ability of metastasis.Citation51 The in silico part of this study identified statistically significant reduced gene expression of epithelial marker genes as well as the enhanced expression of mesenchymal marker genes in the BL cells when compared to the healthy B cells offering putative targets of the EBV-driven malignant transformation. However, these results might be also translatable to solid EBV-associated tumor entities, for which EMT and MET processes are of higher relevance than for BLs. In fact, various EBV-encoded molecules are known to enhance EMT, in particular ebv-miR-BART7-3p,Citation24 ebv-miR-BART8-3p,Citation76 ebv-miR-BART11,Citation77 ebv-miR-BART1Citation31 and LMP1.Citation78
The combination of several other cancer hallmarks is reflected by the analysis of the gene expression pattern of factors involved in cell proliferation, cell cycle regulation as well as of known oncogenes. Many proliferation markers and known oncogenes exerted a statistically significant altered gene expression. A connection between EBV infection and higher NRAS gene expression has been published,Citation79 while an increased PDGFB expression has been reported in NPC, independently of EBV.Citation80 Together with the EBV-encoded LMP2A, SYK plays a role in cell migration of epithelial cells.Citation81 The proliferation marker and cell cycle regulator E2F1 is induced by the EBV-encoded BZLF1 protein.Citation82
In this study, a total of 161 genes with relevance for immune surveillance/immune evasion as well as neoplastic transformation have been screened for their deregulation upon EBV infection. A total of 24 of these 161 genes analyzed have already been linked to EBV infection and for some cases, the underlying molecular mechanisms have been identified. Multifaceted cancer-related processes were detected to be deregulated upon EBV infection and the induced gene expression could be explained by regulatory mechanisms, such as enhanced PIP3-AKT and ERK signaling, caused by EBV-encoded molecules.
A follow-up in silico analysis also highlighted interesting EBV-miR candidates caused a statistically significant downregulation in 5 out of the 24 deregulated target genes, thereby representing putative direct targets of EBV-miRs. Interestingly, 3/5 downregulated genes are involved in IFN signaling. However, a direct interaction between these candidate target genes and the respective EBV-miRs has not yet been proven, while other EBV-mediated effects could directly or indirectly cause the observed downregulation. In addition, 41 tumor biological relevant target genes exerted an upregulation upon EBV infection with a majority of them already described in the literature in the context of EBV as exemplarily described above.
Next to this, it has been demonstrated that various EBV-encoded miRs are expressed and detectable at high levels during latency with heterogeneous levels and a tissue-specific expression patterns in infectious mononucleosis, B cell lymphomas, NPCs as well as in EBV-positive B cell lines, but not in healthy tumor-free tissues and EBV-negative B cell lines. Since these EBV-miRs were stable in FFPE specimen, they might represent suitable prognostic markers, in particular when other EBV detection methods lead to unclear results as e.g. demonstrated for the LMP1 staining associated with the phase of EBV infection (). It was an initial intention of this study to underline the possible usage of EBV-miRs as diagnostic and/or prognostic markers. A value of miRs as biomarkers has already been shown for several human endogenous miRs, such as e.g. the oncogenic miR-21.Citation83 However, there is still an urgent need to extend the knowledge to viral-encoded miRs.
The qPCR-based determination of the most abundant EBV-miRs might also represent a possible alternative for EBV detection and give in depth knowledge of EBV-associated diseases. Furthermore, the combination of EBV-miR expression analyses and in silico prediction studies appears to be very promising tool leading to the identification of novel EBV-miR target genes, while avoiding the actual limitations of online target prediction databases lacking EBV-miRs.
The selection of 161 analyzed genes led to the identification of 24 EBV-driven deregulated genes with a heterogenous chromosomal distribution, but with chromosome 12, 9, and 10 harboring the most deregulated genes. This is noteworthy, since abnormalities of these chromosomes frequently occur in hematopoietic malignancies, such as acute myeloid leukemia (AML). Translocations leading to the well-known oncofusion proteins BCR-ABL1 (t(9;22)), DEK-CAN (t(6;9)), and MN1-TEL (t(12;22)),Citation84 but also abnormalities in 11q23 have been linked to an unfavorable outcome in AML patients.Citation85
Therapeutic strategies involving the targeting/inhibition of viral or endogenous oncogenic miRs have already reached preclinical animal studies and first clinical trials with promising results. These include the oncogenic miR-21 in melanoma, which was blocked by locked-nucleic acid miR inhibitors.Citation86 In the case of Miravirsen (miR-34a) and Cobomarsen (miR-155), miR-based therapies even entered clinical phase II and phase I trials for anti-tumoral therapies. In addition, an increasing number of patents are registered based on miRs as therapeutics against several viruses including the Ebola virus, Hepatitis C virus and the human enterovirus 71. Furthermore, miR-based therapies against cardiovascular diseases, renal diseases and other diseases have entered clinical studies, which have been recently reviewed by Chakraborty and co-authors.Citation87
Conclusions
This approach employs an in silico analysis to screen for novel so far unidentified EBV-targeted tumor biology relevant genes in the host cell by circumvention of the limited possibilities of the available databases in the context of EBV-encoded miRs. Numerous EBV-miRs were analyzed in different EBV-positive human (tumor) tissues/cells, leading to the identification of certain abundantly expressed EBV-miRs suitable for EBV detection, which might also have prognostic potential with an advantage to the conventional methods only analyzing one marker. Despite a limited number of genes were used for screening, five novel so far unidentified putative target genes for EBV-encoded miRs were identified, whose gene products were involved in tumor immunologic and tumor biologic related processes. An increased knowledge of the crucial role of EBV-miRs in EBV-associated malignancies and their targets does not only provide insights into the underlying mechanisms, but also represents a rational for the development of therapeutic strategies for these diseases.
Abbreviations
AML, acute myeloid leukemia; BART, BamHI-A rightward transcript; BHRF1, BamHI fragment H rightward open reading frame 1; BL, Burkitt’s lymphoma; CDS, coding sequence; cHL, classical Hodgkin lymphoma; EBER, Epstein–Barr virus encoded RNA; EBER-CISH, Epstein–Barr virus-encoded RNA chromogenic in situ hybridization; EBV, Epstein–Barr-Virus; EMT, epithelial–mesenchymal transition; FFPE, formalin-fixed paraffin-embedded; HHV, human herpes virus; IL1R1, interleukin 1 receptor 1; IM, infectious mononucleosis; IFN, interferon; lnc, long non-coding; MET, mesenchymal–epithelial transition; miR, microRNA; MM, malignant melanoma; NHL, non-Hodgkin lymphoma; NMHC-IIA, non-muscle myosin heavy chain IIA; NPC, nasopharyngeal carcinoma; NRP1, neuropilin 1; nt, nucleotide; qPCR, quantitative polymerase chain reaction; RCC, renal cell carcinoma; TLR, toll-like receptor; ULBP, UL16-binding protein; UTR, untranslated region.
Author contribution
The corresponding author did ensure that the descriptions are accurate and all authors agreed on the final version of the manuscript. Simon Jasinski-Bergner formulated the idea and was involved in the conceptualization and design of the experiments as well as in the writing the original draft. Juliane Blümke was performing the miR expression experiments, their validation and performed together with Saskia Luise Skiebe the statistical analyses. Marcus Bauer performed IHC and CISH analysis. Claudia Wickenhauser provided study materials and reviewed the manuscript together with Ofer Mandelboim. Barbara Seliger was mentoring the team, coordinated the manuscript and was involved in writing the original draft.
Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article [and/or] its supplementary materials.
Disclosure statement
The authors declare that they have no conflicts of interest.
Additional information
Funding
References
- Jasinski-Bergner S, Mandelboim O, Seliger B. Molecular mechanisms of human herpes viruses inferring with host immune surveillance. J Immunother Cancer. 2020;8(2):e000841. doi:10.1136/jitc-2020-000841
- Henle W, Henle G. Evidence for a relation of Epstein-Barr virus to Burkitt’s lymphoma and nasopharyngeal carcinoma. Bibl Haematol. 1970;(36):706–17. doi:10.1159/000391777
- Montes-Mojarro IA, Fend F, Quintanilla-Martinez L. EBV and the Pathogenesis of NK/T Cell Lymphoma. Cancers. 2021;13(6):1414. doi:10.3390/cancers13061414
- Ambinder RF. Epstein-Barr virus and Hodgkin lymphoma. Hematology Am Soc Hematol Educ Program. 2007;2007:204–209. doi:10.1182/asheducation-2007.1.204
- Danilova NV, Malkov PG, Oleynikova NA, Mikhailov IA. [Epstein-Barrvirus-associated gastric adenocarcinoma]. Arkh Patol. 2019;81(3):74–83. doi:10.17116/patol20198103174
- Tsao SW, Tsang CM, Lo KW. Epstein-Barr virus infection and nasopharyngeal carcinoma. Philos Trans R Soc Lond B Biol Sci. 2017;372(1732):20160270. doi:10.1098/rstb.2016.0270
- Tierney RJ, Shannon-Lowe CD, Fitzsimmons L, Bell AI, Rowe M. Unexpected patterns of Epstein-Barr virus transcription revealed by a high throughput PCR array for absolute quantification of viral mRNA. Virology. 2015;474:117–130. doi:10.1016/j.virol.2014.10.030
- Caetano BFR, Jorge BAS, Müller-Coan BG, Elgui de Oliveira D. Epstein-Barr virus microRNAs in the pathogenesis of human cancers. Cancer Lett. 2021;499:14–23. doi:10.1016/j.canlet.2020.11.019
- Zanella L, Riquelme I, Buchegger K, Abanto M, Ili C, Brebi P. A reliable Epstein-Barr Virus classification based on phylogenomic and population analyses. Sci Rep. 2019;9(1):9829. doi:10.1038/s41598-019-45986-3
- Yang HJ, Huang T-J, Yang C-F, Peng L-X, Liu R-Y, Yang G-D, Chu -Q-Q, Huang J-L, Liu N, Huang H-B, et al. Comprehensive profiling of Epstein-Barr virus-encoded miRNA species associated with specific latency types in tumor cells. Virol J. 2013;10:314. doi:10.1186/1743-422X-10-314
- Pratt ZL, Kuzembayeva M, Sengupta S, Sugden B. The microRNAs of Epstein-Barr Virus are expressed at dramatically differing levels among cell lines. Virology. 2009;386(2):387–397. doi:10.1016/j.virol.2009.01.006
- Jasinski-Bergner S, Stoehr C, Bukur J, Massa C, Braun J, Hüttelmaier S, Spath V, Wartenberg R, Legal W, Taubert H, et al. Clinical relevance of miR-mediated HLA-G regulation and the associated immune cell infiltration in renal cell carcinoma. Oncoimmunology. 2015;4(6):e1008805. doi:10.1080/2162402X.2015.1008805
- Nielsen AF, Gloggnitzer J, Martinez J. MicroRNAs cross the line: the battle for mRNA stability enters the coding sequence. Mol Cell. 2009;35(2):139–140. doi:10.1016/j.molcel.2009.07.006
- Broughton JP, Lovci M, Huang J, Yeo G, Pasquinelli A. Pairing beyond the Seed Supports MicroRNA Targeting Specificity. Mol Cell. 2016;64(2):320–333. doi:10.1016/j.molcel.2016.09.004
- Soltani S, Zakeri A, Tabibzadeh A, Zakeri AM, Zandi M, Siavoshi S, Seifpour S, Farahani A. A review on EBV encoded and EBV-induced host microRNAs expression profile in different lymphoma types. Mol Biol Rep. 2021;48(2):1801–1817. doi:10.1007/s11033-021-06152-z
- Kang BW, Choi Y, Kwon OK, Lee SS, Chung HY, Yu W, Bae HI, Seo AN, Kang H, Lee SK, et al. High level of viral microRNA-BART20-5p expression is associated with worse survival of patients with Epstein-Barr virus-associated gastric cancer. Oncotarget. 2017;8(9):14988–14994. doi:10.18632/oncotarget.14744
- Chan JY, Gao W, Ho W-K, Wei WI, Wong T-S. Overexpression of Epstein-Barr virus-encoded microRNA-BART7 in undifferentiated nasopharyngeal carcinoma. Anticancer Res. 2012;32:3201–3210.
- Siebolts U, Varnholt H, Drebber U, Dienes H-P, Wickenhauser C, Odenthal M. Tissues from routine pathology archives are suitable for microRNA analyses by quantitative PCR. J Clin Pathol. 2009;62(1):84–88. doi:10.1136/jcp.2008.058339
- Liu A, Xu X. MicroRNA isolation from formalin-fixed, paraffin-embedded tissues. Methods Mol Biol. 2011;724:259–267.
- Xi Y, Nakajima G, Gavin E, Morris CG, Kudo K, Hayashi K, Ju J. Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraffin-embedded samples. RNA. 2007;13(10):1668–1674. doi:10.1261/rna.642907
- Jasinski-Bergner S, Mandelboim O, Seliger B. The role of microRNAs in the control of innate immune response in cancer. J Natl Cancer Inst. 2014;106(10). doi:10.1093/jnci/dju257
- Eichmuller SB, Osen W, Mandelboim O, Seliger B. Immune Modulatory microRNAs Involved in Tumor Attack and Tumor Immune Escape. J Natl Cancer Inst. 2017;109(10). doi:10.1093/jnci/djx034
- Morris MA, Laverick L, Wei W, Davis A, O’Neill S, Wood L, Wright J, Dawson C, Young L. The EBV-Encoded Oncoprotein, LMP1, Induces an Epithelial-to-Mesenchymal Transition (EMT) via Its CTAR1 Domain through Integrin-Mediated ERK-MAPK Signalling. Cancers. 2018;10(5):130. doi:10.3390/cancers10050130
- Cai LM, Lyu X-M, Luo W-R, Cui X-F, Ye Y-F, Yuan -C-C, Peng Q-X, Wu D-H, Liu T-F, Wang E, et al. EBV-miR-BART7-3p promotes the EMT and metastasis of nasopharyngeal carcinoma cells by suppressing the tumor suppressor PTEN. Oncogene. 2015;34(17):2156–2166. doi:10.1038/onc.2014.341
- Zhang B, Kracker S, Yasuda T, Casola S, Vanneman M, Hömig-Hölzel C, Wang Z, Derudder E, Li S, Chakraborty T, et al. Immune surveillance and therapy of lymphomas driven by Epstein-Barr virus protein LMP1 in a mouse model. Cell. 2012;148(4):739–751. doi:10.1016/j.cell.2011.12.031
- Choi IK, Wang Z, Ke Q, Hong M, Qian Y, Zhao X, Liu Y, Kim H-J, Ritz J, Cantor H, et al. Signaling by the Epstein–Barr virus LMP1 protein induces potent cytotoxic CD4 + and CD8 + T cell responses. Proc Natl Acad Sci USA. 2018;115(4):E686–E695. doi:10.1073/pnas.1713607115
- Albanese M, Tagawa T, Bouvet M, Maliqi L, Lutter D, Hoser J, Hastreiter M, Hayes M, Sugden B, Martin L, et al. Epstein–Barr virus microRNAs reduce immune surveillance by virus-specific CD8 + T cells. Proc Natl Acad Sci USA. 2016;113(42):E6467–E6475. doi:10.1073/pnas.1605884113
- Nachmani D, Stern-Ginossar N, Sarid R, Mandelboim O. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe. 2009;5(4):376–385. doi:10.1016/j.chom.2009.03.003
- Wong TS, Chen S, Zhang M-J, Chan JYW, Gao W. Epstein-Barrvirus-encoded microRNA BART7 downregulates major histocompatibility complex class I chain-related peptide A and reduces the cytotoxicity of natural killer cells to nasopharyngeal carcinoma. Oncol Lett. 2018;16(3):2887–2892. doi:10.3892/ol.2018.9041
- Choy EY, Siu K-L, Kok K-H, Lung RWM, Tsang CM, To K-F, Kwong DLW, Tsao SW, Jin D-Y. An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival. J Exp Med. 2008;205(11):2551–2560. doi:10.1084/jem.20072581
- Cai L, Ye Y, Jiang Q, Chen Y, Lyu X, Li J, Wang S, Liu T, Cai H, Yao K, et al. Epstein-Barrvirus-encoded microRNA BART1 induces tumour metastasis by regulating PTEN-dependent pathways in nasopharyngeal carcinoma. Nat Commun. 2015;6:7353. doi:10.1038/ncomms8353
- Hsu CY, Yi Y-H, Chang K-P, Chang Y-S, Chen S-J, Chen H-C. The Epstein-Barr virus-encoded microRNA MiR-BART9 promotes tumor metastasis by targeting E-cadherin in nasopharyngeal carcinoma. PLoS Pathog. 2014;10(2):e1003974. doi:10.1371/journal.ppat.1003974
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33(20):e179. doi:10.1093/nar/gni178
- Longo NS, Lugar PL, Yavuz S, Zhang W, Krijger PHL, Russ DE, Jima DD, Dave SS, Grammer AC, Lipsky PE, et al. Analysis of somatic hypermutation in X-linked hyper-IgM syndrome shows specific deficiencies in mutational targeting. Blood. 2009;113(16):3706–3715. doi:10.1182/blood-2008-10-183632
- Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3(3):177–185. doi:10.1016/j.cmet.2006.02.002
- Pajic A, Spitkovsky D, Christoph B, Kempkes B, Schuhmacher M, Staege MS, Brielmeier M, Ellwart J, Kohlhuber F, Bornkamm GW, et al. Cell cycle activation by c-myc in a Burkitt lymphoma model cell line. Int J Cancer. 2000;87(6):787–793. doi:10.1002/1097-0215(20000915)87:6<787::AID-IJC4>3.0.CO;2-6
- Ogwang MD, Bhatia K, Biggar RJ, Mbulaiteye SM. Incidence and geographic distribution of endemic Burkitt lymphoma in northern Uganda revisited. Int J Cancer. 2008;123(11):2658–2663. doi:10.1002/ijc.23800
- Bauer M, Jasinski-Bergner S, Mandelboim O, Wickenhauser C, Seliger B. Epstein-BarrVirus-Associated Malignancies and Immune Escape: The Role of the Tumor Microenvironment and Tumor Cell Evasion Strategies. Cancers. 2021;13(20):5189. doi:10.3390/cancers13205189
- Ramshaw IA, Ramsay AJ, Karupiah G, Rolph MS, Mahalingam S, Ruby JC. Cytokines and immunity to viral infections. Immunol Rev. 1997;159:119–135. doi:10.1111/j.1600-065X.1997.tb01011.x
- Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14(4):778–809, table of contents. doi:10.1128/CMR.14.4.778-809.2001
- Kawai T, Akira S. Antiviral signaling through pattern recognition receptors. J Biochem. 2007;141(2):137–145. doi:10.1093/jb/mvm032
- Rehwinkel J, Gack MU. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat Rev Immunol. 2020;20(9):537–551. doi:10.1038/s41577-020-0288-3
- Ruddell A, Harrell MI, Furuya M, Kirschbaum SB, Iritani BM. B lymphocytes promote lymphogenous metastasis of lymphoma and melanoma. Neoplasia. 2011;13(8):748–757. doi:10.1593/neo.11756
- Huang X, Nolte I, Gao Z, Vos H, Hepkema B, Poppema S, van den Berg A, Diepstra A. Epidemiology of classical Hodgkin lymphoma and its association with Epstein Barr virus in Northern China. PLoS One. 2011;6(6):e21152. doi:10.1371/journal.pone.0021152
- Alsafadi N, Alqarni MS, Attar M, Mgarry R, Bokhari A. Nasopharyngeal Cancer: Prevalence, Outcome, and Impact on Health-Related Quality of Life at Princess Norah Oncology Center, Jeddah, Saudi Arabia. Cureus. 2020;12(5):e8199. doi:10.7759/cureus.8199
- Chang ET, Ye W, Zeng Y-X, Adami H-O. The Evolving Epidemiology of Nasopharyngeal Carcinoma. Cancer Epidemiol Biomarkers Prev. 2021;30(6):1035–1047. doi:10.1158/1055-9965.EPI-20-1702
- Tavakoli A, Monavari SH, Solaymani Mohammadi F, Kiani SJ, Armat S, Farahmand M. Association between Epstein-Barr virus infection and gastric cancer: a systematic review and meta-analysis. BMC Cancer. 2020;20(1):493. doi:10.1186/s12885-020-07013-x
- Sun K, Jia K, Lv H, Wang S-Q, Wu Y, Lei H, Chen X. EBV-Positive Gastric Cancer: Current Knowledge and Future Perspectives. Front Oncol. 2020;10:583463. doi:10.3389/fonc.2020.583463
- Hamashima C. The burden of gastric cancer. Ann Transl Med. 2020;8(12):734. doi:10.21037/atm.2020.03.166
- Board A. Adult Hodgkin Lymphoma Treatment (PDQ(R)): Patient Version. In: PDQ Cancer Information Summaries. Bethesda (MD): National Cancer Institute; 2002.
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi:10.1016/j.cell.2011.02.013
- Liberti MV, Locasale JW. The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem Sci. 2016;41(3):211–218. doi:10.1016/j.tibs.2015.12.001
- Sattler UG, Mueller-Klieser W. The anti-oxidant capacity of tumour glycolysis. Int J Radiat Biol. 2009;85(11):963–971. doi:10.3109/09553000903258889
- McFadden K, Hafez AY, Kishton R, Messinger JE, Nikitin PA, Rathmell JC, Luftig MA. Metabolic stress is a barrier to Epstein-Barr virus-mediated B-cell immortalization. Proc Natl Acad Sci USA. 2016;113(6):E782–90. doi:10.1073/pnas.1517141113
- Sommermann TG, O’Neill K, Plas DR, Cahir-mcfarland E. IKKbeta and NF-kappaB transcription govern lymphoma cell survival through AKT-induced plasma membrane trafficking of GLUT1. Cancer Res. 2011;71(23):7291–7300. doi:10.1158/0008-5472.CAN-11-1715
- Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47(D1):D155–D162. doi:10.1093/nar/gky1141
- Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–73. doi:10.1093/nar/gkt1181
- Rehmsmeier M, Steffen P, Höchsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10(10):1507–1517. doi:10.1261/rna.5248604
- Kruger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34:W451–4. doi:10.1093/nar/gkl243
- Marchand V, Motorin Y. [Detection and functions of viral RNA modifications: perspectives in biology and medicine]. Virologie. 2021;25(2):93–110. doi:10.1684/vir.2021.0899
- Herbert KM, Pimienta G. Consideration of Epstein-Barr Virus-Encoded Noncoding RNAs EBER1 and EBER2 as a Functional Backup of Viral Oncoprotein Latent Membrane Protein 1. mBio. 2016;7(1):e01926–15. doi:10.1128/mBio.01926-15
- Yajima M, Kanda T, Takada K. Critical role of Epstein-Barr Virus (EBV)-encoded RNA in efficient EBV-induced B-lymphocyte growth transformation. J Virol. 2005;79(7):4298–4307. doi:10.1128/JVI.79.7.4298-4307.2005
- Ruf IK, Rhyne PW, Yang C, Cleveland JL, Sample JT. Epstein-Barr virus small RNAs potentiate tumorigenicity of Burkitt lymphoma cells independently of an effect on apoptosis. J Virol. 2000;74(21):10223–10228. doi:10.1128/JVI.74.21.10223-10228.2000
- Chen YR, Liu M-T, Chang Y-T, Wu -C-C, Hu C-Y, Chen J-Y. Epstein-Barr virus latent membrane protein 1 represses DNA repair through the PI3K/Akt/FOXO3a pathway in human epithelial cells. J Virol. 2008;82(16):8124–8137. doi:10.1128/JVI.00430-08
- Tagawa T, Albanese M, Bouvet M, Moosmann A, Mautner J, Heissmeyer V, Zielinski C, Lutter D, Hoser J, Hastreiter M, et al. Epstein-Barr viral miRNAs inhibit antiviral CD4+ T cell responses targeting IL-12 and peptide processing. J Exp Med. 2016;213(10):2065–2080. doi:10.1084/jem.20160248
- Skinner CM, Ivanov NS, Barr SA, Chen Y, Skalsky RL. An Epstein-Barr Virus MicroRNA Blocks Interleukin-1 (IL-1) Signaling by Targeting IL-1 Receptor 1. J Virol. 2017;91(21). doi:10.1128/JVI.00530-17
- Xia T, O’Hara A, Araujo I, Barreto J, Carvalho E, Sapucaia JB, Ramos JC, Luz E, Pedroso C, Manrique M, et al. EBV MicroRNAs in Primary Lymphomas and Targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res. 2008;68(5):1436–1442. doi:10.1158/0008-5472.CAN-07-5126
- Huang WT, Lin CW. EBV-encodedmiR-BART20-5p and miR-BART8 inhibit the IFN-gamma-STAT1 pathway associated with disease progression in nasal NK-cell lymphoma. Am J Pathol. 2014;184(4):1185–1197. doi:10.1016/j.ajpath.2013.12.024
- Min K, Kim JY, Lee SK. Epstein-Barr virus miR-BART1-3p suppresses apoptosis and promotes migration of gastric carcinoma cells by targeting DAB2. Int J Biol Sci. 2020;16(4):694–707. doi:10.7150/ijbs.36595
- Lei T, Yuen K-S, Xu R, Tsao SW, Chen H, Li M, Kok K-H, Jin D-Y. Targeting of DICE1 tumor suppressor by Epstein-Barr virus-encoded miR-BART3* microRNA in nasopharyngeal carcinoma. Int J Cancer. 2013;133(1):79–87. doi:10.1002/ijc.28007
- Zheng X, Wang J, Wei L, Peng Q, Gao Y, Fu Y, Lu Y, Qin Z, Zhang X, Lu J, et al. Epstein-Barr Virus MicroRNA miR-BART5-3p Inhibits p53 Expression. J Virol. 2018;92(23). doi:10.1128/JVI.01022-18
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi:10.1126/science.1160809
- Bonglack EN, Messinger JE, Cable JM, Ch’ng J, Parnell KM, Reinoso-Vizcaíno NM, Barry AP, Russell VS, Dave SS, Christofk HR, et al. Monocarboxylate transporter antagonism reveals metabolic vulnerabilities of viral-driven lymphomas. Proc Natl Acad Sci USA. 2021;118(25). doi:10.1073/pnas.2022495118
- Bi XW, Wang H, Zhang -W-W, Wang J-H, Liu W-J, Xia Z-J, Huang H-Q, Jiang W-Q, Zhang Y-J, Wang L, et al. PD-L1 is upregulated by EBV-driven LMP1 through NF-kappaB pathway and correlates with poor prognosis in natural killer/T-cell lymphoma. J Hematol Oncol. 2016;9(1):109. doi:10.1186/s13045-016-0341-7
- Nakano H, Saito M, Nakajima S, Saito K, Nakayama Y, Kase K, Yamada L, Kanke Y, Hanayama H, Onozawa H, et al. PD-L1 overexpression in EBV-positive gastric cancer is caused by unique genomic or epigenomic mechanisms. Sci Rep. 2021;11(1):1982. doi:10.1038/s41598-021-81667-w
- Lin C, Zong J, Lin W, Wang M, Xu Y, Zhou R, Lin S, Guo Q, Chen H, Ye Y, et al. EBV-miR-BART8-3p induces epithelial-mesenchymal transition and promotes metastasis of nasopharyngeal carcinoma cells through activating NF-kappaB and Erk1/2 pathways. J Exp Clin Cancer Res. 2018;37(1):283. doi:10.1186/s13046-018-0953-6
- Song Y, Li Q, Liao S, Zhong K, Jin Y, Zeng T. Epstein-Barrvirus-encoded miR-BART11 promotes tumor-associated macrophage-induced epithelial-mesenchymal transition via targeting FOXP1 in gastric cancer. Virology. 2020;548:6–16. doi:10.1016/j.virol.2020.05.011
- Zuo LL, Zhang J, Liu L-Z, Zhou Q, Du S-J, Xin S-Y, Ning Z-P, Yang J, Yu H-B, Yue W-X, et al. Cadherin 6 is activated by Epstein-Barr virus LMP1 to mediate EMT and metastasis as an interplay node of multiple pathways in nasopharyngeal carcinoma. Oncogenesis. 2017;6(12):402. doi:10.1038/s41389-017-0005-7
- Almeida JFM, Proenca-Modena JL, Bufalo NE, Peres KC, de Souza Teixeira E, Teodoro L, Beck RM, Moraes AP, Tincani AJ, Arns CW, et al. Epstein-Barr virus induces morphological and molecular changes in thyroid neoplastic cells. Endocrine. 2020;69(2):321–330. doi:10.1007/s12020-020-02253-0
- Liang YY, Deng X-B, Lin X-T, Jiang -L-L, Huang X-T, Mo Z-W, Yuan Y-W, Teh M-T. RASSF1A inhibits PDGFB-driven malignant phenotypes of nasopharyngeal carcinoma cells in a YAP1-dependent manner. Cell Death Dis. 2020;11(10):855. doi:10.1038/s41419-020-03054-z
- Lu J, Lin W-H, Chen S-Y, Longnecker R, Tsai S-C, Chen C-L, Tsai C-H. Syk tyrosine kinase mediates Epstein-Barr virus latent membrane protein 2A-induced cell migration in epithelial cells. J Biol Chem. 2006;281(13):8806–8814. doi:10.1074/jbc.M507305200
- Mauser A, Holley-Guthrie E, Zanation A, Yarborough W, Kaufmann W, Klingelhutz A, Seaman WT, Kenney S. The Epstein-Barr virus immediate-early protein BZLF1 induces expression of E2F-1 and other proteins involved in cell cycle progression in primary keratinocytes and gastric carcinoma cells. J Virol. 2002;76(24):12543–12552. doi:10.1128/JVI.76.24.12543-12552.2002
- Feng YH, Tsao CJ. Emerging role of microRNA-21 in cancer. Biomed Rep. 2016;5(4):395–402. doi:10.3892/br.2016.747
- Kumar CC. Genetic abnormalities and challenges in the treatment of acute myeloid leukemia. Genes Cancer. 2011;2(2):95–107. doi:10.1177/1947601911408076
- Schoch C, Schnittger S, Klaus M, Kern W, Hiddemann W, Haferlach T. AML with 11q23/MLL abnormalities as defined by the WHO classification: incidence, partner chromosomes, FAB subtype, age distribution, and prognostic impact in an unselected series of 1897 cytogenetically analyzed AML cases. Blood. 2003;102(7):2395–2402. doi:10.1182/blood-2003-02-0434
- Javanmard SH, Vaseghi G, Ghasemi A, Rafiee L, Ferns GA, Esfahani HN, Nedaeinia R. Therapeutic inhibition of microRNA-21 (miR-21) using locked-nucleic acid (LNA)-anti-miR and its effects on the biological behaviors of melanoma cancer cells in preclinical studies. Cancer Cell Int. 2020;20:384. doi:10.1186/s12935-020-01394-6
- Chakraborty C, Sharma AR, Sharma G, Lee -S-S. Therapeutic advances of miRNAs: A preclinical and clinical update. J Adv Res. 2021;28:127–138. doi:10.1016/j.jare.2020.08.012
