ABSTRACT
Angioimmunoblastic T-cell lymphoma (AITL) is a malignant hematologic tumor arising from T follicular helper (Tfh) cells. High-throughput genomic sequencing studies have shown that AITL is characterized by a novel highly recurring somatic mutation in RHOA, encoding p.Gly17Val (RHOA G17V). However, the specific role of RHOA G17V in AITL remains unknown. Here, we demonstrated that expression of Rhoa G17V in CD4+ T cells increased cell proliferation and induces Tfh cell specification associated with Pon2 upregulation through an NF-κB-dependent mechanism. Further, loss of Pon2 attenuated oncogenic function induced by genetic lesions in Rhoa. In addition, an abnormality of RHOA G17V mutation and PON2 expression is also detected in patients with AITL. Our findings suggest that PON2 associated with RHOA G17V mutation might control the direction of the molecular agents-based AITL and provide a new therapeutic target in AITL.
KEYWORDS:
Introduction
Angioimmunoblastic T-cell lymphoma (AITL) is a subtype of peripheral T-cell lymphoma (PTCL) and accounts for nearly 20% of PTCL cases.Citation1,Citation2 It usually affects older adults with autoimmune manifestations (e.g., hemolytic anemia, rheumatoid arthritis, or autoimmune thyroiditis) and has limited responses to intensive chemotherapy and high relapse rates.Citation1–3 T follicular helper (Tfh) cells, which are a highly plastic subset of CD4+ T cells specialized in regulating germinal center B cell differentiation into plasma cells and memory B cells, have been identified as cell-of-origin of AITL by immunohistochemical analysesCitation4,Citation5 and gene expression profiling studies.Citation6,Citation7 However, the mechanisms that drive Tfh cell transformation in AITL remain largely unclear.
Recently, high-throughput genomic sequencing technologies have shed light on the mutational profile of AITL and revealed frequent mutations in epigenetic regulators, such as TET2, DNMT3A, and IDH2.Citation8–10 Following these discoveries, a novel, highly recurring somatic mutation in RHOA, encoding p.Gly17Val, was identified in almost 70% of AITLs, in a much smaller proportion of peripheral T-cell lymphomas, not otherwise specified (PTCL-NOS), and was not detected in B-cell or myeloid malignancies.Citation9,Citation11 Following the discovery of the RHOA G17V mutation in AITLs, animal modeling, and molecular modeling simulations support the RHOA G17V mutation as a dominant-negative driver of AITL-specific pathogenesis.Citation12–14 Our previous data firstly showed that coexistence of somatic mutations in the RHOA and the 5-methylcytosine oxidase TET2 cooperatively modulated FoxO1 activity and further disrupted peripheral T cell homeostasis, and caused immunoinflammatory responses that are commonly associated with AITL.Citation12 Cortes et al.Citation13 have shown that the coexistence of RHOA G17V and 5-methylcytosine oxidase TET2 cooperatively resulted in CD4+ T-cell transformation and development of AITL in mice. Likewise, Ng et al.Citation14 have demonstrated that the RHOA G17V mutation contributes to neoplastic and paraneoplastic phenotypes similar to those observed in patients with Tfh lymphomas. A novel mouse model for AITL expressing RHOA G17V under the control of murine Lck distal promoter which in the absence of further genetic manipulations develops multiple AITL-like phenotypic traits.Citation15 Additional functional work from RNA sequencing and bioinformatics analysis demonstrated that expression of RHOA G17V in CD4+ T cells induced Tfh cell specification through inducible co-stimulator (ICOS)-mediated phosphoinositide 3-kinase (PI3K) and mitogen-activated protein kinase (MAPK) signaling pathways with highly expressing PON2.Citation13 Despite these findings, the clinical significance of the association between RHOA G17V mutation and abnormal expression of PON2 in AITLs remains unknown.
PON2 is one of the three highly conserved members of the paraoxonase family of enzymes consisting of PON1, PON2, and PON3.Citation16,Citation17 In contrast to PON1 and PON3, PON2 is not present in serum lipoprotein fractions, but in intracellular proteins, particularly in the perinuclear region and endoplasmic reticulum, and plays a key role in organophosphate poisoning, diabetes, obesity, cardiovascular diseases, and innate immunity.Citation18 Recently, several studies observed marked overexpression of PON2 in cancers, which include ovarian cancer,Citation19 pancreatic cancer,Citation20 and skin neoplasm.Citation21 PON2 plays a role in protection against oxidative stress, inhibition of apoptosis, and progression in malignancies.Citation18 It has also been found to be upregulated in hematopoietic neoplasms, such as T-cell lymphoblastic leukemia and pediatric acute lymphoblastic leukemia, and predicts poor outcome prognosis.Citation22 However, the role of PON2-mediated signaling in the tumorigenesis and progression of RHOA G17V-mutated AITLs has not been investigated.
In this study, we showed the biological function of Pon2 and Rhoa G17V mutation in CD4+ T-cell transformation and polarizing toward Tfh cells, which are the cell-of-origin of AITL. In addition, immunohistochemistry (IHC) and DNA sequencing analyses were applied to detect PON2 expression and RHOA G17V mutation, respectively, to rule out the role of the association between them in the progression and survival of patients with AITL.
Materials and methods
Ethics statement
The human tissue specimen study protocol was approved by the Research Ethics Committee of Ethics Committee of Fujian Medical University (2017KY087) and Sun Yat-set University Cancer Center (B2021-467-01), China. Written informed consent requirements were waived for the nature of this study. The handling of human tissue specimens was in strict accordance with the relevant institutional and national guidelines and regulations.
Patient samples
This study retrospectively investigated 46 diagnosed AITL patients at the Union Hospital of Fujian Medical University from January 2011 to September 2018 and 8 AITL patients at Sun Yat-set University Cancer Center from January 2016 to September 2018. AITL tissues were obtained from 54 patients who underwent surgical biopsy, comprising 39 men and 15 women, with age ranging from 40 to 84 years (average age, 66 years). None of the patients received preoperative chemotherapy or radiotherapy. AITL was diagnosed based on histopathological and immunohistochemical examinations conducted by two experienced hematopathologists (Mxu and Szang) according to the 2017 WHO criteria. The Eastern Cooperative Oncology Group performance status (ECOG PS) and international prognostic index (IPI) for PTCL scores were applied to assign risk stratification.Citation23 Forty normal lymph nodes from patients with inflammatory disease at the Union Hospital of Fujian Medical University were used as controls. Specimens were fixed in 10% neutral formalin and embedded in paraffin.
Animals
Four-week-old female C57BL/6 and T cell receptor-transgenic (TCR-tg) donor mice, e.g. OT-II mice in which T cells carry a transgenic TCR recognizing ovalbumin (OVA) were maintained in temperature-controlled clean racks with a 12-h light/dark cycle. The animals were allowed to acclimatize for 1 week before the start of the experiment. All animal procedures were conducted in full accordance with the Guidelines for the Care and Use of Laboratory Animals, and they were approved by the Institutional Animal Care and Use Committees at Sun Yat-set University Cancer Center.
Follow-up
Complete follow-up information for 46 patients was acquired until September 2018, and the median observation time was 8 months (range: 1–69 months). Overall survival (OS) was defined as the interval between AITL diagnosis and death or the last follow-up.
DNA extraction and targeted amplicon sequencing
DNA was extracted from formalin-fixed paraffin-embedded tissues using a silica membrane–based DNA purification method (DNA Micro Kit, Sangon Biotech, and Shanghai, China). All extracted DNA samples were assessed for the quality using PCR amplification. Forty-six AITLs were assessed using targeted amplicon sequencing to detect the RHOA G17V mutation in exon 2 of the RHOA gene (NM_001664). A fragment of RHOA, including the p.Gly17Val site was amplified using DNA extracted from formalin-fixed paraffin-embedded tissue sections, with 5`-GCCCCATGGTTACCAAAGCA-3` and 5`-TATCGAGGTGGATGGAAAGC-3` as sense and antisense primers, respectively. A PCR-amplified product of 244 bp was obtained in all cases, and direct sequencing of these products was performed. The coding DNA position 50 G > T mutation of the RHOA gene predicted a change in the wild-type G (Gly) to the mutant type V (Val).
Immunohistochemistry and evaluation
Immunohistochemistry was performed using the EliVisionTM Plus two-step system (Maixin Biotech, Fuzhou, China) according to the manufacturer’s instructions. The slides were deparaffinized, rehydrated, and immersed in 3% hydrogen peroxide solution for 10 min. They were then boiled in citrate buffer (pH 6.0) for 3 min and cooled at room temperature for 1 h. Blocking was conducted carried out with 10% normal goat serum at 37°C for 30 min. The slides were then incubated in a 1:100 dilution of rabbit monoclonal anti-human/mouse PON2 antibody (Abcam, Cambridge, UK) for 1 h at 37°C, followed by three washes with PBS. The slides were incubated with polymerized horseradish peroxidase anti-rabbit lgG and visualized with DAB. Counterstaining was performed using hematoxylin. The negative control sections were incubated with pre-immune serum. PBS was used for the primary antibody to serve as a blank control. Positive PON2 were evaluated according to the intensity of the tumor cell-specific cytoplasmic staining. Moderate to strong cytoplasmic staining observed in more than 10% of tumor cells was regarded as positive. Faint cytoplasmic staining or moderate to strong cytoplasmic expression observed in less than 10% of tumor cells was regarded as negative, to account for background staining or staining because of non-tumor cells. The staining results were evaluated by two independent pathologists (Szang and Mxu) without knowledge of the clinicopathologic features, and differences in interpretation were resolved by consensus.
Mouse T cell isolation and activation
As described previously,Citation12 mouse CD4+ T-cells were purified using a CD4+ T cell isolation kit (mouse) (Miltenyi Biotec, Germany). In brief, spleens and lymph nodes were isolated from C57BL/6 mice (6–8 weeks-old) and ground with a syringe plunger to release splenocytes and lymphocytes into a culture dish. The homogenized cell suspensions were filtered with a cell strainer (nylon mesh with 70 µM pores) to remove debris. The suspended cells were centrifuged and the red blood cells were lysed with ACK lysis buffer. The residual cells were then purified by CD4+ T cell isolation kits following the manufacturer’s instructions. The purity of the isolated T cells was assessed by flow cytometric analysis. Then, CD4+ T cells were activated with plate-bound anti-CD3 (clone 145–2C11, 10 μg/mL) plus soluble anti-CD28 (clone 37.51, 2 μg/mL) (BD Bioscience, San Jose, CA).
Plasmid construction and retrovirus transduction
A cDNA fragment encoding FLAG-tagged murine Rhoa WT was synthesized (Sangon Biotech, Shanghai, China) and cloned into the MSCV-IRES-GFP vector between the restriction sites of XhoI and EcoRI. Mutagenesis to create the construct encoding Gly17Val mutant of Rhoa was carried out with the Primestar Mutagenesis Basal kit (Takara, Japan) according to the manufacturer's protocol by using MSCV-IRES-GFP-Rhoa as a template. Based on the mouse Pon2 gene sequence in GenBank (NM_183308.3) and shRNA design principles, two sequences (shRNA1 and shRNA2) that specifically inhibited Pon2 expression were designed and cloned into the MSCV-IRES-mCherry vector between the restriction sites of AgeI and EcoRI. A nonspecific shRNA was used as a negative control (NC). All the plasmids were verified by Sanger sequencing (Sangon Biotech, Shanghai, China). The correct plasmids were prepared using PureLink HiPure Plasmid Maxiprep kit (Thermo Fisher Scientific, MA, USA).
Retroviruses encoding the target gene were generated using MSCV and EcoPack plasmids transfected into the potent retrovirus packaging cell line, Plat-E (Cell Biolabs, USA). Retrovirus-containing supernatants were collected from transfected Plat-E cells and then concentrated using ultracentrifugation (Beckman SW28 rotor, 20,000 rpm for 2 hr at 4°C). Concentrated retroviruses were re-suspended, aliquoted, and stored in −80°C up to 2 months.
After 12–24 h stimulation of CD4+ T cells, concentrated retroviruses at optimized titers, along with polybrene (6 µg/mL, EMD Millipore, MA, USA), were added to cultured T cells, followed by centrifugation (2000 rpm at 37°C for 90 min). The transduction efficiency of retroviruses was examined by flow cytometry 24–48 h after transduction.
Adoptive T-cell transfer
As described previously,Citation12 CD4+ T cells isolated from OT-II mice were transferred into 6 to 8 week-old C57BL/6 female recipients by retro-orbital injection. We immunized the recipient mice by standard foot pad immunization using 50 μg NP14-OVA (Biosearch Technologies, Berlin, Germany) precipitated in alum adjuvant (Thermo Fisher Scientific, MA, USA).
Flow cytometry analysis
Cells were re-suspended in FACS buffer (PBS with 1% BSA, 2 mM EDTA) and incubated with Fc blocker for 10 min on ice. After washing with FACS buffer, cells were incubated with the desired antibodies at optimal concentrations for 20 min on ice in the dark. Cells were then washed with FACS buffer twice and re-suspended in 200 µL FACS buffer for flow cytometry analysis (LSRII, BD Biosciences, NJ, USA). For intracellular staining, cells were stained with surface markers as described above, treated with cell fixation/permeabilization kit reagents (BD Biosciences, NJ, USA) and then incubated with antibodies for desired intracellular markers. CD4-PE, CD4-APC, or CD4-eFluor 450 was purchased from BD Biosciences. Tfh cell staining was performed using a three-step staining protocol.Citation24 Annexin V-pacific Blue (Biolegend, CA, USA) was used to assess the apoptotic status of cells. CellTrace Violet (CTV) (Thermo Fisher Scientific, MA, USA) was applied to identify cell proliferation status, and the proliferation index (P.I.) at each division peak was calculated following the manufacturer’s instructions. Flow cytometry analysis was performed using a LSRII platform (BD Biosciences, NJ, USA) and data were analyzed using FlowJo software 7.6 (TreeStar, CA, USA).
In vitro pharmacological treatments
To test the effect of a molecular inhibitor of PI3K on suppressing Pon2 expression in Rhoa G17V-expressing CD4 + T cells, we used a double PI3Kδ and PI3Kγ inhibitor, Duvesilib (MedChemExpress LLC, Shanghai, China) which was dissolved in a solution containing 5% DMSO and 30% PEG 400 in phosphate buffered saline (PBS). Briefly, Rhoa G17V-expressing CD4+ T cells were plated in 6 well plates (1 × 106 per well) and treated with 25 nM Duvelisib for 24 h. After treatment, cells were assayed by western blot as indicated. Rhoa G17V activation of NF-κB was inhibited pharmacologically with 10 μM BAY 11–7082 (BAY) (Tocris Bioscience, Missouri, United Kingdom) or by transduction with the murine S32/36A mutant SR-IκBα that had been inserted into the pLZRS retroviral vector downstream of an CMV promoter.
Western blotting
Cells were lysed with RIPA buffer (150 mM NaCl, 50 mM Tris-HCl, pH 8.0, 1% Triton X-100, 0.5% sodium deoxycholate and 0.1% SDS) supplemented with protease inhibitor cocktail (GenDEPOT, TX, USA) and incubated on ice for 20 min. Cell debris was removed by centrifuging at 12,000 rpm for 10 min at 4°C. Protein concentrations were measured using a Pierce BCA protein assay kit (Thermo Fisher Scientific, MA, USA). Samples were mixed with SDS sample buffer at 95°C for 10 min. Whole cell lysates were resolved by 10% or 15% SDS-PAGE and transferred to nitrocellulose membranes. Proteins were detected by immunoblotting in TBST (150 mM NaCl, 10 mM Tris-Cl, pH 8.0, 0.5% Tween-20) containing 5% low-fat milk, followed by incubation with rabbit monoclonal anti-human/mouse PON2 antibody (1:1000 dilution, Cell Signaling Technology, MA, USA), rabbit monoclonal anti-human/mouse Phospho-IKKα/β (Ser176/180) antibody (1:500 dilution, Cell Signaling Technology, MA, USA), rabbit monoclonal anti-human/mouse IKKβ antibody (1:1000 dilution, Cell Signaling Technology, MA, USA), rabbit monoclonal anti-mouse IκBα antibody (1:1000 dilution, Cell Signaling Technology, MA, USA), rabbit monoclonal anti-mouse IκBβ antibody (1:1000 dilution, Cell Signaling Technology, MA, USA), rabbit polyclonal anti-human/mouse NF-κB p65 antibody (1:1000 dilution, Cell Signaling Technology, MA, USA), or rabbit monoclonal anti-mouse β-Actin antibody (1:1,000 dilution, Sigma, CA, USA) at RT for 1 hr. Then, the membrane was incubated with HRP-conjugated secondary antibody (goat anti-mouse IgG HRP, Sigma, CA, USA) and proteins were detected using the West-Q Pico Dura ECL kit (GenDEPOT, TX, USA).
Immunofluorescent staining
Cell suspensions were attached to microscope slides through low-speed cytospin centrifugation (400 g, 5 min) to get a uniform monolayer of cells. Cells were fixed in 2% paraformaldehyde for 20 min at room temperature and permeabilized in 0.5% Triton X-100 for 10 min in PBS. Slides were incubated with primary antibody p65 (1:500 dilution, Cell Signaling Technology, MA, USA) for 60 min. Nuclei were stained with DAPI (Sigma-Aldrich, CA, USA). Immune complexes were stained with secondary antibody conjugated to Alexa Fluor 488 or Alexa Fluor 546 (1:200 dilution, Molecular Probes, Invitrogen, USA) and were viewed with an Olympus IX71 confocal microscope.
Multiplexed fluorescence immunohistochemistry and analysis
Multiplexed fluorescence immunohistochemistry (mFIHC) was performed to assess CD4 and PON2 stains in the AITL samples by using TSA-RM staining kit (Panovue Biological Technology, Beijing, China) according to the manufacturer’s instructions. FFPE sections were dewaxed and rehydrated through a series of xylene to alcohol washes before incubating in distilled water. FIHC staining was then performed after heat-induced antigen retrieval followed by cooling and Tris-buffer treatment. First step: the slides were incubated in a 1:100 dilution of rabbit monoclonal anti-human/mouse PON2 antibody (Abcam, Cambridge, UK) for 30 min, then incubated with Anti-Rabbit HRP (K4001; Dako) for 10 min, and amplified with TSA FITC for 10 min. Second step: the slides were incubated with mouse monoclonal anti-human anti-CD4 in a 1:400 dilution (Maixin, Fuzhou, China) for 30 min, followed by Anti-Mouse HRP incubation for 10 min and amplification with TSA Cy5 for 10 min. Nuclei were subsequently visualized with DAPI (1:2000), and the section was coverslipped using Vectashield Hardset mounting media. Specimens were scored positive for PON2 (Green) or CD4 (Red) if strong membrane and cytoplasmic staining in any percentage of tumor cells was present. Any membrane and cytoplasmic staining in tumor cells stained in yellow was considered positive for colocalization of PON2 and CD4. DAPI (Blue) is for nuclei staining.
Statistical analysis
Statistically significant differences between groups were assessed using Fisher’s exact test or one-way analysis of variance with the statistical software SPSS (version 19.0; SPSS Inc., Armonk, NY, USA). Survival rates were calculated using the Kaplan–Meier method, and comparisons of survival curves were performed using the log-rank test. Differences were considered statistically significant at P values <.05.
Results
Pon2 is upregulated by Rhoa G17V in CD4+ T cells
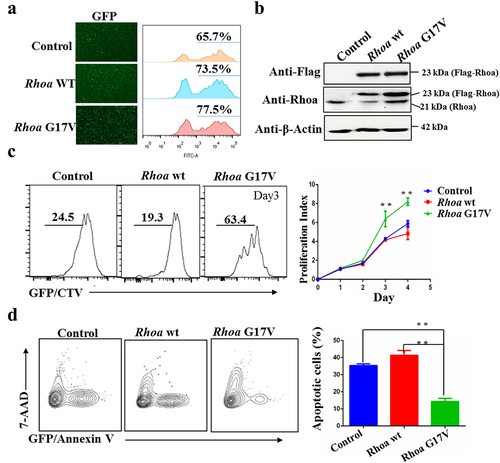
To investigate the role of RHOA G17V mutation in T cell development and the pathogenesis of AITL, we first tested the proliferative response of RhoA G17V-expressing murine CD4+ T cells to stimulation with anti-CD3 and anti-CD28 antibodies. We isolated total CD4+ T cells from lymph nodes and spleens of C57BL/6 mice and then transduced them with a retrovirus encoding wild-type Flag-Rhoa (WT) or Flag-Rhoa G17V or with an empty vector (control) expressing GFP as an internal control for 24 hours in media with anti-CD3 and anti-CD28 antibodies. After flow cytometry analysis showed a retroviral transduction efficiency of 60– 80% in all the experimental groups, as indicated by GFP signals (), we labeled the cells with CellTrace Violet (CTV) and cultured them for additional 4 days. The heterologous expression of Flag-Rhoa, Flag-Rhoa G17V, and endogenous Rhoa was further confirmed by western blotting (). The results revealed a marked increase in cell division of Rhoa G17V-expressing CD4+ T cells as evaluated by CTV staining compared to the control CD4+ T cells and WT groups (). In parallel, we monitored cell apoptosis in Rhoa G17V-expressing CD4+ T cells. In comparison with WT and control CD4+ T cells, Rhoa G17V-expressing CD4+ T cells showed significant reductions in cell apoptosis (annexin V staining) ().
Figure 1. Rhoa G17V expression in CD4 + T cells increases cell proliferation. (a) Retroviral Flag-Rhoa WT, Flag-Rhoa G17V expression vectors, or GFP control vectors was transduced into mouse CD4+ T cells. Transduction efficiency was indicated as GFP signals by flow cytometry analysis. (b) Flag-Rhoa WT, Flag-Rhoa G17V, and endogenous Rhoa expression in CD4+ T cells transduced with retroviral Flag-Rhoa WT, Flag-Rhoa G17V expression vectors, or GFP control vectors were detected by western blot analysis. β-Actin served as a loading control. (c) In vitro CellTrace Violet (CTV) proliferation assay of CD4+ T cells transduced with retroviral Flag-Rhoa WT, Flag-Rhoa G17V expression vectors, or GFP control vectors for four days. (d) Annexin V and 7-ADD staining of CD4+ T cells transduced with retroviral Flag-Rhoa WT, Flag-Rhoa G17V expression vectors, or GFP control vectors on day three were analyzed by flow cytometric analysis. The P value in (C) and (D) was calculated by ANOVA followed by Dunnett’s test using triplicate samples from two independent experiments. Columns indicate means; bars are the standard error. **P ≤ .01.
To further assess how Rhoa G17V affected the fate of CD4+ T cells, a significant upregulation of Pon2 in Rhoa G17V-expressing CD4+ T cells was observed by western blot (). As reported before,Citation12,Citation13 expression of Rhoa G17V in CD4+ T cells dramatically increased PI3K/Akt signaling (). In order to test whether Rhoa G17V regulated Pon2 expression through PI3K/Akt signaling, we focused on the phosphorylated IκB kinase (IKK) complex, which is most prominently known to regulate the IκB degradation in the cytosol as well as the release of NK-ĸB that can then translocate to the nucleus with the ability to induce gene transcription, including PON2 transcription. As illustrated in , Rhoa G17V-expressing CD4+ T cells exhibited a higher level of the phosphorylated IKK complex and lower expression of IκBα and IκBβ than in both WT and control CD4+ T cells. Thus, we hypothesized that RHOA G17V in CD4+ T cells activated PI3K/Akt/IKK signaling to stimulate NF-ĸB, which is the key transcription factor of PON2. In parallel, we performed an immunofluorescence assay and observed a significant increase in the percentage of cells with nuclear NF-κB p65 in Rhoa G17V-expressing CD4+ T cells compared to WT and control CD4+ T cells (P = .0001; ). To confirm the immunofluorescence data, we examined the levels of NF-κB p65 in nuclear fractions by using western blot analysis. The results showed an increase of NF-κB p65 in the nuclear fraction in Rhoa G17V-expressing CD4+ T cells, as compared to WT and control CD4+ T cells (). We next applied for the duvelisib (IPI-145), a clinically approved PI3kδ/γ inhibitor with high activity in follicular lymphoma and chronic lymphocyte leukemia,Citation25 as well as AITL in a phase 1 trial,Citation26 to test whether a molecular inhibitor of PI3K could suppress Pon2 expression in Rhoa G17V-expressing CD4+ T cells. Our data show that duvelisib decreased the phosphorylated IκB kinase (IKK) complex, inactivated the NF-κB p65, and perturbed the expression of Pon2 ().
Figure 2. Rhoa G17V induces Pon2 expression through activation of the PI3K/Akt/NF-κB pathway in CD4+ T cells. (a) Immunoblots showing expression of Pon2, phosphorylated Akt, phosphorylated IκB kinase, IκKβ, IκBα, and IκBβ in CD4+ T cells transduced with retroviral Flag-Rhoa WT, Flag-Rhoa G17V expression vectors, or GFP control vectors. β-Actin served as a loading control. (b) Representative confocal images showing immunostaining of NF-κB p65 (Green) in CD4+ T cells transduced with retroviral Flag-Rhoa WT, Flag-Rhoa G17V expression vectors, or GFP control vectors. DAPI (Blue) was used as a nuclear counterstain. Original magnification, ×400. The bar graphs on the bottom are showing the quantification of NF-κB p65 immunostaining in at least 200 counted cells, presented as percentage ± SEM. ** P < .01. (c) Immunoblots showing an increase of NF-κB p65 in the nucleus from Rhoa G17V-expressing CD4+ T cells. Whole-cell lysates (WCL) and nuclear lysates from CD4+ T cells of the indicated groups were immunoblotted with antibodies as indicated. Histone H3 (His H3) is nuclear, while tubulin are cytosolic. (d) Immunoblots showing Duvelisib suppressed the expression of Pon2, decreased the phosphorylated IκB kinase (IKK) complex, and activated the NF-κB p65 in Rhoa G17V-expressing CD4+ T cells. β-Actin served as a loading control. (e) Immunoblots showing BAY and Iκ-B super repressor inhibited NF-κB p65 activation and Pon2 expression in Rhoa G17V-expressing CD4+ T cells. β-Actin served as a loading control.
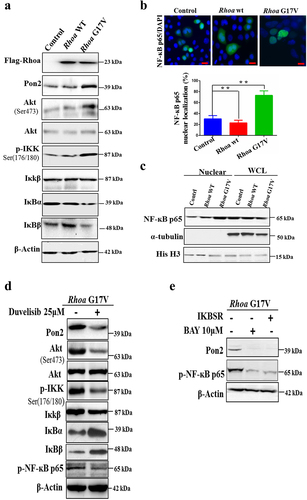
Based on our results showing Rhoa G17V mediated NF-κB activity by regulating the PI3K/Akt/IKK pathway and increased Pon2 expression, we hypothesized that Pon2 expression is dependent on NF-κB activation in Rhoa G17V-expressing CD4+ T cells. In order to test this hypothesis, we used BAY 11–7082 (BAY), a specific NF-κB pathway inhibitor or IκB super-repressor plasmid. Our data showed, as expected, that both BAY and the IκB super-repressor completely abolished the expression of phospho-NF-κB p65 and Pon2 (). Given these findings, we conclude that genetic defects in RHOA upregulated the abnormal expression of PON2 to promote TCR-driven proliferation through the activation of the PI3K/Akt/NF-κB pathway in CD4+ T cells.
Knockdown of Pon2 in CD4+ T cells suppresses Rhoa G17V-induced cell proliferation
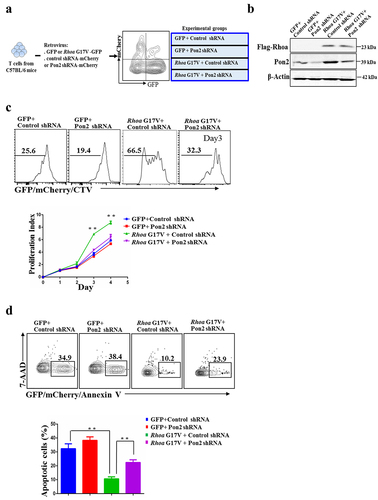
Expression of Rhoa G17V in CD4+ T cells increased cell proliferation with upregulation of Pon2. Here, we tested whether knockdown of Pon2 would functionally impact Rhoa G17V mutation in CD4+ T cells in vitro. We first transduced Rhoa G17V-expressing or control CD4+ T cells with a retrovirus encoding either Pon2-shRNA-IRES-mCherry or nonspecific shRNA control (). Retrovirus Pon2 shRNA expression vectors and the nonspecific shRNA control vector are listed in Supplemental Table 1. The inhibition of Pon2 protein expression by Pon2-shRNA in Rhoa G17V-expressing CD4+ T cells was confirmed using western blot analysis (). Since the protein level of Pon2 in the normal CD4+ T cells is quite low, knockdown of Pon2 in the normal CD4+ T cells was unable to exert an obvious effect on T cell development. In striking contrast, knockdown of Pon2 antagonized the effects of genetic lesions in Rhoa by suppressing the proliferation () and promoting apoptosis () of Rhoa G17V-expressing CD4+ T cells during 4 days of in vitro stimulation. These data demonstrate the crucial role of Pon2-mediated signaling in the Rhoa G17V-induced T cell development and pathogenesis.
Figure 3. Knockdown of Pon2 in CD4+ T cells suppresses Rhoa G17V-induced cell proliferation. (a) The experimental scheme of knockdown of Pon2 in control or Rhoa G17V-expressing CD4+ T cells. GFP was used as a marker to monitor the expression of Rhoa G17V or GFP as control; whereas mCherry signals indicated the expression of Pon2 shRNA-mCherry or mCherry as control. GFP and mCherry double positive cells were gated for further immunophenotypic analysis. (b) Knockdown of Pon2 in control or Rhoa G17V-expressing CD4+ T cells was confirmed by western blot analysis. β-Actin served as a loading control. (c) In vitro CellTrace Violet (CTV) proliferation assay of GFP and mCherry double positive CD4+ T cells of indicated groups for four days. (d) Annexin V and 7-ADD staining of GFP and mCherry double positive CD4+ T cells of the indicated groups on day three were analyzed by flow cytometric analysis. The P value in (c) and (d) was calculated by ANOVA followed by Dunnett’s test using triplicate samples from two independent experiments. Columns indicate means; bars are the standard error. **P ≤ .01.
Knockdown of Pon2 in CD4+ T cells impairs Tfh cell polarization and function
To further investigate the role of PON2 in T cell specification and the pathogenesis of RHOA G17V-induced AITL, we performed adoptive T cell transfer experiments since Tfh cell cannot be successfully induced in vitro. We first isolated naïve CD4+ T cells from OT-II mice and then transduced with Rhoa G17V-expressing or control OT-II CD4+ T cells with a retrovirus encoding either Pon2-shRNA-IRES-mCherry or nonspecific control-shRNA-IRES-mCherry in vitro to yield four groups (GFP-control shRNA, GFP-Pon2 shRNA, or Rhoa G17V-control shRNA, Rhoa G17V-Pon2 shRNA). We then retro-orbitally injected them into 4-week-old female C57BL/6 mice, which were subsequently immunized with OVA conjugated to 4-hydroxy-3-nitrophenylacetyl (NP-OVA) (). After the follow-up period of 3 months, we found that the inguinal lymph nodes from the mice injected with Rhoa G17V-expressing CD4+ T cells with Pon2 knockdown were much smaller than the Rhoa G17V-expressing CD4+ T cells group (). H&E staining further showed that the inguinal lymph nodes isolated from Pon2 knockdown Rhoa G17V-expressing CD4+ T cells have a marked decrease in the numbers of secondary lymphoid follicles compared to the control knockdown group (). We then monitored Tfh polarization by analyzing the expression of Tfh surface markers. Flow cytometric analysis revealed a most notable increase in the population of Tfh cells (CD4+CXCR5+Bcl6+) in Rhoa G17V-expressing CD4+ T cells. Expectedly, knockdown of Pon2 in CD4+ T cells suppressed Rhoa G17V-induced Tfh cell polarization ().
Figure 4. Knockdown of Pon2 in CD4+ T cells impairs Rhoa G17V-induced Tfh cell polarization and function. (a) Schematic representation of experimental design for adoptive T cell transfer experiments. Knockdown of Pon2 in control or Rhoa G17V-expressing OT-II CD4+ T cells were transferred into C57BL/6 mice via retro-orbital injection to characterize the functional consequences. (b) Representative images of the inguinal lymph nodes from the indicated groups one month after adoptive transfer of CD4+ T. (c) H&E staining of the inguinal lymph nodes isolated from the indicated groups. The bar graphs on the right are showing the number of follicles of the inguinal lymph nodes in six mice. ** P < .01. Original magnification: ×40. Scale bar: 200 μm. (d and e) Tfh cells (d) and germinal center B cells (e) from C57BL/6 mice after adoptive transfer of the indicated groups were analyzed by flow cytometry. (f) ELISA showing the detection of serum levels of cytokines from C57BL/6 mice after adoptive transfer of the indicated groups. The P values in (C, D, E, and F) were calculated by ANOVA followed by Dunnett’s test using triplicate samples of four mice of each group in two independent experiments. Columns indicate means; bars are the standard error. **P ≤ .01.
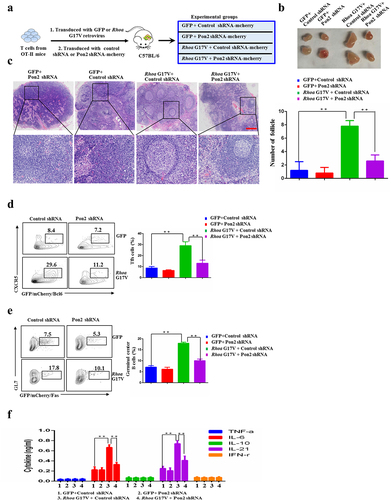
Furthermore, we observed a decrease in the population of Tfh cell-stimulated germinal center B cells (B220+GL7+Fas+) when Pon2 was knockdown in Rhoa G17V-expressing CD4+ T cells (). We also observed the decreased serum levels of Tfh cell-produced proinflammatory IL-6 and IL-21 cytokines by ELISA analysis in Rhoa G17V-expressing CD4+ T cells with Pon2 knockdown (). After 1-year period of follow-up, the amount of Rhoa G17V-expressing CD4+ T cells with Pon2 knockdown in recipient mice gradually decreased (data not shown). However, Rhoa G17V-expressing CD4+ T cells in the recipient mice continuously expanded for the long-term observation (data not shown), indicating Rhoa mutation maintained a malignant proliferation of CD4+ T cells. In summation, these data imply that Pon2 might constitute one of the critical targets subjected to modulation by Rhoa-associated pathways in CD4 + T cells to play an important role in the initial step of the development of Tfh-associated malignancies, such as AITL.
PON2 expression is associated with RHOA G17V mutation in AITLs
Given the close relationship of Pon2 with Rhoa G17V mutation in the mouse Rhoa G17V–expressing CD4+ T cells, which are the cell-of-origin of AITL,Citation13 we explored whether PON2 immunostaining was associated with the RHOA G17V mutation in patients with AITL. We successfully performed targeted amplicon sequencing of exon 2 of the RHOA gene in 46 AITLs with available tissues and confirmed the presence of a substitution mutation of Gly to Val in codon 17(p.G17V) in 24 (52.2%) AITL biopsies (Supplemental Table 2), which is similar to the rate reported by other investigators.Citation8,Citation9
In normal lymph nodes, immunohistochemical staining of PON2 was detected in a few cells in the germinal center, but the signal was quite weak (). There was some PON2 signal in the paracortex region of the lymph nodes (). Among the 54 AITLs assessed using IHC, 33 tumor samples showed strong staining for PON2, located in the cytosol of tumor cells (). Compared with AITLs without the RHOA G17V mutation (), AITLs harboring the RHOA G17V mutation showed robust expression of PON2 (). To further confirm that the PON2 positivity is really coming from the neoplastic T cells, we performed the Multiplexed fluorescence immunohistochemistry (mFIHC) to assess CD4 and PON2 stains in the AITL samples. clearly showed CD4 and PON2 colocalized in the same cells, which proved PON2 expression is in AITL cells.
Figure 5. PON2 expression is associated with RHOA G17V mutation in AITLs. (a–c) Immunohistochemical analysis of PON2 distribution in normal human lymph nodes (a) and lymphoma biopsies isolated from patients with AITLs bearing RHOA G17V mutation (b) or without RHOA G17V mutation (c). EliVision Plus two-step immunohistochemical technique with 3–3’ diaminobenzidine (DAB) staining was used. Original magnification: × 400. scale bar: 10 µm. (d) Detection of PON2 in AITL cells by Multiplexed fluorescence immunohistochemistry. Left upper panel shows conventional DAPI staining. Left middle panel shows the single PON2 staining in green. Left lower panel shows the single CD4 staining in red. Right panel shows the multiplexed fluorescence staining of PON2 and CD4. Original magnification: × 200; scale bar: 100 µm. (e) The histogram illustrates the percentage of PON2 immunostaining in AITLs bearing RHOA G17V mutation or without RHOA G17V mutation. AITLs with RHOA G17V mutation showed significantly higher PON2 expression levels than AITLs with RHOA wild-type (WT) (chi-square test, P = .002). (f) The histogram illustrates the percentage of PON2 immunostaining in AITLs bearing IDH2 R172 mutation or without IDH2 R172 mutation. There was no difference in PON2 immunostaining between AITLs with IDH2 R172 mutation or without IDH2 R172 mutation (chi-square test, P = .908).
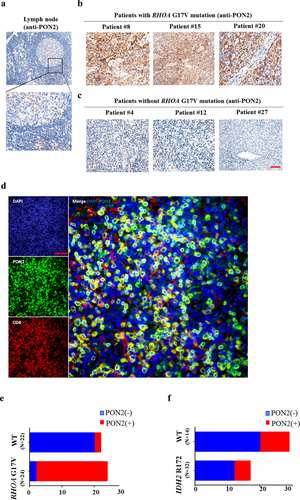
Among these 46 AITLs with available tissue for sequencing of the RHOA gene, PON2 was expressed in 63.0% (29/46) of the patients. Approximately 31.82% (7/22) of AITLs without RHOA G17V mutation showed expression of PON2 (). In striking contrast, approximately 91.67% (22/24) of AITLs harboring the RHOA G17V mutation showed expression of PON2 (). There was a positive association between the percentage of RHOA G17V mutation and PON2 expression in patients with AITL (; , P = .000). In order to confirm that this association between RHOA G17V mutation and PON2 expression was specific, we also tested the connection between PON2 expression and the mutation status of IDH2, which is also frequently mutated in AITLs and contributes to the Tfh phenotype of AITL. Among the 46 patients with AITL, 14 patients (~30.4%) had IDH2 R172 mutations (Supplemental Table 3). Nonetheless, patients with AITL with IDH2 R172 mutation showed heterogeneous PON2 staining and there was no correlation between PON2 immunostaining and the mutation status of IDH2 in 46 patients with AITL (; Supplemental Table 4, P = .908). Thus, our data imply that PON2 might be closely associated with RHOA-associated pathways but not IDH2 to play an important role in the development of AITL.
Table 1. Relationship between PON2 expression and RHOA G17V mutation in patients with AITL.
PON2 expression correlates with aggressive clinicopathological features and poor prognosis of AITL
Pathologic features (growth pattern, follicular dendritic cell proliferation, microvessel density, EBV infection, and Tfh phenotype) were assessed for each case. Interestingly, the mean microvessel density and number of Tfh markers appeared to correlate with the RHOA G17V mutation status and PON2 expression (P < .05; Supplemental Table 5). Other pathologic features, such as growth pattern, follicular dendritic cell proliferation, and EBV infection were not associated with the RHOA G17V mutation status and PON2 expression (P > .05; Supplemental Table 5). Notably, AITLs with RHOA G17V mutation or PON2 expression have classic pathological features of the Tfh phenotype (Supplemental Figure 1).
shows the relationship between the PON2 expression and selected clinicopathological parameters. PON2 expression did not vary significantly with age, sex, LDH level, splenomegaly, or systemic symptoms. However, patients with PON2 expression were prone to have an advanced tumor stage (P = .001), higher ECOG PS (P = .028), presence of BM invasion (P = .002), and higher IPI (P = .001), as well as the presence of RHOA G17V mutation (). Thus, the expression of PON2 is associated with multiple malignant characteristics of AITL.
Table 2. Relationship between PON2 expression and clinicopathologic characteristics of patients with AITL.
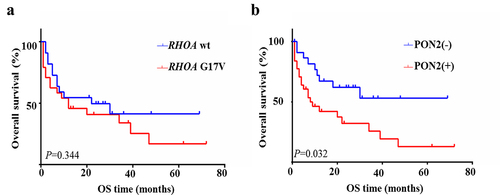
We subsequently assessed the influence of RHOA G17V mutation and PON2 expression on clinical outcomes in patients with AITL. From the results of the Kaplan–Meier statistical analysis, our data showed no difference in overall survival between patients with RHOA G17V-mutated AITL versus wildtype AITL (χ2 = 0.896, P = .344, ), which is similar to the studies reported by other investigators,Citation9,Citation27,Citation28 and one recent meta-analysis.Citation29 The survival rates of the group with or without PON2 expression both declined progressively. However, Kaplan–Meier analysis showed that patients with PON2-positive tumors experienced significantly shorter overall survival than those with PON2-negative tumors (χ2 = 5.545, P = .019, ). This data implies that PON2 might be closely associated with RHOA-associated pathways to play a cumulative effect in the development and progression of RHOA G17V-mutated AITLs.
Discussion
The combination of whole-exome sequencing, RNAseq, and deep targeted sequencing recently revealed a new highly prevalent RHOA mutation (RHOA G17V) in almost 60% of AITL cases, most commonly associated with loss-of-function mutations in the TET2 tumor suppressor gene.Citation8,Citation9 RHOA is a small GTPase involved in diverse cellular processes by cycling between an inactive GDP-bound state and an active GTP-bound state.Citation30,Citation31 It is required for thymocyte development and controls the adhesion, polarization, and migration of T cells.Citation31,Citation32 Inhibition of RHOA function in the thymus by overexpressing the bacterial toxin C3 led to aggressive thymic lymphoma of T-cell origin, suggesting that RHOA-mediated signaling is critical for the transformation of T cells.Citation33 Although some of the analyses carried out in vitro have provided data consistent with the proposed effects of RHOA G17V mutation as a dominant-negative driver of AITL-specific pathogenesis, animal models carrying this mutation showing AITL like phenotypes were attained only upon combining with homozygous null mutation in Tet2 gene.Citation13,Citation14 However, a novel mouse model for AITL expressing human RHOA G17V mutation under the control of murine Lck distal promoter which in the absence of further genetic manipulations develops multiple AITL-like phenotypic traits.Citation15 This is in clear contrast to the previous murine model of AITL.Citation12–14 Obviously, the role of RHOA G17V-mediated signaling in the tumorigenesis and progression of AITLs remains unclear.
In the present study, we have collected compelling evidence to support the notion that genetic defects in RHOA upregulated the abnormal expression of PON2 to promote TCR-driven proliferation in vitro and in vivo through activation of the PI3K/Akt/NF-κB pathway in CD4+ T cells. In CD4+ T cells, RHOA G17V mutation activates the PI3K/Akt pathway, resulting in the production of the second messenger phosphatidylinositol-3,4,5 trisphosphate, activation of the serine/threonine kinases Akt and SGK1, and the phosphorylation of different substrates actively participating in the malignant transformation of T-cells.Citation12,Citation13,Citation34 Among these substrates, IκB kinase (IKK) phosphorylation by Akt triggers the IκB degradation in the cytosol and the release of NK-ĸB that can then translocate to the nucleus with the ability to induce gene transcription.Citation34 In this study, Rhoa G17V-expressing CD4+ T cells exhibited a higher level of the phosphorylated IKK complex and lower expression of IκBα than in both WT and EV-control CD4+ T cells. Compared to control CD4+ T cells, in Rhoa G17V-expressing CD4+ T cells, we also observed higher nuclear expression and localization of NK-ĸB p65, which is the key transcription factor of PON2. In addition, the duvelisib (IPI-145), a clinically approved PI3kδ/γ inhibitor, perturbed the expression of PON2, decreased the phosphorylated IκB kinase (IKK) complex, and inactivated the NF-κB p65. Thus, RHOA G17V mediated PI3K/Akt/NF-κB activity by regulating IKK pathway and increased PON2 expression in CD4+ T cells.
We further show that inhibition of the NF-κB pathway significantly abolished the expression of phospho-NF-κB and Pon2 in Rhoa G17V-expressing CD4+ T cells. Therefore, Pon2 expression is dependent on NF-κB activation in Rhoa G17V-expressing CD4+ T cells. Given these findings, we conclude that genetic defects in RHOA upregulated the abnormal expression of PON2 to promote TCR-driven proliferation truly through activation of the NF-κB pathway in CD4+ T cells. Furthermore, loss of Pon2 has been shown to repress the cell proliferation and induced cell apoptosis in Rhoa G17V-expressing CD4+ T cells. These results suggest that PON2 activation arising from genetic defects in RHOA could lead to oncogenic function and have a cumulative effect in the initial development step of Tfh-associated malignancies, such as AITL. Despite these findings, the clinical significance of the association between RHOA G17V mutation and abnormal expression of PON2 in AITLs remains unknown.
In this study, the RHOA G17V mutation was detected in 52.2% of AITLs. Interestingly, we observed that 91.67% of patients with RHOA G17V mutation showed PON2 expression, which was correlated with the RHOA G17V mutation but no IDH2 mutation in AITLs. Despite the established and dominant role of paraoxonases in cardiovascular diseases and relevant parameters, more recent studies revealed antioxidant and antiapoptotic activities of PON2 in cancers.Citation18,Citation35 Several studies have indicated that PON2 is commonly upregulated in several tumor types, including pancreatic cancer,Citation20,Citation36 hepatocellular carcinoma (HCC),Citation37 skin neoplasm,Citation21,Citation38 prostate cancer,Citation39 and acute lymphoblastic leukemia.Citation40,Citation41 A study reported that PON2 promotes cancer growth and metastasis in pancreatic cells by increasing cellular glucose uptake by modulating the GLUT1-mediated transport.Citation20 PON2 knockdown in pancreatic cancer cells in vitro and a mouse model inhibited cell growth and induced apoptosis through the AMPK-FOXO3A-PUMA pathway.Citation20 Pan et al.Citation40 identified aberrant expression of PON2 in B-cell acute lymphoblastic leukemia (B-ALL) as a mechanism to bypass metabolic gatekeeper functions. Other authors have demonstrated that PON2 is among a small group of upregulated genes that characterize pediatric ALL patients with very poor outcome prognosis.Citation42 PON2 was also identified in an outcome-specific gene expression signature of primary imatinib-resistant chronic myeloid leukemia patients.Citation43 These findings indicate an oncogenic role for PON2 in tumorigenesis. However, whether PON2 expression and the RHOA G17V mutation have a cumulative effect on the tumorigenesis and progression of AITLs is not clearly understood.
We retrospectively investigated how PON2 abnormalities with a RHOA G17V mutation can promote the initiation and progression of AITL. IHC immunostaining of PON2 was very weak in Tfh cells in the germinal center of the lymph nodes. In contrast, we found that PON2 expression was detected in approximately 63% of AITLs. We categorized AITL samples into PON2 expression and PON2 negative expression cohorts. Patients with PON2 expression tended to have an advanced stage, high ECOG PS, presence of BM invasion, and high PIT score. Notably, this study shows that PON2 is expressed in AITLs at a high frequency, which ultimately supports the tumorigenic potential of PON2 as a potential tumor marker. Furthermore, PON2 was associated with RHOA G17V mutation and the combined effect of the two proteins and genetic biomarker might control the direction of the molecular agents-based AITL.
Tfh cells are a specialized subset of CD4+ T cells that migrate into germinal centers to help B cells maturation via stimulation of FDC proliferation. The close relationship between AITL cells and Tfh cells implies that aberrant deregulated genes in AITL are likely relevant for normal Tfh development and function. Conversely, Tfh differentiation and proliferation pathways may provide important growth and survival cues contributing to the pathogenesis of AITL. In our mouse studies, expression of Rhoa G17V in CD4 + T cells significantly increases differentiation toward the Tfh lineage, as showed by increased numbers of CD4+ CXCR5+ PD1+ cells, suggesting an instructive role for this mutation in the establishment of the Tfh features characteristic of AITL. Due to the lack of the cell lines and animal models of AITL, we fail to know how the associations of PON2 and RHOA G17V mutation exactly promote disease progression in patients with AITL. Although further analyses will be necessary to understand how PON2 could influence CD4+ T cells and AITL cell metabolism and phenotype, our results suggest that PON2 expression resulting from RHOA G17V mutation provides an additional basis for devising new diagnosis, prognosis, and therapeutics for lymphoma.
Supplemental Material
Download MS Word (3.7 MB)Acknowledgments
This work was supported by Natural Science Foundation of Guangdong Province (Grant No. 2019A1515011354). The authenticity of this article has been validated by uploading the key raw data onto the Research Data Deposit public platform (www.researchdata.org.cn), with the approval RDD number as RDDB2022568543.
Disclosure statement
No potential conflicts of interest are disclosed.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/2162402X.2022.2134536
Additional information
Funding
References
- Lemonnier F, Gaulard P, de Leval L. New insights in the pathogenesis of t-cell lymphomas. Curr Opin Oncol. 2018;30(5):277–15. doi:10.1097/CCO.0000000000000474.
- Jaffe ES, Nicolae A, Pittaluga S. Peripheral t-cell and nk-cell lymphomas in the who classification: pearls and pitfalls. Mod pathol. 2013;26(Suppl 1):S71–87. doi:10.1038/modpathol.2012.181.
- de Leval L, Gisselbrecht C, Gaulard P. Advances in the understanding and management of angioimmunoblastic t-cell lymphoma. Br J Haematol. 2010;148(5):673–689. doi:10.1111/j.1365-2141.2009.08003.x.
- Dupuis J, Boye K, Martin N, Copie-Bergman C, Plonquet A, Fabiani B, Baglin A-C, Haioun C, Delfau-Larue M-H, Gaulard P, et al. Expression of cxcl13 by neoplastic cells in angioimmunoblastic t-cell lymphoma (aitl): a new diagnostic marker providing evidence that aitl derives from follicular helper t cells. Am J Surg Pathol. 2006;30(4):490–494. doi:10.1097/00000478-200604000-00009.
- Grogg KL, Attygalle AD, Macon WR, Remstein ED, Kurtin PJ, Dogan A. Expression of cxcl13, a chemokine highly upregulated in germinal center t-helper cells, distinguishes angioimmunoblastic t-cell lymphoma from peripheral t-cell lymphoma, unspecified. Mod pathol. 2006;19(8):1101–1107. doi:10.1038/modpathol.3800625.
- de Leval L, Rickman DS, Thielen C, Reynies A, Huang YL, Delsol G, Lamant L, Leroy K, Brière J, Molina T, et al. The gene expression profile of nodal peripheral t-cell lymphoma demonstrates a molecular link between angioimmunoblastic t-cell lymphoma (aitl) and follicular helper t (tfh) cells. Blood. 2007;109(11):4952–4963. doi:10.1182/blood-2006-10-055145.
- Piccaluga PP, Agostinelli C, Califano A, Carbone A, Fantoni L, Ferrari S, Gazzola A, Gloghini A, Righi S, Rossi M, et al. Gene expression analysis of angioimmunoblastic lymphoma indicates derivation from t follicular helper cells and vascular endothelial growth factor deregulation. Cancer Res. 2007;67(22):10703–10710. doi:10.1158/0008-5472.CAN-07-1708.
- Odejide O, Weigert O, Lane AA, Toscano D, Lunning MA, Kopp N, Kim S, van Bodegom D, Bolla S, Schatz JH, et al. A targeted mutational landscape of angioimmunoblastic t-cell lymphoma. Blood. 2014;123(9):1293–1296. doi:10.1182/blood-2013-10-531509.
- Yoo HY, Sung MK, Lee SH, Kim S, Lee H, Park S, Kim SC, Lee B, Rho K, Lee J-E, et al. A recurrent inactivating mutation in rhoa GTPase in angioimmunoblastic t cell lymphoma. Nat Genet. 2014;46(4):371–375. doi:10.1038/ng.2916.
- Iqbal J, Amador C, McKeithan TW, Chan WC. Molecular and genomic landscape of peripheral t-cell lymphoma. Cancer Treat Res. 2019;176:31–68.
- Butzmann A, Sridhar K, Jangam D, Kumar J, Sahoo MK, Shahmarvand N, Warnke R, Rangasamy E, Pinsky BA, Ohgami RS, et al. A comprehensive analysis of rhoa mutation positive and negative angioimmunoblastic t-cell lymphomas by targeted deep sequencing, expression profiling and single cell digital image analysis. Int J Mol Med. 2020;46(4):1466–1476. doi:10.3892/ijmm.2020.4686.
- Zang S, Li J, Yang H, Zeng H, Han W, Zhang J, Lee M, Moczygemba M, Isgandarova S, Yang Y, et al. Mutations in 5-methylcytosine oxidase tet2 and rhoa cooperatively disrupt t cell homeostasis. J Clin Invest. 2017;127(8):2998–3012. doi:10.1172/JCI92026.
- Cortes JR, Ambesi-Impiombato A, Couronné L, Quinn SA, Kim CS, da Silva Almeida AC, West Z, Belver L, Martin MS, Scourzic L, et al. Rhoa g17v induces t follicular helper cell specification and promotes lymphomagenesis. Cancer Cell. 2018;33(259–273.e7):259–273.e7. doi:10.1016/j.ccell.2018.01.001.
- Ng SY, Brown L, Stevenson K, deSouza T, Aster JC, Louissaint A, Weinstock DM. Rhoa g17v is sufficient to induce autoimmunity and promotes t-cell lymphomagenesis in mice. Blood. 2018;132(9):935–947. doi:10.1182/blood-2017-11-818617.
- Lee GJ, Jun Y, Yoo HY, Jeon YK, Lee D, Lee S, Kim J. Angioimmunoblastic T-cell lymphoma-like lymphadenopathy in mice transgenic for human RHOA with p.Gly17Val mutation. Oncoimmunology. 2020;9(1):1746553. doi:10.1080/2162402X.2020.1746553.
- Bacchetti T, Ferretti G, Sahebkar A. The role of paraoxonase in cancer. Semin Cancer Biol. 2019;56:72–86. doi:10.1016/j.semcancer.2017.11.013.
- Furlong CE, Marsillach J, Jarvik GP, Costa LG. Paraoxonases-1, −2 and −3: what are their functions? Chem Biol Interact. 2016;259:51–62. doi:10.1016/j.cbi.2016.05.036.
- Witte I, Altenhöfer S, Wilgenbus P, Amort J, Clement AM, Pautz A, Li H, Förstermann U, Horke S. Beyond reduction of atherosclerosis: pon2 provides apoptosis resistance and stabilizes tumor cells. Cell Death Dis. 2011;2(1):e112. doi:10.1038/cddis.2010.91.
- Devarajan A, Su F, Grijalva V, Yalamanchi M, Yalamanchi A, Gao F, Trost H, Nwokedi J, Farias-Eisner G, Farias-Eisner R, et al. Paraoxonase 2 overexpression inhibits tumor development in a mouse model of ovarian cancer. Cell Death Dis. 2018;9(3):392. doi:10.1038/s41419-018-0395-2.
- Nagarajan A, Dogra SK, Sun L, Gandotra N, Ho T, Cai G, Cline G, Kumar P, Cowles RA, Wajapeyee N, et al. Paraoxonase 2 facilitates pancreatic cancer growth and metastasis by stimulating glut1-mediated glucose transport. Mol Cell. 2017;67(685–701.e6):685–701.e6. doi:10.1016/j.molcel.2017.07.014.
- Bacchetti T, Salvolini E, Pompei V, Campagna R, Molinelli E, Brisigotti V, Togni L, Lucarini G, Sartini D, Campanati A, et al. Paraoxonase-2: a potential biomarker for skin cancer aggressiveness. Eur J Clin Invest. 2021;51(5):e13452. doi:10.1111/eci.13452.
- Ross ME, Zhou X, Song G, Shurtleff SA, Girtman K, Williams WK, Liu H-C, Mahfouz R, Raimondi SC, Lenny N, et al. Classification of pediatric acute lymphoblastic leukemia by gene expression profiling. Blood. 2003;102(8):2951–2959. doi:10.1182/blood-2003-01-0338.
- Gallamini A, Stelitano C, Calvi R, Bellei M, Mattei D, Vitolo U, Morabito F, Martelli M, Brusamolino E, Iannitto E, Zaja F. Peripheral t-cell lymphoma unspecified (ptcl-u): a new prognostic model from a retrospective multicentric clinical study. Blood. 2004;103(7):2474–2479. doi:10.1182/blood-2003-09-3080.
- Chen R, Bélanger S, Frederick MA, Li B, Johnston RJ, Xiao N, Liu Y-C, Sharma S, Peters B, Rao A, et al. In vivo RNA interference screens identify regulators of antiviral cd4(+) and cd8(+) t cell differentiation. Immunity. 2014;41(2):325–338. doi:10.1016/j.immuni.2014.08.002.
- Patel K, Danilov AV, Pagel JM. Duvelisib for cll/sll and follicular non-Hodgkin lymphoma. Blood. 2019;134(19):1573–1577. doi:10.1182/blood.2019001795.
- Horwitz SM, Koch R, Porcu P, Oki Y, Moskowitz A, Perez M, Myskowski P, Officer A, Jaffe JD, Morrow SN, et al. Activity of the pi3k-δ,γ inhibitor duvelisib in a phase 1 trial and preclinical models of t-cell lymphoma. Blood. 2018;131(8):888–898. doi:10.1182/blood-2017-08-802470.
- Ondrejka SL, Grzywacz B, Bodo J, Makishima H, Polprasert C, Said JW, Przychodzen B, Maciejewski JP, Hsi ED. Angioimmunoblastic t-cell lymphomas with the rhoa p.Gly17val mutation have classic clinical and pathologic features. The American. J Surg Pathol. 2016;40(3):335–341. doi:10.1097/PAS.0000000000000555.
- Wang C, McKeithan TW, Gong Q, Zhang W, Bouska A, Rosenwald A, Gascoyne RD, Wu X, Wang J, Muhammad Z, et al. IDH2 R172 mutations define a unique subgroup of patients with angioimmunoblastic T-cell lymphoma. Blood. 2015;126(15):1741–1752. doi:10.1182/blood-2015-05-644591.
- Nguyen PN, Tran NTB, Nguyen TPX, Ngo TNM, Lai DV, Deel CD, Hassell LA, Vuong HG. Clinicopathological implications of rhoa mutations in angioimmunoblastic t-cell lymphoma: a meta-analysis: rhoa mutations in aitl. Clin Lymphoma Myeloma Leuk. 2021;21(7):431–438. doi:10.1016/j.clml.2021.03.002.
- Joo E, Olson MF, Gerges NZ. Regulation and functions of the rhoa regulatory guanine nucleotide exchange factor gef-h1. Small GTPases. 2020;11(1):1–14. doi:10.1080/21541248.2017.1361898.
- Bros M, Haas K, Moll L, Grabbe S. Rhoa as a key regulator of innate and adaptive immunity. Cells. 2019;8(7):733. doi:10.3390/cells8070733.
- Rougerie P, Delon J. Rho GTPases: masters of t lymphocyte migration and activation. Immunol Lett. 2012;142(1–2):1–13. doi:10.1016/j.imlet.2011.12.003.
- Cleverley SC, Costello PS, Henning SW, Cantrell DA. Loss of rho function in the thymus is accompanied by the development of thymic lymphoma. Oncogene. 2000;19(1):13–20. doi:10.1038/sj.onc.1203259.
- Harriague J, Bismuth G. Imaging antigen-induced pi3k activation in t cells. Nat Immunol. 2002;3(11):1090–1096. doi:10.1038/ni847.
- Manco G, Porzio E, Carusone TM. Human paraoxonase-2 (pon2): protein functions and modulation. Antioxidants. 2021;10(2):256. doi:10.3390/antiox10020256.
- Tsegaye MA, Schafer ZT. Collapsing the metabolic pon2zi scheme in pancreatic ductal adenocarcinoma. Trends Cell Biol. 2017;27(11):785–786. doi:10.1016/j.tcb.2017.09.003.
- Sukketsiri W, Porntadavity S, Phivthong-ngam L, Lawanprasert S. Lead inhibits paraoxonase 2 but not paraoxonase 1 activity in human hepatoma hepg2 cells. J Appl Toxicol. 2013;33(7):631–637. doi:10.1002/jat.1789.
- Campagna R, Bacchetti T, Salvolini E, Pozzi V, Molinelli E, Brisigotti V, Sartini D, Campanati A, Ferretti G, Offidani A, et al. Paraoxonase-2 silencing enhances sensitivity of a375 melanoma cells to treatment with cisplatin. Antioxidants. 2020;9(12):1238. doi:10.3390/antiox9121238.
- Ribarska T, Ingenwerth M, Goering W, Engers R, Schulz WA. Epigenetic inactivation of the placentally imprinted tumor suppressor gene tfpi2 in prostate carcinoma. Cancer Genomics Proteomics. 2010;7:51–60.
- Pan L, Hong C, Chan LN, Xiao G, Malvi P, Robinson ME, Geng H, Reddy ST, Lee J, Khairnar V, Cosgun KN. Pon2 subverts metabolic gatekeeper functions in b cells to promote leukemogenesis. Proc Natl Acad Sci U S A. 2021;118(7):e2016553118.
- Hui PY, Chen YH, Qin J, Jiang XH. Pon2 blockade overcomes dexamethasone resistance in acute lymphoblastic leukemia. Hematology. 2022;27(1):32–42. doi:10.1080/16078454.2021.2009643.
- Kang H, Chen IM, Wilson CS, Bedrick EJ, Harvey RC, Atlas SR, Devidas M, Mullighan CG, Wang X, Murphy M, et al. Gene expression classifiers for relapse-free survival and minimal residual disease improve risk classification and outcome prediction in pediatric b-precursor acute lymphoblastic leukemia. Blood. 2010;115(7):1394–1405. doi:10.1182/blood-2009-05-218560.
- Frank O, Brors B, Fabarius A, Li L, Haak M, Merk S, Schwindel U, Zheng C, Müller MC, Gretz N, et al. Gene expression signature of primary imatinib-resistant chronic myeloid leukemia patients. Leukemia. 2006;20(8):1400–1407. doi:10.1038/sj.leu.2404270.