ABSTRACT
Because DNA polymerase cannot replicate telomeric DNA at linear chromosomal ends, eukaryotes have developed specific telomere maintenance mechanisms (TMMs). A major TMM involves specialized reverse transcriptase, telomerase. However, there also exist various telomerase-independent TMMs (TI-TMMs), which can arise both in pathological conditions (such as cancers) and during evolution. The TI-TMM in cancer cells is called alternative lengthening of telomeres (ALT), whose mechanism is not fully understood. We generated stably maintained telomerase-independent survivors from C. elegans telomerase mutants and found that, unlike previously described survivors in worms, these survivors “mobilize” specific internal sequence blocks for telomere lengthening, which we named TALTs (templates for ALT). The cis-duplication of internal genomic TALTs produces “reservoirs” of TALTs, whose trans-duplication occurs at all chromosome ends in the ALT survivors. Our discovery that different TALTs are utilized in different wild isolates provides insight into the molecular events leading to telomere evolution.
Alternative means of telomere lengthening in cancers and in nature
Without exception, eukaryotic nuclear genomes are contained in linear chromosomes.Citation1 The ends of linear chromosomes are called “telomeres,” after the Greek “telo,” meaning “end,” and “mere,” meaning “part.” Telomeres could not be replicated by conventional DNA polymerase.Citation2 Unless this problem were solved, chromosomes would gradually become shorter. Thus, some other type of DNA replication is clearly responsible for maintaining chromosome integrity. The “end-replication problem” is solved by various mechanisms such as recombination, retro-transposition and reverse-transcription to add sequences to the ends of chromosomes.Citation3 Many eukaryotic cells, including human cells, use a specialized reverse transcriptase called telomerase.Citation4
Not all cells consistently avoid the end-replication problem. For example, in humans, telomerase is expressed only in germ cells and stem cells.Citation5 The telomeres of somatic cells gradually shorten. If telomeres shrink below a certain threshold, cells enter replicative senescence and stop dividing.Citation6 An exception to this situation occurs in cancer cells, which can divide indefinitely without losing telomeres. To achieve unlimited proliferation, cancer cells must overcome the replicative senescence resulting from the lack of telomerase activity. Thus, most cancer cells re-express telomerase.Citation7 However, approximately 15% of cancer cells do not show telomerase activity, and this mechanism of telomere maintenance is called alternative lengthening of telomeres (ALT).Citation8 Some types of cancer show a predominance of ALT.Citation9 ALT can be activated under telomerase-deficient conditions, and may therefore serve as a back-up mechanism in telomerase-positive cells.
There are interesting cases in nature in which telomerase-independent telomere maintenance mechanisms (TI-TMMs) act (summarized in ). During the evolution of animals and plants, telomerase has been independently lost several times.Citation10 The loss of telomerase must have first caused a crisis at the chromosomal end sequences. Many pieces of evolutionary evidence show that telomeres were replaced with a tandem array of DNA elements instead of simple telomeric repeats. The most well-studied case involves the order Diptera, which lost telomerase more than 5 times as early as 260 million years ago.Citation11 Drosophila is a representative case.Citation12 Drosophila uses the end-specific retrotransposons HeT-A, TART and TAHRE (collectively called HTT) for telomere maintenance. In plants, some Solanaceae subgroups do not have telomerase activity. The Cestrum subgroup of Solanaceae has satellite repeat sequences at the telomeres.Citation13 Alliaceae species also have unusual telomere sequences. The telomeres of Allium cepa contain satellite sequences and rDNA repeats.Citation14
Figure 1. Telomerase-independent telomere maintenance mechanisms. After telomerase loss, alternative mechanisms maintain telomere length. In cancer cells, variant telomere repeats are interspersed within telomeres after recombination-dependent lengthening. Retrotransposon is adopted in many organisms. An example is Drosophila. In Allium cepa, telomeres are protected by other genomic sequences such as minisatellite and rDNA.
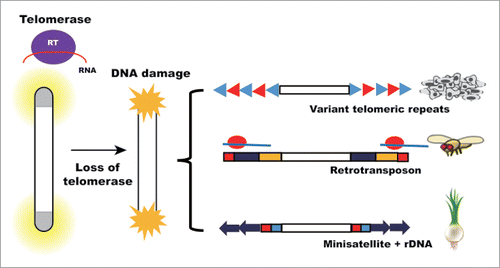
ALT in cancer cells and TI-TMM in natural cases share the fact that they all survive by telomerase-independent telomere lengthening mechanisms, and the “gold standard” of TI-TMM including ALT is the lack of telomerase activity. Although TI-TMM can be achieved by diverse means, it is also possible that ALT in cancer and TI-TMM in nature share some mechanistic details.
Internal genomic region used by worm telomerase-independent survivors
As described above, in cases of defective telomerase activity or gene loss, sequences other than the canonical telomeric repeats have frequently been used as alternatives in telomere maintenance. Because ALT studies have primarily focused on cancer cases in mammals, ALT is typically viewed as a problem for human health rather than a natural adaptive mechanism for cell survival. Most cellular studies have not considered the organismal perspective. Our research Seo et al. has focused on exploring the possible mechanisms of TI-TMM that provides a selective advantage at the organismal level after telomerase loss.
Telomerase-independent survivors were isolated from 2 natural isolates (N2, CB4856) of C. elegans treated with the alkylating agent EMS.Citation15 While the brood size of telomerase deficient mutant strain gradually decreased and became sterile after limited number of generations (usually 15–20), the survivors could be maintained by transferring 10 first larval stage (L1) worms at each generation. Unexpectedly, the telomere sequences of these survivors were cut by a restriction enzyme that does not cut the canonical telomere sequence. In addition, when hybridized with telomeric repeat DNA probe, the restricted telomere showed band patterns distinct from the canonical telomere smear pattern. Therefore, we predicted that specific non-telomeric units might be inserted into the telomeres in ALT survivors.
To identify these inserted sequences, we performed whole-genome sequencing analysis. From the paired-end sequencing data, we collected those reads that contained telomeric repeats in either of the paired-end reads and aligned the reads to the reference N2 genome. Interestingly, these reads aligned to an internal genomic region with multiple copies in both survivors. In N2 survivors, these reads were located in the telomere-adjacent region on the left arm of chromosome I. In CB4856 survivors, these reads were located in a subtelomeric internal region of chromosome V. We named these amplified sequences TALTs (Templates for ALT). We experimentally confirmed that these special sequences co-localize with telomere signals.
The CB4856-type TALT was named TALT1, and the N2-type TALT was named TALT2. We found that amplified TALT1 sequences, but not the background read sequences, contained 2 SNPs. However, we were unable to find any SNP differences between the 2 strains by Sanger sequencing of the internal regions of chromosome V. Interestingly, the experiments showed that another sequence containing these SNPs was already located at a telomere-adjacent region of the right arm of chromosome V in CB4856, but not in N2. The reason for the misalignment was a structural variation of CB4856 in this telomere-adjacent region, which was absent in the reference genome. Collectively, our results show that TALT sequences located in telomere-adjacent regions produced by cis-duplicated TALT can be mobilized for telomere lengthening after telomerase loss in C. elegans (“trans-duplication” of TALT).
Unique structure of TALT sequences
The two TALTs that we identified in wild isolates share a particular feature: a unique sequence flanked by telomeric repeats. The TALT1-specific sequence includes the promoter and one exon of the T26H2.5 ORF, and TALT2 includes an intergenic region. TALTs are probably duplicated as units to chromosome ends of the survivors in a repetitive head-to-tail pattern.
Although the expression of TALT-unique sequences is slightly increased, we do not expect that any functional protein would be translated from TALTs. However, we cannot exclude the possibility that TALT transcripts can be used as templates for reverse transcription, as in Drosophila. Another possibility is that a TALT sequence could be used as a docking site for specific proteins such as heterochromatin factors or DNA binding-proteins, as in fission yeast.Citation16
Flanking sequences, the other components of TALTs, include canonical and variant telomeric repeats. In ALT cells, the canonical telomere sequence is replaced by a variant telomere sequences in an interspersed pattern.Citation17 The insertion of variant telomeres is thought to alter telomere-binding proteins and recruit nuclear orphan receptors (COUP-TF2 and TR4) and the nucleosome remodelling deacetylase (NuRD complex).Citation18 We anticipate that variant repeats of TALTs may also recruit other binding proteins, as in human ALT cells.
The DNA damage response (DDR) should be suppressed within telomeres to avoid chromosomal fusion and to maintain integrity. In normal mammalian telomeres, shelterin complexes perform this protective role. However, in ALT tumor cells, the density of shelterin is decreased and variant repeat-binding proteins cannot fully suppress DDR.Citation19 These altered telomere proteins reportedly suppress non-homologous end joining, but not homologous recombination, thereby allowing ALT. The imperfectly protected telomere state described above is called the “intermediate” state of telomeres. It is conceivable that TALTs in worms may recruit binding proteins that differ from canonical telomere-binding proteins, as in ALT tumor cells. Two pieces of evidence show that TALTs may induce a condition similar to the “intermediate” state of telomeres. First, the mRNA expression of DDR genes is specifically up-regulated in TALT survivors. Second, TALTs integrated into telomeres can be self-sustained without additional stimuli, even when canonical telomere sequences are provided by mating ALT survivors with telomerase-positive worms. Therefore, TALTs at telomeres are likely to be in an intermediate state that could induce transposition or recombination ().
Figure 2. Possible mechanisms of TALT duplication in telomere maintenance. Each TALT has its own unique sequence flanked by telomere repeats. While the protein products encoded by unique sequences do not seem to be critical players in regulating telomere maintenance, specific DNA binding proteins may have functional roles such as inducing heterochromatin, upregulating transposition activity and enhancing recombination events.
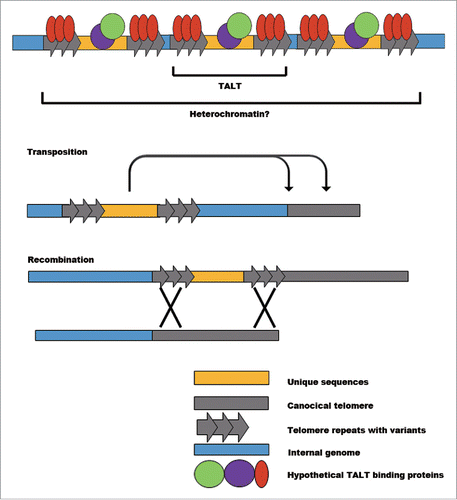
These findings raise the following question: what causes TALT insertion into telomeres? We suspected that EMS would induce TALT insertion because TALT duplication emerged only in the EMS-treated condition. EMS is commonly used as an inducer of point mutations in genetic studies of C. elegans. However, all the EMS-induced DNA changes of the original survivors disappeared after a few rounds of genetic outcrosses, strongly indicating that there is no specific point mutation that is responsible for TALT insertion into telomeres in the survivors. Then, we hypothesized that the DNA damage caused by EMS might induce ALT. Interestingly, we were able to induce TALT insertion by using gamma irradiation, which is a double strand break (DSB) inducer (C. Kim, S. Sung, B. Seo, and J. Lee, unpublished data). Telomeres have been reported to be more sensitive to DNA damage because of their G-rich nature.Citation20 Therefore, the accumulation of DNA damage in the telomere region might make telomeres prone to TALT insertion by an as-of-yet unknown mechanism. One possibility is that damaged telomeres have more recombinogenic features that allow them to easily invade nearby telomere-like sequences such as TALTs that are located in telomere-adjacent regions.
In previous studies, 2 different approaches have been used to identify ALT survivors in C. elegans.Citation21,22 One method is based on maintaining telomerase mutants by large-scale transfer of animals at each generation, in which very rare natural survivors can arise. The exact mechanism of escaping sterility in these natural survivors has not been elucidated. The other is a candidate approach, in which mutations of telomere-related genes were tested for extending the transgenerational lifespan of telomerase deficient animals. It was reported that pot-1 or pot-2 single mutants alone showed long and heterogeneous telomere length, which had been regarded as an ALT-related feature, and importantly, trt-1; pot-1 or trt-1; pot-2 double mutants showed a longer trans-generational lifespan than trt-1 single mutant animals. There were a large amount of C-circles, which are extrachromosomal, single-stranded C-rich telomeric circular DNA molecules that might be a template of ALT recombination, thus indicating a recombination-dependent ALT mechanism.Citation23 However, the stable maintenance of survivors has not always been successful by small-scale transfer. Uniquely, our TALT survivors can be stably maintained by 10 L1 worms transfer. In addition, TALT survivors use complex tandem repeats to cope with telomere loss. Given that many organisms use non-telomeric tandem repeats to preserve telomeres, TALT insertion is reminiscent of an evolutionarily conserved mechanism that has repeatedly been used to overcome telomerase loss.
cis-duplication of TALTs as a snapshot of the evolution of subtelomeres
The telomere-adjacent regions of N2 and CB4856 have been independently duplicated with their own TALT during evolution. In other organisms, telomere-adjacent regions are called subtelomeres. Subtelomeres are highly polymorphic regions because they have high rates of mutation and recombination.Citation24 Although the definition of subtelomeres is not intuitively clear, the subtelomeres of many organisms have certain features in common. Subtelomeres are composed of various repeat elements, including variant telomeric repeats, but the extent of repeat duplication and divergence varies greatly within and between species.Citation25 Because of their highly variable and repetitive features, the complete assembly of the subtelomeric regions has not been achieved thoroughly. Therefore, the structure and function of subtelomeres have remained mainly unanswered.
Subtelomeric regions can be used as templates for telomere lengthening under telomerase-deficient conditions. For example, the Y' element of budding yeast, located in the subtelomere, can be utilized by a recombination-mediated pathway.Citation26 The fission yeast Schizosaccharomyces pombe uses the subtelomere as a template for TI-TMM activation.Citation27 Additionally, in human ALT cells, telomere variant repeats are enriched in subtelomeres.Citation28 TALT amplification clearly shows that subtelomeric regions can be amplified for TI-TMM activation, perhaps representing first description of this phenomenon in a multicellular organism. We speculate that TALTs may be found in other organisms, including humans, if read lengths and the cost of whole-genome sequencing are improved so as to obtain a completely assembled map of subtelomeres.
We have found that internal genomic regions, TALTs, can be duplicated to the telomere and stabilize the genome after telomerase loss in multicellular organisms. TALT duplication might represent the re-activation of an ancient mechanism that existed before telomerase evolved. Before TALT activation, chromosomes could be fused by telomere defects. The altered karyotype can still be stably maintained after TALT activation. Thus, our survivors might suggest one partial case of chromosome evolution.
TALT-like DNA structures also exist in the genome of SV40, which is used for cancer transformation. These sequences can also be duplicated in the telomere during tumor transformation.Citation29 Thus, TALTs may represent an evolutionarily conserved structure for telomere maintenance. However, we do not know which parts are important for TALT movement. It would be interesting to identify the necessary parts of TALT for movement, such as the unique sequence of TALT or the telomere-like repeats, via the clustered regularly interspaced short palindromic repeat (CRISPR) system.
The trans-duplication of TALTs can overcome telomerase defect. Currently, the mechanism of trans-duplication remains unknown. We hypothesize that TALTs may have other binding proteins in addition to telomere-binding proteins. These proteins may initiate some type of signal that recruits the duplication machinery. Proteomic approaches will enable the identification of TALT-binding proteins. In addition, to identify the duplication machinery, we conducted candidate RNAi experiments with recombination and DNA-damage response factors. However, we did not identify the TALT duplication machinery in that screen, thus suggesting that unknown machinery may regulate trans-duplication.
The cis-duplication of TALTs can occur without telomerase loss. Interestingly, in the phylogenetic tree of wild C. elegans isolates, the cis-duplication of TALT2 has independently occurred many times. In nature, certain environmental conditions may apply selective pressure for TALT cis-duplication. Therefore, a cis-duplicated TALT is a “scar” of natural selection and a reservoir for trans-duplication. In addition, we have found multiple copies of TALT1 in the telomere-adjacent region in the right arm of chromosome V in CB4856. This duplication event is similar to the subtelomere formation process of other organisms. Consistently, in humans, duplicated units in the subtelomere may be used for ALT cancer formation. Therefore, understanding mechanisms underlying TALTs will provide valuable information for ALT cancer therapy.
Abbreviations
| TMM | = | telomere maintenance mechanism |
| ALT | = | alternative lengthening of telomeres |
| TALT | = | templates for ALT |
| TI-TMM | = | telomerase-independent telomere maintenance mechanism |
| EMS | = | ethyl methanesulfonate |
| DDR | = | DNA damage response |
| DSB | = | double strand break |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. Beomseok Seo and Eunkyeong Kim (Seoul National University) for discussion on the manuscript.
Funding
This research was supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number : HI14C1277)
References
- Blackburn EH. Structure and function of telomeres. Nature 1991; 350:569-73; PMID:1708110; http://dx.doi.org/10.1038/350569a0
- Olovnikov AM. A theory of marginotomy: the incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J Theor Biol 1973; 41:181-90; PMID:4754905; http://dx.doi.org/10.1016/0022-5193(73)90198-7
- Fajkus J, Sýkorová E, Leitch AR. Telomeres in evolution and evolution of telomeres. Chromosome Res 2005; 13:469-79; PMID:16132812; http://dx.doi.org/10.1007/s10577-005-0997-2
- Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 1985; 43:405-13; PMID:3907856; http://dx.doi.org/10.1016/0092-8674(85)90170-9
- Shay JW, Wright WE. Telomeres and telomerase in normal and cancer stem cells. FEBS Lett 2010; 584:3819-25; PMID:20493857; http://dx.doi.org/10.1016/j.febslet.2010.05.026
- Campisi J. Cellular senescence as a tumor-suppressor mechanism. Trends Cell Biol 2001; 11:S27-S31; PMID:11684439; http://dx.doi.org/10.1016/S0962-8924(01)82148-6
- Shay J, Bacchetti S. A survey of telomerase activity in human cancer. Eur J Cancer 1997; 33:787-91; PMID:9282118; http://dx.doi.org/10.1016/S0959-8049(97)00062-2
- Cesare AJ, Reddel RR. Alternative lengthening of telomeres: models, mechanisms and implications. Nat Rev Genet 2010; 11:319-30; PMID:20351727; http://dx.doi.org/10.1038/nrg2763
- Heaphy CM, Subhawong AP, Hong S-M, Goggins MG, Montgomery EA, Gabrielson E, Netto GJ, Epstein JI, Lotan TL, Westra WH. Prevalence of the alternative lengthening of telomeres telomere maintenance mechanism in human cancer subtypes. Am J Pathol 2011; 179:1608-15; PMID:21888887; http://dx.doi.org/10.1016/j.ajpath.2011.06.018
- Mason JM, Reddy HM, Frydrychova RC. Telomere maintenance in organisms without telomerase. INTECH Open Access Publisher, 2011 :323-346. http://dx.doi.org/10.5772/19348; http://www.intechopen.com/books/dna-replication-current-advances/telomere-maintenance-in-organisms-without-telomerase
- Mason JM, Randall TA, Frydrychova RC. Telomerase lost? Chromosoma July 11, 2015:1-9; PMID:26162505; http://dx.doi.org/10.1007/s00412-015-0528-7
- Garavís M, González C, Villasante A. On the Origin of the Eukaryotic Chromosome: The Role of Noncanonical DNA Structures in Telomere Evolution. Genome Biol Evolution 2013; 5:1142-50; http://dx.doi.org/10.1093/gbe/evt079
- Sykorova E, Lim KY, Chase MW, Knapp S, Leitch IJ, Leitch AR, Fajkus J. The absence of Arabidopsis‐type telomeres in Cestrum and closely related genera Vestia and Sessea (Solanaceae): first evidence from eudicots. Plant J 2003; 34:283-91; PMID:12713535; http://dx.doi.org/10.1046/j.1365-313X.2003.01731.x
- Pich U, Schubert I. Terminal heterochromatin and alternative telometric sequences in Allium cepa. Chromosome Res 1998; 6:315-22; PMID:9688522; http://dx.doi.org/10.1023/A:1009227009121
- Seo B, Kim C, Hills M, Sung S, Kim H, Kim E, Lim DS, Oh H-S, Choi RMJ, Chun J, et al. Telomere maintenance through recruitment of internal genomic regions. Nat Commun 2015; 6:8189; PMID:26382656; http://dx.doi.org/10.1038/ncomms9189
- Schoeftner S, Blasco MA. A “higher order” of telomere regulation: telomere heterochromatin and telomeric RNAs. EMBO J 2009; 28:2323-36; PMID:19629032; http://dx.doi.org/10.1038/emboj.2009.197
- Conomos D, Stutz MD, Hills M, Neumann AA, Bryan TM, Reddel RR, Pickett HA. Variant repeats are interspersed throughout the telomeres and recruit nuclear receptors in ALT cells. J Cell Biol 2012; 199:893-906; PMID:23229897; http://dx.doi.org/10.1083/jcb.201207189
- O'Sullivan RJ, Almouzni G. Assembly of telomeric chromatin to create ALTernative endings. Trends Cell Biol 2014; 24(11):675-85; PMID:25172551
- Conomos D, Pickett HA, Reddel RR. Alternative lengthening of telomeres: remodeling the telomere architecture. Front Oncol 2013; 3:27; PMID:23429284; http://dx.doi.org/10.3389/fonc.2013.00027
- Hewitt G, Jurk D, Marques FD, Correia-Melo C, Hardy T, Gackowska A, Anderson R, Taschuk M, Mann J, Passos JF. Telomeres are favoured targets of a persistent DNA damage response in ageing and stress-induced senescence. Nat Commun 2012; 3:708; PMID:22426229; http://dx.doi.org/10.1038/ncomms1708
- Cheng C, Shtessel L, Brady MM, Ahmed S. Caenorhabditis elegans POT-2 telomere protein represses a mode of alternative lengthening of telomeres with normal telomere lengths. Proc Natl Acad Sci 2012; 109:7805-10; http://dx.doi.org/10.1073/pnas.1119191109
- Lackner DH, Raices M, Maruyama H, Haggblom C, Karlseder J. Organismal propagation in the absence of a functional telomerase pathway in Caenorhabditis elegans. EMBO J 2012; 31:2024-33; PMID:22425786; http://dx.doi.org/10.1038/emboj.2012.61
- Henson JD, Cao Y, Huschtscha LI, Chang AC, Au AY, Pickett HA, Reddel RR. DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nat Biotechnol 2009; 27:1181-5; PMID:19935656; http://dx.doi.org/10.1038/nbt.1587
- Linardopoulou EV, Williams EM, Fan Y, Friedman C, Young JM, Trask BJ. Human subtelomeres are hot spots of interchromosomal recombination and segmental duplication. Nature 2005; 437:94-100; PMID:16136133; http://dx.doi.org/10.1038/nature04029
- Mefford HC, Trask BJ. The complex structure and dynamic evolution of human subtelomeres. Nat Rev Genet 2002; 3:91-102; PMID:11836503; http://dx.doi.org/10.1038/nrg727
- Lundblad V, Blackburn EH. An alternative pathway for yeast telomere maintenance rescues est1− senescence. Cell 1993; 73:347-60; PMID:8477448; http://dx.doi.org/10.1016/0092-8674(93)90234-H
- Nakamura TM, Cooper JP, Cech TR. Two modes of survival of fission yeast without telomerase. Science 1998; 282:493-6; PMID:9774280; http://dx.doi.org/10.1126/science.282.5388.493
- Varley H, Pickett HA, Foxon JL, Reddel RR, Royle NJ. Molecular characterization of inter-telomere and intra-telomere mutations in human ALT cells. Nat Genet 2002; 30:301-5; PMID:11919561; http://dx.doi.org/10.1038/ng834
- Marciniak RA, Cavazos D, Montellano R, Chen Q, Guarente L, Johnson FB. A novel telomere structure in a human alternative lengthening of telomeres cell line. Cancer Res 2005; 65:2730-7; PMID:15805272; http://dx.doi.org/10.1158/0008-5472.CAN-04-2888
