ABSTRACT
Previously, we identified a group of nuclear hormone receptors (NHRs) that promote longevity in the nematode Caenorhabditis elegans following germline-stem cell (GSC) loss. This group included NHR-49, the worm protein that performs functions similar to vertebrate PPARα, a key regulator of lipid metabolism. We showed that NHR-49/PPARα enhances mitochondrial β-oxidation and fatty acid desaturation upon germline removal, and through the coordinated enhancement of these processes allows the animal to retain lipid homeostasis and undergo lifespan extension. NHR-49/PPARα expression is elevated in GSC-ablated animals, in part, by DAF-16/FOXO3A and TCER-1/TCERG1, two other conserved, pro-longevity transcriptional regulators that are essential for germline-less longevity. In exploring the roles of the other pro-longevity NHRs, we discovered that one of them, NHR-71/HNF4, physically interacted with NHR-49/PPARα. NHR-71/HNF4 did not have a broad impact on the expression of β-oxidation and desaturation targets of NHR-49/PPARα. But, both NHR-49/PPARα and NHR-71/HNF4 were essential for the increased expression of DAF-16/FOXO3A- and TCER-1/TCERG1-downstream target genes. In addition, nhr-49 inactivation caused a striking membrane localization of KRI-1, the only known common upstream regulator of DAF-16/FOXO3A and TCER-1/TCERG1, suggesting that it may operate in a positive feedback loop to potentiate the activity of this pathway. These data underscore how selective interactions between NHRs that function as nodes in metabolic networks, confer functional specificity in response to different physiological stimuli.
Introduction
Different stages of an animal's life, such as development, sexual maturity, peak fertility, and reproductive senescence require distinct metabolic programs. During these transitions, physiological homeostasis needs to be maintained, so a metabolic network must be both robust and flexible. This is brought about by the precise orchestration of numerous gene-expression networks. Understanding how these networks operate across multiple cells, tissues and organ systems to alter feeding, energy expenditure, lipid storage and length of life, depending on reproductive cues, is a significant undertaking. This is an important problem because the ability to respond to the persistent adaptive challenges, such as those created by changing procreational demands, is critical for an organism's survival. Indeed, multiple aging-associated debilitations and diseases in humans are attributed to loss of metabolic homeostasis with age.Citation1
Recently, research in the nematode Caenorhabditis elegans has revealed novel insights into the relationship between metabolic status, reproduction and aging.Citation2-4 In worms, as in many other organisms, mating has damaging consequences for the female, including reducing its size, stress-resistance and lifespan.Citation5,6 Alternatively, removing the germline increases worm lifespan significantly,Citation7 in accordance with the observation that reduced fertility is often associated with increased lifespan in many species.Citation8-11 However, recent studies into the molecular basis of these phenomena have shown that the relationship between reproduction and aging is far more nuanced than previously appreciated reviewed in reference Citation12. For instance, while it is true that in some animals mating is detrimental to the health of the female, it is also well documented that in many others, males transfer ‘nuptial gifts’ along with the sperm that provide direct benefits to the recipient female. These ‘nuptial gifts’ vary from energy supplies to glandular products that increase longevity.Citation13 Similarly, simply making C. elegans sterile (by removing the differentiated gametes alone, for instance) does not make them live longer. It is only upon elimination of germline-stem cells (GSCs), a population of totipotent proliferative cells that give rise to the entire adult germline, that lifespan is extended.Citation14 Little is understood about what causes some reproductive signals to impede health and others to improve it. However, it is evident that reproductive transitions require metabolic adaptability, and this in turn may determine whether procreative events produce beneficial or detrimental consequences to the animal.
Removal of GSCs in C. elegans is accompanied by the activation of a group of transcription factors that collectively mediate gene-expression changes resulting in the enhancement of protective cellular processes such as proteasomal function, autophagy and stress resistance (reviewed inCitation2). These conserved proteins are essential for the longevity of GSC-less animals and include DAF-16/FOXO3A, TCER-1/TCERG1, PHA-4/FOXA, HLH-30/TFEB, HSF-1/HSF1, SKN-1/NRF2.Citation7,15-19 Additionally, three members of the nuclear hormone receptor (NHR) family of transcription regulators, DAF-12/FXR, NHR-80/HNF4 and NHR-49/PPARα are important for GSC-less longevity.Citation7,20-22 Multiple lines of evidence have indicated that these factors link lipid metabolism to reproductive stimuli and aging. DAF-16/FOXO3A, TCER-1/TCERG1, SKN-1/NRF2 and DAF-12/FXR targets include lipid-metabolic genes,Citation15,19,20,23-25 PHA-4/FOXA- and HLH-30/TFEB-mediated enhancement of autophagy contributes to increased lipase activityCitation17,18 and NHR-80/HNF4 promotes fatty-acid desaturation in long-lived, GSC-less adults.Citation21 However, the gene-regulatory network (GRN) in which these factors operate is poorly defined and it remains unclear how their activities allow an animal to not only survive the physiological disruption created by loss of GSCs but to convert it into a beneficial enhancement of health and lifespan.
Discovery of the role of NHR-49/PPARα in lipid homeostasis and longevity of germline-less animals
NHRs are of particular relevance in the context of lipid metabolism and aging since many of them are lipid-sensing factors whose ligands are fatty-acid moieties and steroid signals, and because they have pivotal roles in metabolic homeostasis.Citation26 In C. elegans, the NHR family has expanded to include 284 members, many of which are derived from a single ancestor, the hepatocyte nuclear factor 4 (HNF4).Citation27,28 In a recent study, we identified a group of NHRs that are essential for the longevity of germline-ablated worms. Of these, NHR-49 produced the most striking lifespan phenotypes.Citation22 NHR-49 is sequentially similar to HNF4α but performs functions undertaken by peroxisome proliferator activated receptor α (PPARα) in vertebrates.Citation29,30 The degree of equivalence between NHR-49 and PPARα is currently unclear, but in view of their similar activities, and for clarity and convenience, NHR-49 is referred to as the functional homolog of PPARα, here and in other studies.Citation27,31-33 PPARα is a key regulator of fatty-acid β-oxidation and a member of the PPAR family that also includes other modulators of energy metabolism.Citation34 nhr-49 was first identified as a regulator of mitochondrial β-oxidation genes during development and for inducing the expression of some β-oxidation while repressing others during short-term fasting.Citation29,30 In subsequent studies, it was found to be essential for adult reproductive diapause (ARD), an adaptive mechanism by which sexually mature adults delay reproduction and retain a small group of GSCs that are used to repopulate the gonad and continue reproduction when feeding is resumed.Citation35 Our study disclosed that NHR-49/PPARα was critical for any lifespan increment following GSC removal.Citation22 Based on its previously-described functions, it is plausible that NHR-49/PPARα facilitates endurance under diverse conditions that pose physiological challenges to the animal such as nutritional limitation or loss of reproductive capacity.
Using quantitative PCRs (Q-PCRs) and a transgenic strain expressing GFP-tagged NHR-49/PPARα protein under control of its endogenous promoter, we showed that NHR-49/PPARα was transcriptionally upregulated upon germline loss, in part through the activities of DAF-16/FOXO3A and TCER-1/TCERG1. Strikingly, in an independent genomic study, we identified nhr-49, as well as multiple β-oxidation and desaturation genes, as being upregulated by DAF-16/FOXO3A- and/or TCER-1/TCERG1 in GSC-less adults (Amrit et al.Citation36). Based on this evidence, and the previously-documented regulation of these processes by NHR-49/PPARα, we asked if NHR-49/PPARα enhanced mitochondrial β-oxidation and/or fatty-acid desaturation in germline-less animals. Indeed, the expression levels of 12 genes predicted to function in mitochondrial β-oxidation that we tested were all elevated in long-lived, germline-less adults. Of these, the upregulation of seven genes was NHR-49/PPARα dependent. These genes encode enzymes that together represent all the individual steps of mitochondrial β-oxidation necessary for the breakdown of fatty acids into acetyl CoA. RNAi-mediated knockdown of many of these genes shortened the longevity of germline-ablated animals. Our observations indicated that NHR-49/PPARα enhances the breakdown of fatty acids through mitochondrial β-oxidation following germline loss, and this augmentation is important for the consequent adaptation and longevity.Citation22
nhr-49 mutants exhibit altered expression of enzymes that mediate fatty-acid desaturation, during development as well as fasting.Citation29,30 Fatty-acid desaturation is also regulated by NHR-80 that is structurally similar to HNF4Citation37 (we refer to NHR-80 as NHR-80/HNF4 in keeping with the terminology used in worm literatureCitation21,27). In a previous study, Hugo Aguilaniu's laboratory reported that, in germline-less mutants too, NHR-80/HNF4 elevates the expression of enzymes responsible for converting a saturated fatty acid (SFA), stearic acid (SA, C16:0) into a mono-unsaturated fatty acid (MUFA), oleic acid (OA, C18:1n9).Citation21 We found that NHR-49/PPARα similarly controlled the expression of these desaturases in germline-less adults. Through lipidomic analyses, we further discovered that GSC-depleted adults showed an increase in unsaturated fatty acid (UFA) levels and a concomitant decrease in SFAs.Citation22,36 The conversion of SA to OA, as well as that of many other SFAs to MUFAs, was prevented and the overall levels of MUFAs and PUFAs were demonstrably reduced in the absence of NHR-49/PPARα.Citation22 Thus, it appears that NHR-49/PPARα mediates the simultaneous enhancement of mitochondrial β-oxidation as well as widespread desaturation of fatty acids following GSC removal. How it modulates disparate biochemical pathways and what other functions are undertaken by the protein upon germline removal remain unknown.
Results
NHR-49/PPARα potentiates DAF-16/FOXO3A and TCER-1/TCERG1 activity in germline-less adults
NHRs have modular ligand-binding domains (LBDs) that can be used to bind co-factors, to homodimerize or to heterodimerize with other NHRs. Through these dimerizations, NHRs, including NHR-49/PPARα, control the transcription of different subsets of targets.Citation37-39 Since our screen identified multiple NHRs that were essential for germline-less longevity, we asked if NHR-49/PPARα exhibited similar heterodimeric interactions with the other NHRs too. Using yeast 2 hybrid (Y2H) assays, we tested the potential interactions of 6 NHRs (genes encoding 5 of these, nhr-60, nhr-71, nhr-81, nhr-212 and nhr-213, were identified in our screenCitation22) with NHR-49/PPARα, DAF-16/FOXO3A and two of five TCER1/TCERG1 transcripts, TCER-1a and TCER-1d). We found that one of the NHRs we previously identified, NHR-71, () interacted with NHR-49/PPARα (). NHR-71 shows structural homology to HNF4 although its precise function has not been described so far, so we refer to it as NHR-71/HNF4. Interestingly, NHR-71/HNF4 and NHR-49/PPARα were reported to exhibit reciprocal interactions with each other in a previous report that characterized transcriptional networks in C. elegans.Citation39 To address the mechanistic relevance of this interaction in GSC-less animals, we asked if NHR-71/HNF4 is also required for any of the molecular events controlled by NHR-49/PPARα upon germline ablation, i.e., β−oxidation, and desaturation. In temperature-sensitive glp-1 mutants, a widely used genetic surrogate for the longevity caused by GSC removal,Citation14 nhr-71 RNAi moderately impaired the expression of acs-2 (), but did not cause a consistent reduction in the expression of other β−oxidation genes upregulated by NHR-49/PPARα upon germline loss (eg., acs-17, cpt-5, ech-7 and acdh-11; data not shown). The upregulation of the desaturase genes, fat-5 and fat-6, was not hampered by nhr-71 RNAi either (). This suggested that NHR-71/HNF4 may not partner with NHR-49/PPARα to regulate the expression of the latter's β-oxidation and desaturation target genes upon GSC loss.
Since both nhr-49 and nhr-71 were identified in our screen based on a necessity for upregulation of a DAF-16/FOXO3A and TCER-1/TCERG1 target gene, stdh-1/dod-8,Citation15 we tested if these factors may also influence the expression of other DAF-16/FOXO3A and TCER-1/TCERG1 targets. nhr-49;glp-1 mutants were significantly impaired in the induction of sod-3, a well-known, direct DAF-16/FOXO3A targetCitation40,41 as well as mdt-15Citation42 and lipl-4,Citation24 two other genes upregulated by DAF-16/FOXO3ACitation25,43 in glp-1 mutantsCitation23,24 () suggesting that NHR-49/PPARα feeds back positively to potentiate the DAF-16/TCER-1 pathway. The upregulation of mdt-15 and sod-3 was also attenuated upon nhr-71 RNAi ().
The only known common upstream regulator of DAF-16/FOXO3A and TCER-1/TCERG1 is KRI-1, an intestinal Ankyrin-repeat protein that is essential for DAF-16/FOXO3A nuclear localization and TCER-1/TCERG1 upregulation in germline-less adults.Citation15,44 Based on the strong influence of nhr-49 inactivation on upregulation of DAF-16/FOXO3A and TCER-1/TCERG1 targets, we asked if NHR-49/PPARα acts in a positive feedback loop to potentiate the activities of these factors. We reasoned that it is likely to do so through KRI-1 regulation, which was also identified as being downregulated in nhr-49 mutants in a previous genomic study.Citation38 Indeed, we found that a GFP-tagged KRI-1 protein reporter that shows diffuse cytoplasmic and nuclear expression in intestinal cells of germline-ablated animals was highly membrane-localized in germline-less nhr-49 mutants (). These data support the possibility that NHR-49/PPARα positively feeds back into the DAF-16/TCER-1 pathway by directing KRI-1 sub-cellular localization. It may do so by hetero-dimerizing with NHR-71/HNF4, but the possibility remains to be experimentally tested ().
Figure 1. NHR-49/PPARα collaborates with NHR-71/HNF4 to promote longevity of germline-less animals. A: Effect of nhr-71 RNAi on the longevity of glp-1 mutants. Adult glp-1 mutants were subjected to feeding RNAi by culturing them on bacteria expressing dsRNA targeting daf-16 (blue; m = 16.8 ± 0.2; n = 106/106; P vs. control <0.0001), tcer-1 (brown; m = 19.9 ± 0.7; n = 108/108; P vs. control <0.0001), nhr-49 (maroon; m = 16.5 ± 0.1; n = 116/117; P vs. control <0.0001) and nhr-71 (red; m = 19.8 ± 0.5; n = 103/103; P vs. control 0.0001). Lifespan curve of control animals grown on empty vector is shown in green (m = 25.1 ± 0.1; n = 94/117). Y-axis shows the fraction of animals alive at any time point, X-axis the days of adulthood. Data is shown as mean lifespan in days (m) ± standard error of the mean (SEM). ‘n’ refers to the number of worms analyzed divided by total number of worms tested in the experiment. P values were calculated using the log rank (Mantel Cox) method. Similar results were obtained in 2 additional trials. B: Interaction of NHR-49/PPARα and NHR-71/HNF4 in Yeast-Two-Hybrid (Y2H) assay. Small scale matrix to test Y2H interactions between NHRs that influence glp-1 longevity as well as DAF-16/FOXO3A and TCER-1/TCERG1, by growth on 20 mM 3AT and by LacZ expression. Growth on permissive –Leu –Trp plates is provided as a control. Gal4-activation domain (AD) and DNA-binding domain fusions (DB) for the NHR-49/PPARα ligand binding domain (LBD) and the indicated full-length NHRs, a DAF-16/FOXO3A full-length, TCER-1/TCERG1a full length and a C-terminally truncated isoform (TCER-1/TCERG1d) were tested pairwise. AD and DB are empty vector controls and an NHR-25-DB-SMO-1 AD interaction was used as a positive control. The NHR-49 ligand-binding domain (LBD) bait included amino acids 120-474 encoded by the nhr-49a transcript. All other baits were full-length proteins. As seen here, DAF-16/FOXO3A shows auto-activation that has not been reported before. The NHR-49 LBD bait did not exhibit homo-dimerization, in keeping with previous reports where the full-length protein was found to be essential for homodimerizationCitation38,39
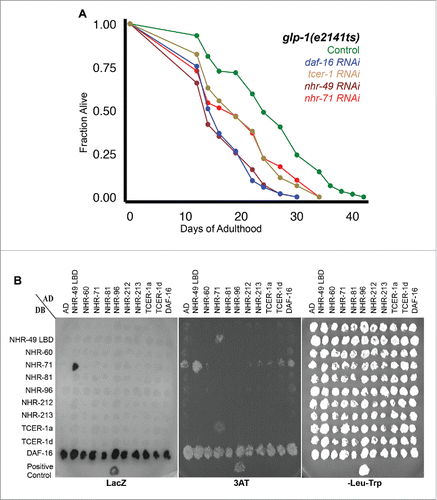
Figure 2. nhr-49 and nhr-71 are required for upregulation of DAF-16/FOXO3A target genes in germline-less adults. A-D: mRNA levels of acs-2 (A), fat-5 (B), fat-6 (C) and the NHR-49/PPARα co-regulator and DAF-16/FOXO3A target gene, mdt-15 (D), were measured in day 2 glp-1 adults grown on control vector (green) and those fed RNAi bacteria targeting nhr-49 (red) and nhr-71 (pink). X-axis represents the RNAi treatments and Y-axis the fold change in expression (normalized to ‘housekeeping’ gene rpl-32). (E) Quantitative PCR (Q-PCR) analysis of mRNA levels of DAF-16/FOXO3A target lipl-4 compared between wild-type worms (wt, gray) and nhr-49 (blue), glp-1 (green) and nhr-49;glp-1 (red) day 2 adults. In A-E, error bars display standard error of the mean, and asterisks depict the statistical significance of the observed differences in an unpaired, 2-tailed t-test with P values 0.05 (*) and 0.005 (**). (F) Expression of DAF-16/FOXO3A direct target sod-3 examined using a transcriptional GFP reporter in glp-1 mutants (glp-1;Psod-3::gfp). Day 2 adults subjected to RNAi treatments shown on the X-axis were classified as high (blue), medium (orange) and low (gray) based on the level of intestinal GFP. ‘n’ signifies the total number of worms examined in three independent trials. Error bars represent the standard error of the mean
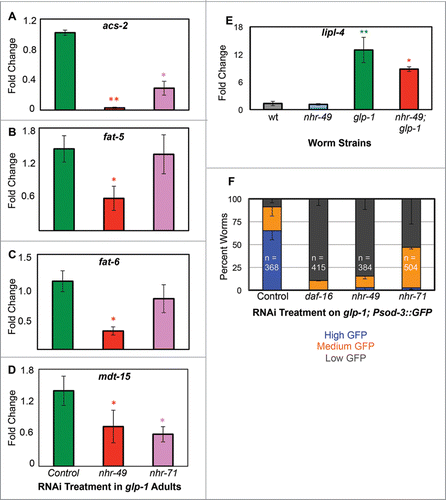
Figure 3. NHR-49/PPARα potentiates the DAF-16/FOXO3A and TCER-1/TCERG1 pathway by influencing the sub-cellular localization of KRI-1. A-E: KRI-1 is predominantly membrane-restricted in intestinal nuclei of germline-less worms in the absence of NHR-49/PPARα. Pkri-1::GFP::TAP::kri-1 expression examined in day 2 adults of glp-1 (A, B) and nhr-49;glp-1 (C, D). GFP is largely diffuse in glp-1 mutants, but in nhr-49;glp-1 adults there is a distinct relocation to membranes, especially near the lumen (indicated by arrows). Quantification of the data is shown in E. Y-axis represents the percent of worms that showed membrane localization (green) or diffuse cytoplasmic or nuclear expression (gray) of GFP in one or more intestinal cells. ‘n’ indicates total number of worms examined
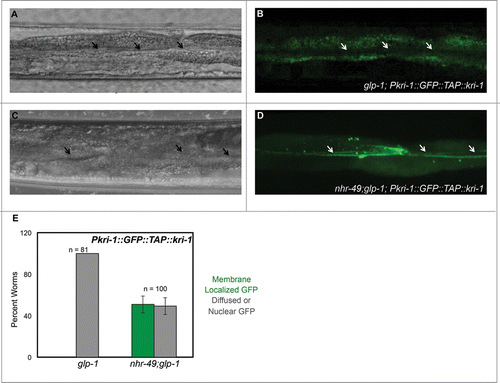
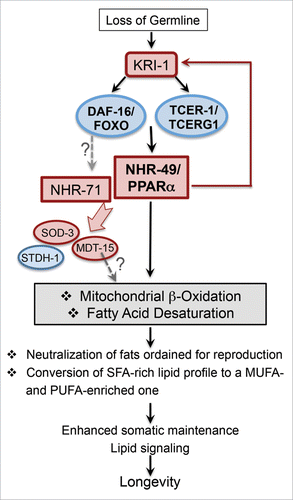
Figure 4. Schematic representation of the proposed model to explain NHR-49/PPARα function in promoting the longevity of germline-less animals (modified from Ratnappan et al.,Citation22). Following germline loss, NHR-49/PPARα is upregulated by the joint activity of DAF-16/FOXO3A and TCER-1/TCERG1. NHR-49/PPARα, in turn, mediates the upregulation of genes involved in fatty-acid β−oxidation and desaturation. The synchronized enhancement of these processes allows the animal to adapt to loss of fertility and orchestrate a lipid-homeostasis profile that supports longevity. NHR-49/PPARα and NHR-71/HNF4, another NHR essential for germline-less longevity, are required for the upregulation of DAF-16/FOXO3A targets previously shown to be elevated upon germline loss, such as sod-3, stdh-1 and mdt-15. Loss of nhr-49 causes membrane relocation of KRI-1, the only known common upstream regulator of DAF-16/FOXO3A and TCER-1/TCERG1, suggesting that NHR-49/PPARα may operate in a feedback loop to potentiate the activity of this pathway by regulating KRI-1 sub-cellular localization. Our data suggest that NHR-71/HNF4, similar to NHR-49/PPARα, may be upregulated by DAF-16/FOXO3A and TCER-1/TCERG1. MDT-15, a known co-regulator of NHR-49/PPARα, may also contribute to the increased expression of β-oxidation genes. Regulatory steps for which evidence is provided by our data are indicated by red arrows. Putative relationships suggested by our observations are represented by gray, stippled arrows.
Discussion
The coordination of disparate lipid-metabolic pathways by NHR-49/PPARα
Germline loss not only increases lifespan, it also increases fat accumulation in intestinal cells of the sterile animals. Surprisingly, they also exhibit enhanced expression of mitochondrial β-oxidation genes, and these genes are required for lifespan extension. These observations suggest that fatty-acid β-oxidation is elevated following GSC removal and contributes to the consequent longevity.Citation22 So, what purpose does enhanced β-oxidation serve in these animals? Similarly, why does GSC removal cause the animal to simultaneously augment β-oxidation and desaturation- lipid-degradative and lipid-synthetic pathways, respectively? We posit that the simultaneous elevation of these antagonistic processes reflects increased lipid turnover, which in turn allows the organism to adapt to the physiological catastrophe of lost fertility by (a) breaking down fats that would have been utilized in reproduction, and (b) converting the lipids to unsaturated forms that are more amenable for storage and cellular maintenance.
This hypothesis, of enhanced lipid turnover as an adaptive response to germline loss, may also explain a dramatic and surprising fat loss displayed by GSC-less nhr-49 mutants. Since β−oxidation is a lipolytic pathway, we expected nhr-49;glp-1 mutants to exhibit further increase in fat levels. However, using the lipid-labeling dye oil red O (ORO) as well as gas chromatography/mass spectrometry (GC/MS), we found that nhr-49;glp-1 mutants had lower fat levels than glp-1 adults, and this fat was rapidly lost with age.Citation22 This could be explained in part by the fact that NHR-49/PPARα also contributed to de novo lipid synthesis,Citation22 but it does not account for the dramatic age-related fat depletion seen in the absence of NHR-49/PPARα. We propose that this drastic fat depletion is a consequence of accumulation of free fatty acids (FFAs), due to simultaneous inhibition of β−oxidation and desaturation in the absence of NHR-49/PPARα, which causes lipotoxicity. FFAs stimulate insulin release and serve as key signaling molecules. But their chronic accrual causes deregulated insulin secretion and apoptosis in pancreatic β−cellsCitation45; in muscle and liver cells it leads to insulin resistance.Citation46 Impaired fatty-acid oxidation is a primary cause of lipotoxicity in human cells, and desaturation is critical for protecting β-cells and endothelial cells from toxic effects of SFA accumulation.Citation47,48 Long-chain, non-metabolized SFAs are implicated as the primary causative agents underlying lipotoxicity.Citation49,50 We observed significant elevation of these species, including SA, in germline-less nhr-49 mutants. SA is also a poor substrate for triglyceride (TAG) synthesis and a potent inducer of pro-inflammatory responses and cell death in human cells.Citation51 FFA accumulation also prevents de novo lipid synthesis by inhibiting the function of fat-synthesis enzymes.Citation52 Hence, it is conceivable that in the absence of NHR-49/PPARα, the lipid turnover that would facilitate adaptation to loss of fertility is prevented, and the resultant accumulation of toxic FFAs causes lipotoxicity that gets aggravated over a short time and causes lipid degeneration and death.
The healthy obesity of long-lived, germline-less animals
Germline-ablated animals are remarkable because they manifest an apparently paradoxical healthy obesity, along with longer life, as compared to their leaner, fertile counterparts.Citation22,53 This discrepancy is not unique to C. elegans either. Similar observations have been made in other model organisms and in the natural world.Citation11,54,55 Even in human populations, while obesity is generally associated with disease, ‘Metabolically Healthy Obese’ individuals are noteworthy because they retain excessive weight without developing metabolic disorders.Citation56 Hence, there is accumulating evidence that absolute fat level is not the deterministic factor for health and longevity. Instead, the composition of lipids has a significant bearing on metabolic health. Lipids with higher UFAs are generally associated with improved cellular maintenance.Citation57 In hibernating animals, fatty acid composition of stored lipids affects the depth and duration of torpor. Diets rich in MUFAs and PUFAs have a beneficial impact on mammalian torpor, whereas diets rich in SFAs have a negative effect.Citation58,59 Alternatively, SFAs are critical for reproductive health. They make up >70% of human oocytes, of which SA and PA are the most abundant, whereas, MUFAs make up <15%.Citation60-62 Thus, activation of NHR-49/PPARα may help transform the SFA-rich, reproduction-oriented lipid profile of a fertile adult into a UFA-rich profile that is more conducive for fat storage and cellular maintenance.
Feedback loops and NHR partnerships triggered by germline signals
Our observations, that NHR-49/PPARα influences the subcellular localization of KRI-1, the common upstream regulator of DAF-16/FOXO3A and TCER-1/TCERG1, and is required for increased expression of DAF-16/FOXO3A and TCER-1/TCERG1 targets reveal the existence of a positive feedback loop between NHR-49/PPARα and its upstream regulatory factors (). Since NHR-71/HNF4 is also required for the expression of some of these genes, and since NHR-49/PPARα binds NHR-71/HNF4 in Y2H assays, it is plausible that the two NHRs partner with each other to accomplish this positive feedback regulation, however this remains to be experimentally tested. NHR-71:NHR-49 binding has been observed before, and in the same report NHR-49/PPARα was also found to bind NHR-274, one of the pro-longevity NHRs identified in our screen, but that we could not test in the Y2H assays.Citation39 Partnerships between NHR-49/PPARα and many NHRs have been reported in other contexts. It physically interacts with NHR-66 and both nhr-49 and nhr-66 are required for repression of 3 genes involved in sphingolipid metabolism. Similarly, it binds NHR-80/HNF4 and both genes facilitate lipid desaturation, possibly working together.Citation38 Additionally, NHR-49/PPARα interactions with NHR-79 and NHR-234 have been described.Citation37 As observed in normal, fertile animals, NHR-49/PPARα may dimerize with NHR-80/HNF4 to regulate desaturase gene expression in GSC-less adults too. However, the factors that it would interact with to regulate β-oxidation remain unknown. One or more of the other pro-longevity NHRs we identified are likely candidates to fulfill these roles, though we did not notice binding in our Y2H assays. It is possible that heterodimerization is triggered specifically upon germline ablation that could not be recapitulated in a Y2H experimental setup. Regardless, these data provide insights that underscore how NHRs such as NHR-49/PPARα, that function as key nodes of large metabolic networks, achieve functional specificity through selective interactions with other constituents of the network, depending on the physiological stimulus.
Perspective
PPAR proteins were the first identified genetic sensors for fat and their study has revolutionized our understanding of energy metabolism.Citation34 PPARα is implicated in metabolic diseases such as dyslipidemia and CVD and PPARα agonists like Fibrates are widely used lipid-lowering drugs.Citation50,63 Interestingly, a role for PPARα in mammalian female reproduction has been revealed recently,Citation64 and in ovariectomized rats Fenofibrate helps maintain bone-mass.Citation65 Given this context, and accruing evidence that reproductive signals alter lifespan of diverse species, it is enticing to speculate if PPARα and other NHRs may influence vertebrate longevity. Since molecular dissection of metabolic pathways is challenging in mammalian systems, understanding how NHR-49/PPARα and its partners promote C. elegans longevity is likely to provide new mechanistic insights and therapeutic targets for regulating lipid homeostasis and aging in humans.
Materials and methods
Worm culture and lifespan analysis
The following worm strains were used in this study: N2 (wild type), CF1903 {glp-1(e2141)III}, AGP12a {nhr-49(nr2041)I}, AGP22 {nhr-49(nr2041)I;glp-1(e2141ts)III)}, CF1929 {glp-1(e4141ts)III; muIs84[Psod-3::GFP]} AGP116 muIs162 [Pkri-1::GFP::TAP::kri-1];nhr-49(nr2041)I;glp-1(e2141ts)V and CF3352 muIs162 [Pkri-1::GFP::TAP::kri-1];glp-1(e2141ts)III. All strains were maintained by standard techniques at 20°C. Lifespan experiments were conducted as described previously and have been discussed in detail elsewhere.Citation66 For all experiments that involved the glp-1 genetic background, eggs were incubated at 20°C for 2–6 h, transferred to 25°C to eliminate germ cells, then shifted back to 20°C on day 1 of adulthood (˜72 h later) for the rest of their lifespan.
Q-PCRs
Worm RNA was isolated using the mirVana miRNA Isolation Kit (Ambion, AM1561), DNAse treated (DNase kit, Sigma #AMPD1) and reverse transcribed into cDNA (Protoscript m-MuLV First Strand cDNA Synthesis kit, NEB #E6300S) according to the manufacturer's instructions. Quantitative real-time PCRs were performed using an Applied Bio Systems 7300 Real Time PCR System employing the Sybr Green chemistry (SensiMix SYBR Hi-ROX kit, Bioline #QT-605). Gene expression data were normalized to the housekeeping gene rpl-32. Data reported here were obtained by combining results from 3 biological replicates, each comprising 2–4 technical repeats.
GFP assays
Eggs were transferred to freshly-seeded Escherichia coli OP50 plates (or E. coli HT115 RNAi plates targeting the appropriate gene), incubated at 20°C for 2–6 h, transferred to 25°C (to eliminate germ cells in strains containing glp-1 mutation), then shifted back to 20°C on day 1 of adulthood. GFP assays were conducted on day 2 of adulthood, using a Leica MZ16F stereomicroscope. All assays were performed blind after initial familiarization of GFP levels in control plates by the experimenter.
Yeast two-hybrid (Y2H) assay
The cDNAs used in yeast 2-hybrid experiments were Gateway cloned (Invitrogen) into pDONR221 vector to generate entry clones and then moved into pAD-dest and pDB-dest vectors, which contain the Gal4 activation domain and DNA binding domain, respectively.Citation67 Yeast transformations and Y2H assays were carried out as described by Deplancke et al.Citation68
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This study was funded by the Ellison Medical Foundation's New Scholars in Aging award (AG-NS-0879-12) to AG and a grant from the National Institutes of Health (CA020535) to KRY.
References
- Storlien L, Oakes ND, Kelley DE. Metabolic flexibility. Proc Nutrit Soc 2004; 63:363-8; PMID:15294056; http://dx.doi.org/10.1079/PNS2004349
- Ghazi A. Transcriptional networks that mediate signals from reproductive tissues to influence lifespan. Genesis 2013; 51:1-15; PMID:22945891; http://dx.doi.org/10.1002/dvg.22345
- Hansen M, Flatt T, Aguilaniu H. Reproduction, fat metabolism, and life span: what is the connection? Cell Metabol 2013; 17:10-9; PMID:23312280; http://dx.doi.org/10.1016/j.cmet.2012.12.003
- Kenyon C. A pathway that links reproductive status to lifespan in Caenorhabditis elegans. Ann N Y Acad Sci 2010; 1204:156-62; PMID:20738286; http://dx.doi.org/10.1111/j.1749-6632.2010.05640.x
- Maures TJ, Booth LN, Benayoun BA, Izrayelit Y, Schroeder FC, Brunet A. Males shorten the life span of C. elegans hermaphrodites via secreted compounds. Science 2014; 343:541-4; PMID:24292626; http://dx.doi.org/10.1126/science.1244160
- Shi C, Murphy CT. Mating induces shrinking and death in Caenorhabditis mothers. Science 2014; 343:536-40; PMID:24356112; http://dx.doi.org/10.1126/science.1242958
- Hsin H, Kenyon C. Signals from the reproductive system regulate the lifespan of C. elegans. Nature 1999; 399:362-6; PMID:10360574; http://dx.doi.org/10.1038/20694
- Kirkwood TB. Evolution of ageing. Nature 1977; 270:301-4; PMID:593350; http://dx.doi.org/10.1038/270301a0
- Le Cunff Y, Baudisch A, Pakdaman K. Evolution of aging: individual life history trade-offs and population heterogeneity account for mortality patterns across species. J Evol Biol 2014; 27:1706-20; PMID:24925106; http://dx.doi.org/10.1111/jeb.12423
- Min KJ, Lee CK, Park HN. The lifespan of Korean eunuchs. Curr Biol 2012; 22:R792-3; PMID:23017989; http://dx.doi.org/10.1016/j.cub.2012.06.036
- Tatar M, Kopelman A, Epstein D, Tu MP, Yin CM, Garofalo RS. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science 2001; 292:107-10; PMID:11292875; http://dx.doi.org/10.1126/science.1057987
- Keith SA, Ghazi A. Recent discoveries in the reproductive control of aging. Curr Genet Med Rep 2014; 3:26-34; http://dx.doi.org/10.1007/s40142-014-0060-8
- Gwynne DT. Sexual conflict over nuptial gifts in insects. Annu Rev Entomol 2008; 53:83-101; PMID:17680720; http://dx.doi.org/10.1146/annurev.ento.53.103106.093423
- Arantes-Oliveira N, Apfeld J, Dillin A, Kenyon C. Regulation of life-span by germ-line stem cells in Caenorhabditis elegans. Science 2002; 295:502-5; PMID:11799246; http://dx.doi.org/10.1126/science.1065768
- Ghazi A, Henis-Korenblit S, Kenyon C. A transcription elongation factor that links signals from the reproductive system to lifespan extension in Caenorhabditis elegans. PLoS Genet 2009; 5:e1000639; PMID:19749979; http://dx.doi.org/10.1371/journal.pgen.1000639
- Hansen M, Hsu AL, Dillin A, Kenyon C. New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet 2005; 1:119-28; PMID:16103914; http://dx.doi.org/10.1371/journal.pgen.0010017
- Lapierre LR, De Magalhaes Filho CD, McQuary PR, Chu CC, Visvikis O, Chang JT, Gelino S, Ong B, Davis AE, Irazoqui JE, et al. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat Commun 2013; 4:2267; PMID:23925298
- Lapierre LR, Gelino S, Melendez A, Hansen M. Autophagy and lipid metabolism coordinately modulate life span in germline-less C. elegans. Curr Biol 2011; 21:1507-14; PMID:21906946; http://dx.doi.org/10.1016/j.cub.2011.07.042
- Steinbaugh MJ, Narasimhan SD, Robida-Stubbs S, Moronetti Mazzeo LE, Dreyfuss JM, Hourihan JM, Raghavan P, Operana TN, Esmaillie R, Blackwell TK. Lipid-mediated regulation of SKN-1/Nrf in response to germ cell absence. Elife 2015; 4; PMID:26196144; http://dx.doi.org/10.7554/eLife.07836
- Gerisch B, Rottiers V, Li D, Motola DL, Cummins CL, Lehrach H, Mangelsdorf DJ, Antebi A. A bile acid-like steroid modulates Caenorhabditis elegans lifespan through nuclear receptor signaling. Proc Natl Acad Sci U S A 2007; 104:5014-9; PMID:17360327; http://dx.doi.org/10.1073/pnas.0700847104
- Goudeau J, Bellemin S, Toselli-Mollereau E, Shamalnasab M, Chen Y, Aguilaniu H. Fatty acid desaturation links germ cell loss to longevity through NHR-80/HNF4 in C. elegans. PLoS Biol 2011; 9:e1000599; PMID:21423649; http://dx.doi.org/10.1371/journal.pbio.1000599
- Ratnappan R, Amrit FR, Chen SW, Gill H, Holden K, Ward J, Yamamoto KR, Olsen CP, Ghazi A. Germline signals deploy NHR-49 to modulate fatty-acid beta-oxidation and desaturation in somatic tissues of C. elegans. PLoS Gen 2014; 10:e1004829; PMID:25474470; http://dx.doi.org/10.1371/journal.pgen.1004829
- McCormick M, Chen K, Ramaswamy P, Kenyon C. New genes that extend Caenorhabditis elegans' lifespan in response to reproductive signals. Aging Cell 2012; 11:192-202; PMID:22081913; http://dx.doi.org/10.1111/j.1474-9726.2011.00768.x
- Wang MC, O'Rourke EJ, Ruvkun G. Fat metabolism links germline stem cells and longevity in C. elegans. Science 2008; 322:957-60; PMID:18988854; http://dx.doi.org/10.1126/science.1162011
- Zhang P, Judy M, Lee SJ, Kenyon C. Direct and indirect gene regulation by a life-extending FOXO protein in C. elegans: roles for GATA factors and lipid gene regulators. Cell Metabol 2013; 17:85-100; PMID:23312285; http://dx.doi.org/10.1016/j.cmet.2012.12.013
- Evans RM, Mangelsdorf DJ. Nuclear Receptors, RXR, and the Big Bang. Cell 2014; 157:255-66; PMID:24679540; http://dx.doi.org/10.1016/j.cell.2014.03.012
- Antebi A. Nuclear receptor signal transduction in C. elegans. Worm Book 2015:1-49; PMID:26069085; http://dx.doi.org/10.1895/wormbook.1.64.2
- Taubert S, Ward JD, Yamamoto KR. Nuclear hormone receptors in nematodes: evolution and function. Mol Cell Endocrinol 2011; 334:49-55; PMID:20438802; http://dx.doi.org/10.1016/j.mce.2010.04.021
- Van Gilst MR, Hadjivassiliou H, Jolly A, Yamamoto KR. Nuclear hormone receptor NHR-49 controls fat consumption and fatty acid composition in C. elegans. PLoS Biol 2005; 3:e53; PMID:15719061; http://dx.doi.org/10.1371/journal.pbio.0030053
- Van Gilst MR, Hadjivassiliou H, Yamamoto KR. A Caenorhabditis elegans nutrient response system partially dependent on nuclear receptor NHR-49. Proc Natl Acad Sci U S A 2005; 102:13496-501; PMID:16157872; http://dx.doi.org/10.1073/pnas.0506234102
- Burkewitz K, Morantte I, Weir HJ, Yeo R, Zhang Y, Huynh FK, Ilkayeva OR, Hirschey MD, Grant AR, Mair WB. Neuronal CRTC-1 governs systemic mitochondrial metabolism and lifespan via a catecholamine signal. Cell 2015; 160:842-55; PMID:25723162; http://dx.doi.org/10.1016/j.cell.2015.02.004
- Folick A, Oakley HD, Yu Y, Armstrong EH, Kumari M, Sanor L, Moore DD, Ortlund EA, Zechner R, Wang MC. Aging. Lysosomal signaling molecules regulate longevity in Caenorhabditis elegans. Science 2015; 347:83-6; PMID:25554789; http://dx.doi.org/10.1126/science.1258857
- Ma DK, Li Z, Lu AY, Sun F, Chen S, Rothe M, Menzel R, Sun F, Horvitz HR. Acyl-CoA Dehydrogenase Drives Heat Adaptation by Sequestering Fatty Acids. Cell 2015; 161:1152-63; PMID:25981666; http://dx.doi.org/10.1016/j.cell.2015.04.026
- Poulsen L, Siersbaek M, Mandrup S. PPARs: fatty acid sensors controlling metabolism. Semin Cell Dev Biol 2012; 23:631-9; PMID:22273692; http://dx.doi.org/10.1016/j.semcdb.2012.01.003
- Angelo G, Van Gilst MR. Starvation protects germline stem cells and extends reproductive longevity in C. elegans. Science 2009; 326:954-8; PMID:19713489; http://dx.doi.org/10.1126/science.1178343
- Amrit FRG, Steenkiste EM, Ratnappan R, Chen S-W, McClendon TBH, Kostka D, Yanowitz JL, Olsen CP, & Ghazi A. (2016). DAF-16 and TCER-1 facilitate adaptation to germline loss by restoring lipid homeostasis and repressing reproductive physiology in C. elegans. PLoS Genet. 10(12):e1005788; PMID: 26862916; http://dx.doi./org/10.1371/journal.pgen.1005788
- Brock TJ, Browse J, Watts JL. Genetic regulation of unsaturated fatty acid composition in C. elegans. PLoS genetics 2006; 2:e108; PMID:16839188
- Arda HE, Taubert S, MacNeil LT, Conine CC, Tsuda B, Van Gilst M, Sequerra R, Doucette-Stamm L, Yamamoto KR, Walhout AJ. Functional modularity of nuclear hormone receptors in a Caenorhabditis elegans metabolic gene regulatory network. Mol Syst Biol 2010; 6:367; PMID:20461074; http://dx.doi.org/10.1038/msb.2010.23
- Pathare PP, Lin A, Bornfeldt KE, Taubert S, Van Gilst MR. Coordinate regulation of lipid metabolism by novel nuclear receptor partnerships. PLoS genetics 2012; 8:e1002645; PMID:22511885; http://dx.doi.org/10.1371/journal.pgen.1002645
- Reece-Hoyes JS, Pons C, Diallo A, Mori A, Shrestha S, Kadreppa S, Nelson J, Diprima S, Dricot A, Lajoie BR, et al. Extensive rewiring and complex evolutionary dynamics in a C. elegans multiparameter transcription factor network. Mol Cell 2013; 51:116-27; PMID:23791784; http://dx.doi.org/10.1016/j.molcel.2013.05.018
- Libina N, Berman JR, Kenyon C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell 2003; 115:489-502; PMID:14622602; http://dx.doi.org/10.1016/S0092-8674(03)00889-4
- Oh SW, Mukhopadhyay A, Dixit BL, Raha T, Green MR, Tissenbaum HA. Identification of direct DAF-16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nat Genet 2006; 38:251-7; PMID:16380712; http://dx.doi.org/10.1038/ng0406-398
- Taubert S, Van Gilst MR, Hansen M, Yamamoto KR. A Mediator subunit, MDT-15, integrates regulation of fatty acid metabolism by NHR-49-dependent and -independent pathways in C. elegans. Genes Dev 2006; 20:1137-49; PMID:16651656; http://dx.doi.org/10.1101/gad.1395406
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 2003; 424:277-83; PMID:12845331; http://dx.doi.org/10.1038/nature01789
- Berman JR, Kenyon C. Germ-cell loss extends C. elegans life span through regulation of DAF-16 by kri-1 and lipophilic-hormone signaling. Cell 2006; 124:1055-68; PMID:16530050; http://dx.doi.org/10.1016/j.cell.2006.01.039
- Zhou YP, Grill V. Long term exposure to fatty acids and ketones inhibits B-cell functions in human pancreatic islets of Langerhans. J Clin Endocrinol Metab 1995; 80:1584-90; PMID:7745004
- Kraegen EW, Cooney GJ. Free fatty acids and skeletal muscle insulin resistance. Curr Opin Lipidol 2008; 19:235-41; PMID:18460913; http://dx.doi.org/10.1097/01.mol.0000319118.44995.9a
- Busch AK, Gurisik E, Cordery DV, Sudlow M, Denyer GS, Laybutt DR, Hughes WE, Biden TJ. Increased fatty acid desaturation and enhanced expression of stearoyl coenzyme A desaturase protects pancreatic beta-cells from lipoapoptosis. Diabetes 2005; 54:2917-24; PMID:16186393; http://dx.doi.org/10.2337/diabetes.54.10.2917
- Peter A, Weigert C, Staiger H, Rittig K, Cegan A, Lutz P, Machicao F, Haring HU, Schleicher E. Induction of stearoyl-CoA desaturase protects human arterial endothelial cells against lipotoxicity. Am J Physiol Endocrinol Metab 2008; 295:E339-49; PMID:18523127; http://dx.doi.org/10.1152/ajpendo.00022.2008
- Rockenfeller P, Ring J, Muschett V, Beranek A, Buettner S, Carmona-Gutierrez D, Eisenberg T, Khoury C, Rechberger G, Kohlwein SD, et al. Fatty acids trigger mitochondrion-dependent necrosis. Cell Cycle 2010; 9:2836-42; PMID:20647757; http://dx.doi.org/10.4161/cc.9.14.12346
- Unger RH, Clark GO, Scherer PE, Orci L. Lipid homeostasis, lipotoxicity and the metabolic syndrome. Biochim Biophys Acta 2010; 1801:209-14; PMID:19948243; http://dx.doi.org/10.1016/j.bbalip.2009.10.006
- Pai T, Yeh YY. Stearic acid unlike shorter-chain saturated fatty acids is poorly utilized for triacylglycerol synthesis and beta-oxidation in cultured rat hepatocytes. Lipids 1996; 31:159-64; PMID:8835403; http://dx.doi.org/10.1007/BF02522615
- Nikawa J, Tanabe T, Ogiwara H, Shiba T, Numa S. Inhibitory effects of long-chain acyl coenzyme A analogues on rat liver acetyl coenzyme A carboxylase. FEBS Lett 1979; 102:223-6; PMID:37122; http://dx.doi.org/10.1016/0014-5793(79)80005-8
- O'Rourke EJ, Soukas AA, Carr CE, Ruvkun G. C. elegans major fats are stored in vesicles distinct from lysosome-related organelles. Cell metabolism 2009; 10:430-5; PMID:19883620; http://dx.doi.org/10.1016/j.cmet.2009.10.002
- Berryman DE, List EO, Coschigano KT, Behar K, Kim JK, Kopchick JJ. Comparing adiposity profiles in three mouse models with altered GH signaling. Growth Horm IGF Res 2004; 14:309-18; PMID:15231300; http://dx.doi.org/10.1016/j.ghir.2004.02.005
- Judd ET, Wessels FJ, Drewry MD, Grove M, Wright K, Hahn DA, Hatle JD. Ovariectomy in grasshoppers increases somatic storage, but proportional allocation of ingested nutrients to somatic tissues is unchanged. Aging Cell 2011; 10:972-9; PMID:21834847; http://dx.doi.org/10.1111/j.1474-9726.2011.00737.x
- Naukkarinen J, Heinonen S, Hakkarainen A, Lundbom J, Vuolteenaho K, Saarinen L, Hautaniemi S, Rodriguez A, Fruhbeck G, Pajunen P, et al. Characterising metabolically healthy obesity in weight-discordant monozygotic twins. Diabetologia 2014; 57:167-76; PMID:24100782; http://dx.doi.org/10.1007/s00125-013-3066-y
- Gonzalez-Covarrubias V, Beekman M, Uh HW, Dane A, Troost J, Paliukhovich I, van der Kloet FM, Houwing-Duistermaat J, Vreeken RJ, Hankemeier T, et al. Lipidomics of familial longevity. Aging Cell 2013; 12:426-34; PMID:23451766; http://dx.doi.org/10.1111/acel.12064
- Chauhan V, Sheikh A, Chauhan A, Tsiouris J, Malik M, Vaughan M. Changes during hibernation in different phospholipid and free and esterified cholesterol serum levels in black bears. Biochimie 2002; 84:1031-4; PMID:12504283; http://dx.doi.org/10.1016/S0300-9084(02)00006-8
- Geiser F, McAllan BM, Kenagy GJ. The degree of dietary fatty acid unsaturation affects torpor patterns and lipid composition of a hibernator. J Comp Physiol B 1994; 164:299-305; PMID:7962785; http://dx.doi.org/10.1007/BF00346446
- Ferreira CR, Saraiva SA, Catharino RR, Garcia JS, Gozzo FC, Sanvido GB, Santos LF, Lo Turco EG, Pontes JH, Basso AC, et al. Single embryo and oocyte lipid fingerprinting by mass spectrometry. J Lipid Res 2010; 51:1218-27; PMID:19965589; http://dx.doi.org/10.1194/jlr.D001768
- Matorras R, Ruiz JI, Mendoza R, Ruiz N, Sanjurjo P, Rodriguez-Escudero FJ. Fatty acid composition of fertilization-failed human oocytes. Hum Reprod 1998; 13:2227-30; PMID:9756301; http://dx.doi.org/10.1093/humrep/13.8.2227
- McEvoy TG, Coull GD, Broadbent PJ, Hutchinson JS, Speake BK. Fatty acid composition of lipids in immature cattle, pig and sheep oocytes with intact zona pellucida. J Reprod Fertil 2000; 118:163-70; PMID:10793638; http://dx.doi.org/10.1530/reprod/118.1.163
- Barter PJ, Rye KA. Is there a role for fibrates in the management of dyslipidemia in the metabolic syndrome? Arterioscler Thromb Vasc Biol 2008; 28:39-46; PMID:17717290; http://dx.doi.org/10.1161/ATVBAHA.107.148817
- Yang J, Chen L, Zhang X, Zhou Y, Zhang D, Huo M, Guan Y. PPARs and Female Reproduction: Evidence from Genetically Manipulated Mice. PPAR Res 2008; 2008:723243; PMID:18401459
- Stunes AK, Westbroek I, Gustafsson BI, Fossmark R, Waarsing JH, Eriksen EF, Petzold C, Reseland JE, Syversen U. The peroxisome proliferator-activated receptor (PPAR) alpha agonist fenofibrate maintains bone mass, while the PPAR gamma agonist pioglitazone exaggerates bone loss, in ovariectomized rats. BMC Endocr Disord 2011; 11:11; PMID:21615901; http://dx.doi.org/10.1186/1472-6823-11-11
- Amrit FR, Ratnappan R, Keith SA, Ghazi A. The C. elegans lifespan assay toolkit. Methods 2014; 68:465-75; PMID:24727064; http://dx.doi.org/10.1016/j.ymeth.2014.04.002
- Reboul J, Vaglio P, Rual JF, Lamesch P, Martinez M, Armstrong CM, Li S, Jacotot L, Bertin N, Janky R, et al. C. elegans ORFeome version 1.1: experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat Genet 2003; 34:35-41; PMID:12679813; http://dx.doi.org/10.1038/ng1140
- Deplancke B, Vermeirssen V, Arda HE, Martinez NJ, Walhout AJ. Gateway-compatible yeast one-hybrid screens. CSH Protoc 2006; 2006:pdb.prot4590-pdb.prot4590 PMID:22485967; http://dx.doi.org/10.1 101/pdb.prot4590
