Abstract
The S-system, a set of nonlinear ordinary differential equations and derived from the generalized mass action law, is an effective model to describe various biological systems. Parameters in S-systems have significant biological meanings, yet difficult to be estimated because of the nonlinearity and complexity of the model. Given time series biological data, its parameter estimation turns out to be a nonlinear optimization problem. A novel method, auxiliary function guided coordinate descent, is proposed in this paper to solve the optimization problem by cyclically optimizing every parameter. In each iteration, only one parameter value is updated and it proves that the objective function keeps nonincreasing during the iterations. The updating rules in each iteration is simple and efficient. Based on this idea, two algorithms are developed to estimate the S-systems for two different constraint situations. The performances of algorithms are studied in several simulation examples. The results demonstrate the effectiveness of the proposed method.
1. Introduction
There are many various molecules and productions in a cell. Some of them can regulate others via some mechanisms to achieve specific cellular functions, based on which the cells adapt to the changing environments. These components and their interactions constitute biological systems, such as metabolic pathways and genetic regulatory networks. One task of systems biology is to reveal the interactions and the biological functions those interactions may result in Klipp, Herwig, Kowald, Wierling, and Lehrach (2005). Instead of focusing on individual components, systems biology applies system engineering methods and principles to studying all components and their interactions as parts of a biological system. Such a systematic view provides an insight into the control and optimization of parts of the systems while taking the effects those may have on the whole system into account. It is of help to the discovery of new properties of biological systems and may provide valuable clues and new ideas in practical areas such as disease treatment and drug design (Gardner, di Bernardo, Lorenz, and Collins Citation2003).
One effective way to study the biological system is using mathematical or computational methods. Many mathematical models have been developed to describe the molecular biological systems based on biochemical principles. Most of them are nonlinear in both parameters and state variables (Klipp et al. 2005; Voit 2000). Nonlinear optimization problems are usually formulated for estimating the parameters in those models. However, analytical solutions to those problems are hardly available. One of the most popular models is the S-system which is highly nonlinear and derived from the generalized mass action law (Voit, 2000).
The S-system is an effective mathematical framework to characterize and analyse the molecular biological systems. An S-system consisting of N components is type of power-law formalism and typically a group of nonlinear ordinary differential equations (ODEs),
The parameter estimation and structure identification of the S-systems are extremely difficult tasks. The parameter estimation usually occurs after or in the process of structure identification. As the parameter estimation of the S-systems is essentially a nonlinear problem, in principle, all nonlinear optimization algorithms can be used, e.g. Gauss–Newton method and its variants, such as Box–Kanemasu interpolation method, Levenberg damped least-squares method, and Marquardt's method (Beck & Arnold, Citation1977). However, most of these methods are initial-sensitive and need to calculate the inverse of the Hessian which is computational expensive.
Several numerical methods have been proposed to estimate the parameters in S-systems. Most of them are based on heuristic methods. For example, Kikuchi, Tominaga,Arita, Takahashi, and Tomita (2003) employ a genetic algorithm to infer the S-systems. The effectiveness of the simulated annealing is studied in Gonzalez, Kper, Jung,Naval, and Mendoza (2007). Voit and Almeida (Citation2004) develop an artificial neural network (ANN)-based method to identify and estimate the parameters of S-systems. Ho,Hsieh, Yu, and Huang (2007) develop an intelligent two-stage evolutionary algorithm that combines the genetic algorithm and the simulated annealing. An unified approach has been proposed in Wang, Qian, and Dougherty (2010). Most of these methods are computational expensive and do not sufficiently take advantage of special model structure of the S-systems.
Several methods taking the model structure of S-systems into account have been proposed. Wu and Mu (2009) introduce a separable parameter estimation method which divides the parameters into two groups: one group is linear in model while the other nonlinear. This method has been extended with a genetic algorithm to the case when system topology is unavailable in Liu, Wu, and Zhang (Citation2012b). One can observe that if parameters in one term on the right-hand side of Equation (1) is known, moving it to the left and taking logarithm of both sides, a linear model is obtained. Using this observation, an alternating regression method is proposed by Chou, Martens, and Voit (Citation2006), which reduces the nonlinear optimization into iterative procedures of linear regression. However, the convergence of the iterations cannot be guaranteed. Similarly, an alternating weighted least-squares method is proposed by Liu, Wu, and Zhang(Citation2012a), in which the objective function to be optimized is approximated by a weighted linear regression in each iteration. However, this approximation may fail in some cases.
This study focus on the parameter estimation of S-systems when system topology is known. A novel method, auxiliary function guided coordinate descent (AFGCD), is proposed. After decoupling the S-systems by replacing the derivatives with numerical slops, the parameter estimation is formulated as a nonlinear optimization problem. AFGCD iteratively solves this problem and in each iteration only one parameter is optimized with other parameters fixed. Instead of directly optimizing the objective function, AFGCD takes advantage of a property of the exponential function and constructs an auxiliary function for each kinetic order parameter to optimize. It shows that optimizing the auxiliary function makes the objective function keep nonincreasing. Because the auxiliary function is simple and its optimization has an analytical solution, the parameter updating in each iteration is simple and efficient. Based on this idea, two algorithms are developed to estimate the parameters in S-systems under two different situations. One algorithm is developed for the case when the range of each parameter is known and the other algorithm for the case that the only constraint is the non-negativity of the rate constants.
The remaining of this paper is organized as follows. In Section 2, two algorithms are developed based on the idea of AFGCD and their descent properties are proven. In Section 3, the effectivenesses of the proposed algorithms are studied by several simulation examples. Finally, Section 4 concludes this study and points out some future works along this research.
2. Auxiliary function guided coordinate descent
2.1 Problem statement
Consider a biological system of N molecular species, described by an S-system as Equation (1), and assume that for each molecular species Xi, a time series concentration data measured at n equally spaced time points, are obtained. This study assumes that the topology of the system is available, i.e. the zero-values kinetic orders are known. The purpose is to estimate the parameters of nonzero-valued kinetic orders and rate constants. We substitute the derivative of Xi at time t with the estimated slop, Sit, so that the original coupled ordinary differential equations are decoupled into n×N uncoupled algebraic equations (Vilela et al.,Citation2008; Voit & Almeida, Citation2004):
To determine the parameter values, the sum of least squares is usually employed as an objective function to be minimized, i.e. the parameters of each ODE equation i in Equation (1) is obtained by solving
where δ is a predefined small positive number; Gi and Hi are the sets of indices of molecular species which have effects on the production and degradation of molecular species i, respectively; and
. The minimization of Equation (4) is non-trivial, as it is highly nonlinear and contains a lot of parameters.
2.2 Proposed method
The optimization problem (4) has no analytical solutions and therefore, iterative methods are considered. A coordinate descent strategy is applied in this study, i.e. we cyclically optimize one parameter in one iteration with other parameters fixed such that the objective function keeps nonincreasing in every iteration. More specifically, denote by the parameters of an optimization problem (e.g. for Equation (4),
) at iteration ℓ. After one iteration, the first parameter is updated,
and after k iterations, the first k parameters are updated,
.
Consider the problem (4) and suppose the parameter hik, for some k∈Hi, needs to be updated at the current iteration, one simple way to make sure that the objective function keeps nonincreasing is to use the classical gradient descent method:
In this paper, a smart stepsize is derived. The introduced stepsize is computational efficient and the non-increase of the objective function in each iteration is guaranteed, which is proved in the following subsection. The algorithm for solving the problem (4) is illustrated in Algorithm 1, in which the updates for each parameter are shown in Equations (6)–(9). To check the convergence, the following stopping criteria is used
here η is a preset threshold. In this paper, we set η=10−5.
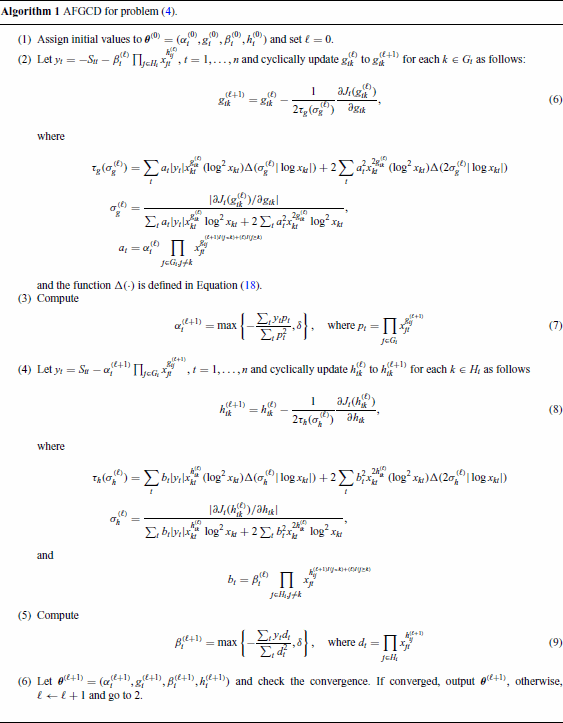
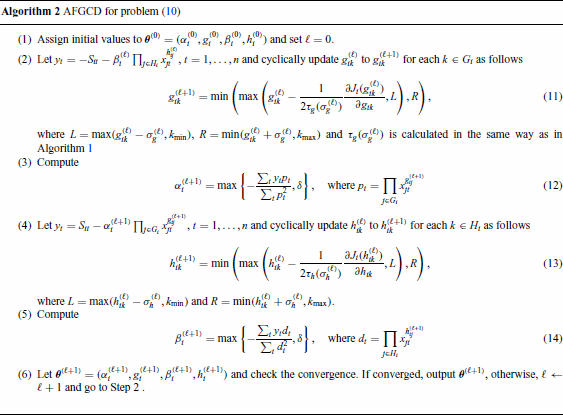
In some situations, from experiments, the range of each parameter is known. Then, more constraints can be added to the problem (4) as follows:
2.3 Descent property
Similar to Seung and Lee (Citation2001) and Li, Wu, Zhang, andWu (2013), we make use of an auxiliary function to prove the nonincrease of the objective function in each iteration.
definition 1
is an auxiliary function of J(h) if there exists
, such that, if
The auxiliary function is useful because of the following lemma.
lemma 1
If F is an auxiliary function, then J is nonincreasing under the update
Proof
Lemma 2 shows a property of the exponential function, which is useful for the following discussions.
lemma 2
For some point h′, the exponential function eh satisfies
Proof
Then, the descent property of Algorithm 1 is shown in the following theorem.
Theorem 1
The updating rules (6)–(9) in Algorithm 1 keep the objective function in problem (4) nonincreasing.
Proof First, we show that the updating rules (6) and (8) can be obtained by minimizing constructed auxiliary functions. Here, we only prove that the rule (8) and the rule (6) can be proved similarly.
Suppose in one iteration, we need to update the parameter to
with all the other parameters fixed. Using the notations in Algorithm 1, the objective function is
For Algorithm 2, we have the same descent property.
Theorem 2
The updating rules (11)–(14) in Algorithm 2 keep the objective function in problem (10) nonincreasing.
Proof Similar to Theorem 1, considering the updating of , the auxiliary function is defined as in Equation (20). The updating rule is obtained by solving
3. Simulation examples
To study the performances of the proposed methods, we apply the algorithms to simulated data and compare the estimated parameters with the corresponding true ones.
3.1 Performances of Algorithm 1
3.1.1 Four-dimensional model
Consider the following S-system of four molecular species (Voit, 2000):
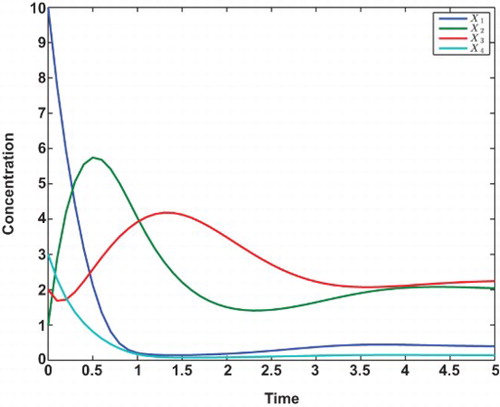
In this example, the data are generated with . The time series data are shown in , from which we can see all states of Xi’s are eventually in the steady states. Algorithm 1 is applied to estimating the parameters from these data with initial values for all parameters chosen by
Table 1. Estimated results of the four-dimensional model from Algorithm 1.
3.1.2 Five-dimensional model
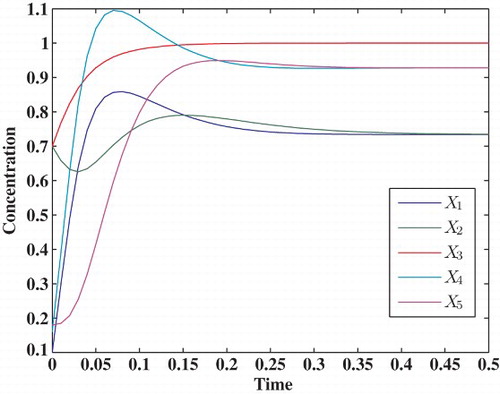
A benchmark five-dimensional model (Vilela et al., Citation2008; Yang, Dent, & Nardini, Citation2012) is considered,
Table 2. Estimated results of the five-dimensional model from Algorithm 1.
3.1.3 Six-dimensional model
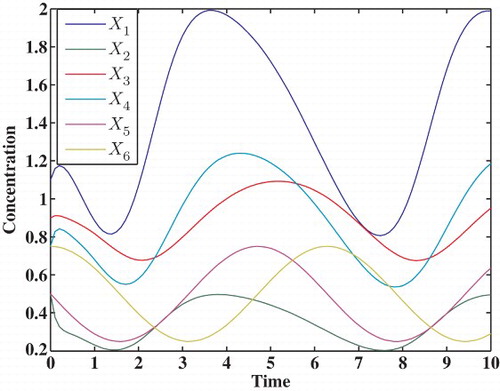
In this example, Algorithm 1 is applied to estimate the parameters in the following six-dimensional S-system Yanget al. (2012):
Table 3. Estimated results of the six-dimensional model from Algorithm 1.
3.2 Performances of Algorithm 2
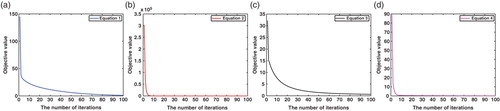
The performances of Algorithm 2 is studied in this example. We apply the algorithm to the data generated from the four-dimensional, five-dimensional, and six-dimensional models, respectively. The time series data we use are exactly the same as above-mentioned examples. Since Algorithm 2 solves the problem (10) which includes a range constraint for each parameter, the initial values for the algorithm are randomly generated in those ranges. In this study, the rate constants are restricted in the range [0.1, 10] and kinetic orders are in the range [−2, 3]. For each model, we run the algorithm for 100 times with different initial values and choose the one with minimum objective value as the final result. The estimated results from Algorithm 2 for each model are reported in the . From the results, it can be seen that estimated parameters for the four-dimensional and six-dimensional models have very small relative errors compared with their corresponding true values. For the five-dimensional model, most parameters are estimated with low errors, while the estimations of the parameters in the third ODE have relatively large errors. This may be due to the limited information contained in the time series data. We can also see that the estimated objective function values for these three models are all very small. All these results show the effectiveness of the proposed algorithm.
Table 4. Estimated results of the four-dimensional model from Algorithm 2.
Table 5. Estimated results of the five-dimensional model from Algorithm 2.
Table 6. Estimated results of the six-dimensional model from Algorithm 2.
4. Conclusions
The S-system is an effective mathematical model to characterize and analyse the molecular biological systems. The parameters in S-systems have significant biological meanings. However, the estimation of these parameters from time series biological data is non-trivial, because of the nonlinearity and complexity of this model. An novel method, the AFGCD, is proposed in this study to estimate the parameters of S-systems. To solve the nonlinear optimization problem involved in the parameter estimation, the proposed method optimizes one parameter at a time. Taking advantage of a property of the exponential function, an auxiliary function is constructed for each kinetic order parameter. We prove that updating the parameter value by optimizing the auxiliary function keeps the objective function nonincreasing. As the auxiliary function is quadratic, the analytical solution exists and therefore the updating rule for each parameter is simple and efficient. Based on this idea, two algorithms are developed: one estimates the parameters in S-system with the only non-negative constraints on rate constants; the other puts constraints both on rate constants and kinetic orders. Their performances are studied in simulation examples, which show that the estimated parameter values from the proposed algorithms are very close to the corresponding true values and the optimized objective functions has very low values. All of these results demonstrate the effectivenesses of the proposed algorithms.
In this study, the topology information of the system is assumed to be known. One direction of the future work is to extend the current method with structure identification method to infer the S-system without knowing the system topology.
This study is supported by Natural Science and Engineering Research Council of Canada (NSERC).
REFERENCES
- Bazaraa, M. S., Sherali, H. D., & Shetty, C. M. (2006). Nonlinear programming: Theory and algorithms. Hoboken, NJ: Wiley-Interscience. ISBN 0471486000.
- Beck, J. V., & Arnold, K. J. (1977). Parameter estimation in engineering and science. New York: Wiley.
- Bertsekas, D. P. (1999). Nonlinear programming. Belmont, MA: Athena Scientific. ISBN 1886529000.
- Chou, I.-C., Martens, H., & Voit, E. (2006). Parameter estimation in biochemical systems models with alternating regression. Theoretical Biology and Medical Modelling, 3(1), 25. ISSN 1742-4682. doi: 10.1186/1742-4682-3-25
- Gardner, T. S., di Bernardo, D., Lorenz, D., & Collins, J. J. (2003). Inferring genetic networks and identifying compound mode of action via expression profiling. Science, 301(5629), 102–105. doi: 10.1126/science.1081900
- Gonzalez, O. R., Kper, C., Jung, K., Naval, P. C., & Mendoza, E. (2007). Parameter estimation using simulated annealing for S-system models of biochemical networks. Bioinformatics, 23(4), 480–486. doi: 10.1093/bioinformatics/btl522
- Ho, S.-Y., Hsieh, C.-H., Yu, F.-C., & Huang, H.-L. (2007). An intelligent two-stage evolutionary algorithm for dynamic pathway identification from gene expression profiles. Computational Biology and Bioinformatics, IEEE/ACM Transactions on, 4(4), 648–704. ISSN 1545-5963. doi: 10.1109/tcbb.2007.1051
- Kikuchi, S., Tominaga, D., Arita, M., Takahashi, K., & Tomita, M. (2003). Dynamic modeling of genetic networks using genetic algorithm and S-system. Bioinformatics, 19(5), 643–650. doi: 10.1093/bioinformatics/btg027
- Klipp, E., Herwig, R., Kowald, A., Wierling, C., & Lehrach, H. (2005, May). Systems biology in practice: Concepts, implementation and application. Weinheim: Wiley-VCH.
- Li, L.-X., Wu, L., Zhang, H.-S., & Wu, F.-X. (2013). A fast algorithm for nonnegative matrix factorization and its convergence. Neural Networks and Learning Systems, IEEE Transactions, PP(99), 1. doi: 10.1109/TNNLS.2013.2280906
- Liu, L.-Z., Wu, F.-X., & Zhang, W.-J. (2012a). Alternating weighted least squares parameter estimation for biological s-systems. 2012 IEEE 6th international conference on systems biology (ISB), Xi'an, China, pp. 6–11. doi: 10.1109/ISB.2012.6314104
- Liu, L.-Z., Wu, F.-X., & Zhang, W. J. (2012b). Inference of biological s-system using the separable estimation method and the genetic algorithm. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 9(4), 955–965. ISSN 1545-5963. doi: 10.1109/TCBB.2011.126
- Seung, D., & Lee, L. (2001). Algorithms for non-negative matrix factorization. Advances in Neural Information Processing Systems, 13, 556–562.
- Vilela, M., Chou, I.-C., Vinga, S., Vasconcelos, A., Voit, E., & Almeida, J. (2008). Parameter optimization in S-system models. BMC Systems Biology, 2(1), 35. ISSN 1752-0509. doi: 10.1186/1752-0509-2-35
- Voit, E. O. (2000, September). Computational analysis of biochemical systems: A practical guide for biochemists and molecular biologists. Cambridge, UK: Cambridge University Press. ISBN 0521785790.
- Voit, E. O., & Almeida, J. (2004). Decoupling dynamical systems for pathway identification from metabolic profiles. Bioinformatics, 20(11), 1670–1681. doi: 10.1093/bioinformatics/bth140
- Wang, H., Qian, L., & Dougherty, E. (2010, March). Inference of gene regulatory networks using S-system: A unified approach. Systems Biology, IET, 4(2), 145–156. ISSN 1751-8849. doi: 10.1049/iet-syb.2008.0175
- Wu, F.-X., & Mu, L. (2009, June). Separable parameter estimation method for nonlinear biological systems. ICBBE 2009. 3rd international conference on bioinformatics and biomedical engineering, Beijing, China, 2009, pp. 1–4. doi: 10.1109/ICBBE.2009.5163379
- Yang, X., Dent, J. E., & Nardini, C. (2012). An S-system parameter estimation method (SPEM) for biological networks. Journal of Computational Biology, 19(2), 175–187. doi: 10.1089/cmb.2011.0269