 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Vaccination against endothelial cells (ECs) lining the tumor vasculature represents one of the most attractive potential cancer immunotherapy options due to its ability to prevent solid tumor growth. Using this approach, target antigens can be derived from ECs and used to develop a universal cancer vaccine. Unfortunately, direct immunization with EC preparations can elicit autoimmune vasculitis in normal tissues. Recently, tumor-induced changes to the human EC surface were described that provided a basis for designing efficient EC-based vaccines capable of eliciting immune responses that targeted the tumor endothelium directly. This review examines these data from the perspective of designing EC-based cancer vaccines for the treatment of all solid tumors, including the antigen composition of vaccine formulations, the selection ECs for antigen derivation, the production and control of antigens, and the method for estimating vaccine efficacy and safety. As the vaccine preparation requires a specifically derived set of natural cell surface antigens, a new vaccine preparation concept was formulated. Antigen compositions prepared according to this concept were named SANTAVAC (Set of All Natural Target Antigens for Vaccination Against Cancer).
Introduction
Despite tremendous progress, effective treatments have yet to be developed for most cancer types. Novel, effective cancer-preventative therapies, especially cancer vaccines, are crucially needed.Citation1 Traditional cancer vaccine formulations are composed of cancer cell antigens.Citation2 However, targeting the tumor endothelium with antiangiogenic vaccines has advantages over targeting cancer cells because the tumor endothelium is genetically stable and has a low probability of acquiring drug resistance.Citation3 The endothelial cell (EC) to cancer cell ratio in tumors can vary between 1:50 and 1:100; hence, the number of targeted cells is much smaller when the tumor endothelium is targeted.Citation4 Destruction of only a few ECs can lead to vascular obstruction and, thus, to the arrest of tumor growth and even tumor destruction because vascular integrity is essential to tumor growth and metastasis.Citation5-8 For these reasons, immunotherapies targeting the tumor vasculature represent a promising approach for preventing solid tumor growth and metastasis.
Among the various approaches used to elicit immunity against tumor endothelium-specific antigens, active immunization with ECs has been the preferred approach over immunotherapies targeting specific epitopes. Cell-based vaccines elicit immune responses targeting a comprehensive array of target cell antigens, including previously undescribed antigens.Citation9–11 Although they have been shown to inhibit the growth of experimental tumors,Citation12–17 cell-based approaches unfortunately also have been associated with the elicitation of autoimmunity in animals and clinical trials.Citation18–23 Experimental autoimmune vasculitis was described after immunization with ECs.Citation24,25 Autoimmune-mediated damage to microvessels (as the primary target of anticancer EC-based vaccination strategies) may lead to systemic damage of the vessels, leading to destruction of the vasculature, internal hemorrhage, and even destruction of internal organs. For these reasons, vaccine specificity is needed for the development of an EC-based vaccine. Specifically, these vaccines must contain distinct antigens from those expressed by ECs in normal tissues, in order to prevent harmful autoimmune responses from being elicited.
EC heterogeneity provides background for vaccine development
EC heterogeneity has been described at the levels of cell morphology, function, gene expression, and antigen composition.Citation26,27 EC phenotypes vary between different organs, as well as between different tissues within the same organ. In addition, the gene expression profile of ECs can be significantly influenced by the tumor.Citation28-30 Tumor cells release growth factors that alter the gene expression profiles of cultured ECs.Citation31-33 Studies have described differences between gene expression profiles in isolated tumor ECs compared to the profiles of ECs harvested from matched tissues.Citation34,35 These data suggest that EC heterogeneity should be considered in the design of EC-based vaccines that can target tumor vessels selectively.
The simplest way to examine the impact of tumor cells on EC heterogeneity is to model the tumor-endothelium interactions in vitro by culturing ECs in the presence of tumor-conditioned medium. Tumor cells release growth factors into the culture medium. These factors can affect the proliferation and protein expression profiles of the ECs. In these experiments, ECs cultured in the presence of supernatants harvested from normal (untransformed) cells are used as controls. Media conditioned by normal cells possess a limited capacity to support cell growth in culture due to a lack of growth factors. Thus, control ECs must be cultured in the presence of endothelial cell growth supplement (ECGS) prepared from brain gland tissue.Citation36
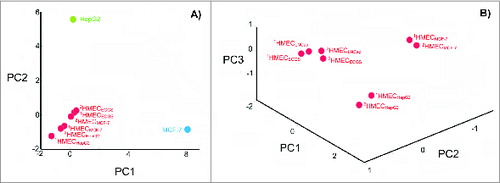
Recently, experiments were performed comparing the expression profiles of cell surface targets between experimental and control cells, which demonstrated that data regarding EC heterogeneity can be applied to vaccine design approaches. Tumor type-specific changes were observed on the surface of cultured human microvascular endothelial cells (HMECs) () in the presence of tumor-conditioned medium collected from different cancer cells.Citation37-40 Changes in the cell surface profiles were characterized by cell proteomic footprinting (CPF), an advanced proteomics approach used to characterize cell phenotypes via mass spectrometric analysis of extracellular surface ().Citation41 Tumor-induced changes in the protein expression profiles of the HMEC surface were estimated on the basis of deviations in the principle component analysis (PCA) plot compared to the typical HMEC phenotype (). The HMEC profiles were grouped together in a distinct location from the profiles of the non-EC controls. Examining the relationships between surface profiles within the HMEC group revealed 3 interesting observations (). First, HMECs from the same tissue had the same surface antigen profile, as indicated by the high similarity between HMEC surface profiles obtained from the same adipose tissue from different donors. Second, tumors induced reproducible tumor type-specific changes in the HMEC surface antigen profile, which ranged from relatively insignificant (e.g., 1HMECLNCap and 2HMECLNCap) to pronounced (e.g., 1HMECHepG2 and 2HMECHepG2). Third, tumor-induced changes in the antigen profile facilitated HMEC escape from cytotoxic T lymphocyte (CTL)-mediated cell death in an in vitro model of human antiangiogenic vaccination.Citation37,39
Figure 1. Endothelial cells (ECs) in cultures. (A) Representative human microvascular ECs (HMECs) isolated from adipose tissue and used to prepare the SANTAVAC preparation. HMECs had numerous cytoplasmic extensions and/or a cobblestone-like morphology typical of adipose-derived microvascular ECs.Citation89 Images were obtained using a Leica DM5000B microscope. Cells were isolated by using magnetic beads linked to anti-CD31 antibodies (visible on the cells). (B) Example of ECs sprouting around cancer cells under angiogenic stimuli. Artwork prepared using data obtained from a multiphoton microscopy image of a tumor spheroid in a collagen matrix.Citation90
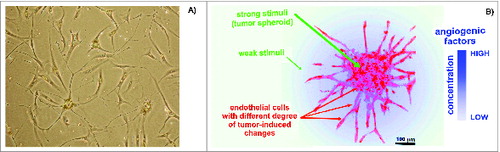
Figure 2. Cell proteomic footprinting. (A) Adherent cell culture after washing away traces of culture medium and subsequently treated with a protease. Released fragments of the cell surface proteins were collected and subjected to mass spectrometry analysis. The set of obtained peptide molecular weights represents the cell culture proteomic footprint. (B) Examples of cell proteomic footprints for non-ECs (MCF-7 and HepG2) and HMECs induced to grow in the presence of stimuli provided from normal tissue (HMECECGS) or cancer cells (HMECHepG2). Adapted from.Citation37,41
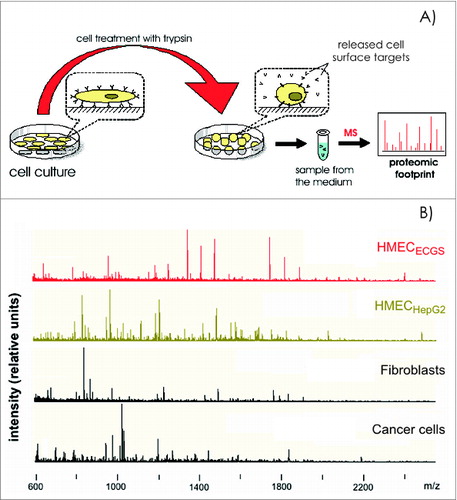
Figure 3. Degree of change in the HMEC surface antigen expression profile after incubation in the presence of tumor-conditioned medium. (A) Principle component analysis (PCA) of cell surface profiles obtained from HMECs and control non-ECs (HepG2 and MCF-7) that were projected in the space of the first 2 principal components. (B) PCA of cell surface profiles obtained only for HMECs projected into the space of the first 3 principal components. Cell surface profiles are shown for HMECs stimulated to grow in the presence of EC growth supplement (1HMECECGS and 2HMECECGS), MCF-7 cell-conditioned medium (1HMECMCF-7 and 2HMECMCF-7), LNCap cell-conditioned medium (1HMECLNCap and 2HMECLNCap), or HepG2 cell-conditioned medium (1HMECHepG2 and 2HMECHepG2). Superscript numbers correspond to different donors used to establish HMEC primary cultures.
Taken together, these findings provide useful information regarding the design of efficient cancer vaccines. Specifically, by constructing vaccines with compositions of antigens divergent from those expressed by normal ECs, one can avoid the elicitation of autoimmune reactions.
Source and composition of antigens used for the development of an EC-based universal cancer vaccine
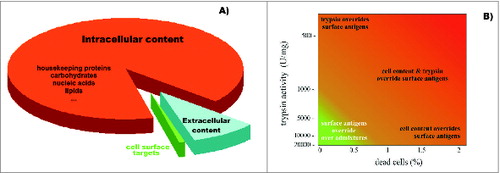
Vaccine design and development processes have focused on using cells as the source of native antigens for eliciting immune responses against target cells.Citation11,42,43 Whole cells possess a set of cell-surface antigens that, ideally, should be prioritized for vaccine design.Citation44,45 However, whole cells also express abundant intracellular antigens that are ubiquitous to all mammalian cells and could elicit various adverse autoimmune responses ().Citation40 Fortunately, immune access to cell surface target antigens (e.g., by antibodies and cytotoxic cells) suggests that these targets would also be similarly accessible to proteases, whose action products can be isolated after in vitro proteolytic cleavage.Citation38,46,47 In a previous study using an in vitro model of cancer vaccination, trypsinizing the surface of cancer cell line (MCF-7) cells yielded a digest containing 0.7% of the total cell protein. When the trypsin digest was used to stimulate a cytotoxic antitumor response in vitro, 10–40% more cancer cells were killed compared to the response stimulated with whole cells.Citation46 Furthermore, the digest's composition, comprising the proteolytically cleaved cell surface targets, were directly related to the killing rate of target cells in a cytotoxicity assay (CTA).Citation41,47
Figure 4. Diagram representing (A) the relative ratio of surface targets that are accessible for humoral and cytotoxic immunity, and the remaining cellular content that is undesired for preparation of cell-based vaccines; and (B) the effects of trypsin impurities and cell death rates on the preparation of cell surface antigens. The lighter region in the lower left corner represents conditions where cell-surface antigen preparations are compatible with the preparation of SANTAVAC vaccines. Trypsin purity is reflected by the levels of enzymatic activity. Adapted from Lokhov.Citation40
These findings suggest that a set of cell surface targets exists, which represents the cell's ‘antigenic essence’ and can be used in vaccine formulations. CPF can be used to characterize the compositions of different sets of cell surface targets for vaccine preparations, in order to target immune responses against antigens related to the tumor vasculature rather than normal tissue.Citation41,47 Indeed, in the above study, the killing rate of target HMECs in the CTA was directly related to the similarity between the CPF results of the target HMECs and the HMECs used to generate antigens for targeting immune responses. This observation serves as the basis for the development of EC-based cancer vaccines with strictly defined characteristics and comprised of proteolytically cleaved HMEC surface targets as a means of eliciting immunity against tumor vasculature-associated antigens.
Findings from previous studies have motivated the use of primary HMECs for generating vaccine targets. The microvasculature was previously shown to be involved in tumor angiogenesis and microvasculature-derived ECs exhibited functional differences compared to large vessel-derived ECs,Citation48,49 including differences in the response to growth stimulatorsCitation50,51 and extracellular protein expression patterns.Citation52–54 To generate natural EC phenotypes and appropriate responses of ECs to growth stimuli, it is essential that primary HMECs be used rather than cell lines. Natural EC phenotypes are difficult to generate when using immortalized cell lines with intrinsic proliferative properties due to virus transfection. Further investigation of the phenotypic response of immortalized HMECs to tumor growth stimuli may highlight the applicability of these cells for vaccine design. Subcutaneous fat tissue seems to be the most suitable source of HMECs, based on observations that HMECs lining tumor vessels are derived from surrounding tumor tissues. However, primary and metastatic tumors can occur in many different tissues throughout the body. Therefore, it is rational to derive HMECs from the abundant and easily accessible subcutaneous fat tissue, which can be obtained easily from biopsy material or waste after liposuction.
Designing universal cancer vaccines with defined safety and efficacy
In the context of designing universal cancer vaccines (UCVs), the most promising finding has been the nonspecific influence of cancer cells on the heterogeneity of HMEC surface antigens. Cancer cells change the HMEC surface antigens through a cancer cell type-independent mechanism, suggesting that HMEC heterogeneity is a result of differences in the strength of growth signals.Citation37 This observation led researchers to hypothesize that the tumor would affect the HMEC surface profile in the same manner in vivo as in vitro, and that tumor-induced changes in the HMEC antigenic profile would be a consequence of the magnitude of the growth stimulus. Distance also influences the tumor's effects on the EC antigenic profile.
If stimuli of different strengths are present simultaneously in tumors in vivo, then due to gradual diminishing growth stimuli as it relates to the distance from the tumor cells, it can be expected that HMECs with different target surface profiles will also be present in the tumor-associated vasculature (). This assumption is very important for vaccine design because the destruction of any type of HMEC at any location in the tumor vasculature or in vessels approaching the tumor should lead to vessel obstruction and arrest of tumor growth. Thus, CTA results involving different target HMECs with tumor-induced phenotypes can be directly attributed to the design of the tumor type-independent HMEC-based vaccine (i.e., UCV design).
In a previous study, HMEC targets, stimulated to grow by human prostate adenocarcinoma (LNCap) cells, were effectively killed through the immunity elicited in response to antigens on the surface of HMECs, stimulated through growth in the presence of human hepatocellular carcinoma (HepG2) cells. These findings supported the in vitro design of an UCV with a targeting efficacy of 2.45.Citation39 Herein, efficacy is defined as the fold difference between the number of killed target HMECs stimulated to grow by tumor cells compared to the number of killed HMEC targets stimulated to grow by normal tissue cells. This efficacy provides a therapeutic window in which tumor HMEC cells could be killed before normal tissue HMECs are adversely affected.
Diverse HMEC targets are present in vivo, and diverse antigen compositions can be prepared from HMEC cultures by providing tumor stimuli of different strengths. Nevertheless, in vitro studies are limited in their ability to describe vaccine efficacy by the availability of specified cell targets and antigens (e.g., CTA). Therefore, for a better characterization of vaccine efficacy, the CTA data were examined using an approximation of the dependence of the target cell killing rate on the similarity of target cell surface profiles to the surface profile of cells used to generate antigens for the target immune response in CTA. These data suggested that an efficient autologous vaccine can be generated by utilizing the surface antigens of HMEC cultures if their tumor-induced cell surface profile and the profile of target HMECs are quite similar (i.e., correlation coefficient for their CPF ≥ 0.82). In this scenario, the efficacy of the autologous vaccine will exceed 18 (i.e., 18 tumor ECs will be destroyed before one EC in normal tissue is destroyed).Citation39
When the similarity of the cell surface profiles of the target HMECs and the HMECs used as a vaccine were plotted against viable target cell counts in a CTA (), points on this plot could be linearly approximated with R2 values equal or almost equal to one. Thus, the cytotoxicity of CTL was predefined by the cell surface profiles and could be described by the equation:
Figure 5. Cytotoxicity assays (CTAs). Data represent the cytotoxicity of effector CTLs against target HMECs, plotted vs. the similarity between surface profiles of target cells and cells used as a source of antigens for eliciting the targeted immune response. Data represent the mean value of 3 independent measurements. The similarity between the cell surface profiles is presented as the correlation coefficient r between corresponding proteomic footprints. 1▸2 – First letter corresponds to HMECs used as a source of antigens for eliciting immune response in the CTA, and the second letter corresponds to the target HMECs used in the same CTA. Red and brown letters correspond to autologous and allogeneic antigens, respectively. Different letters are used to identify HMECs stimulated to grow in the presence of an EC growth supplement (G), MCF-7 cell-conditioned medium (M), LNCap cell-conditioned medium (L), or HepG2 cell-conditioned medium (H). Data were scaled to bring all controls to equal values (25,000 cells, see CONTROL line). Dashed lines show examples of linear dependence (n = k*r + b) between CTA data n and r. All data in the plot were described by linear equations, and the variations of the coefficients (k and b) were interdependent.Citation37
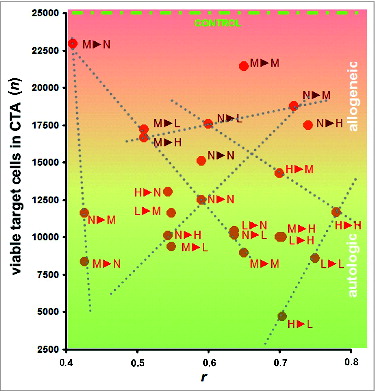
An additional consideration in vaccine preparation is whether auto- or alloantigens should be used. Although autoantigens elicit high killing rates of the target cells after vaccination, using allogeneic antigens allows the patient's own biomaterial to be excluded from vaccine preparation, thereby simplifying research and development activities and facilitating vaccine implementation in clinical practice. The data presented in indicate that targeting immune response with the alloantigens may be sufficient for vaccine efficacy, even with the low target cell killing rates. Correctly prepared alloantigen compositions that induce low killing rates also exhibited low HMEC killing rates when HMECs were stimulated to proliferate by normal cells. In one case, a high killing rate was observed for HepG2-stimulated target HMECs, which targeted the immune response to HMEC allogenic surface antigens that were stimulated to grow in the presence of human breast adenocarcinoma (MCF-7) cells. Moreover, the target cell killing rate was directly related to the in vitro design of the UCV, with a targeting efficacy of 4 (variables for the equation are derived from ):
This efficacy provides a therapeutic window in which tumor HMEC cells could be killed before HMECs of normal tissues are adversely affected.
The CTA data obtained for alloantigens can be approximated to define the dependence of target cell killing rates on the similarity between the surface profiles of the target cells and the cells used to generate alloantigens. However, the CTA data supporting this approach are currently insufficient. If additional CTA and CPF research were to be conducted, then CTA data for alloantigens could be better estimated to define the maximum efficacy for alloantigens.
SANTAVAC concept
Compositions of HMEC antigens intended for vaccination are based on a specifically derived set of defined natural cell surface antigens. These formulations are referred to as the Set of All Natural Target Antigens for Vaccination Against Cancer (SANTAVAC). SANTAVAC-based UCV formulations can be mixed with different adjuvants, and their immunogenicity and safety can be tested in vivo. The unique SANTAVAC vaccine design approach is defined by the following basic statements:
Cancer vaccines that target tumor endothelium have advantages over vaccines targeting cancer cells because cancer cells demonstrate a high capacity for escaping immune responses.
Intracellular antigens are less accessible to humoral and cytotoxic immune responses; therefore, cell surface targets are prioritized for preparing cancer vaccines.
Cancer vaccines should represent the set of cell surface targets representing the ‘antigenic essence’ of cells.
The decision to target immune responses to tumor vasculature-associated antigens, rather than normal tissue vasculature-associated antigens, is based on differences between EC surface targets.
To identify differences between EC surface targets, CPF is utilized; the resulting footprints represent snapshots of cell surface targets.
Immune access to cell surface targets (e.g., by antibodies and cytotoxic cells) suggests that these targets would also be similarly accessible to proteases whose action products can be isolated after in vitro proteolytic cleavage.
Cell surface targets that are proteolytically cleaved and collected from live HMECs with tumor-induced phenotypes represent antigen compositions consisting of a comprehensive set of natural antigens prioritized for anti-cancer vaccination and named SANTAVAC.
The efficacy and safety of SANTAVAC vaccines are directly defined by the cell surface profile of the HMECs used to generate the antigens.
To design SANTAVAC vaccines with defined efficacy and safety, CPF and CTA must be performed on the target HMECs and HMECs used to generate antigens usable in vaccine formulations.
Although autoantigens induce high target cell killing rates after vaccination, alloantigens are also highly efficient through the lower killing rate of HMECs present in normal tissues.
The in vitro-measured SANTAVAC efficacy may be increased in vivo because the destruction of only one EC in the tumor vasculature may lead to destruction of up to 100 cancer cells in a tumor.
SANTAVAC represents sets of antigenic compositions intended for preparing a family of UCVs to be administered with adjuvants, for which the dosage and vaccination schedules need to be defined.
The antigenic composition represents the essence of any vaccine and defines the targets to which immune response should be induced. Numerous approaches can be taken to prepare vaccines containing the same antigenic composition that differ only in the antigen dose and adjuvants used. Different protective effects and tumor responses can be achieved, depending on the vaccine regimen, patient, and tumor type/stage. Moreover, the SANTAVAC concept covers basic principals for the design of SANTAVAC vaccines; therefore, the immunogenicity, and especially safety in relation to vasculature of normal tissues should be obligatory tested in subsequent in vivo studies for each SANTAVAC vaccine.
The SANTAVAC approach can be considered as a universal solution for preparing UCVs that differ from each other and in the intended target cancer. However, several important peculiarities of vaccine development involving the use of proteolytically cleaved cell surface antigens need to be emphasized. Although the protease-based isolation of cell surface moleculesCitation55–58 and the use of cancer cells for vaccination have long been described, the approach has some important limitations in terms of protease purity, protease-mediated cellular damage, and antigen preparations.
SANTAVAC contamination associated with protease impurity
One of the earliest studies examining the effect of trypsin on cells was conducted on tumor cells in 1958. This study demonstrated a significant loss of cell mass (up to 20%) without any apparent change in cell viability.Citation59 Subsequent studies examining the effect of protease treatment on cell integrity used cells in suspensionCitation60–62 or from intact tissues.Citation63,64 In both cases, the integrity of the treated cells after trypsinization was sufficient to maintain the cell viability.Citation59,62,65,66 However, other methods for treating intact tissues with proteases have been demonstrated to cause considerable damage to the treated cells.Citation63 A study by Anghilery and Dermietzel demonstrated that trypsinization freed up to 10% of the cell material, including significant amounts of lipid and nucleic acids from intracellular lipo- and nucleoproteins.Citation57 When a mild treatment of cells with 0.1% trypsin was performed, up to 10.4% of the cellular RNA and 11.4% of the cellular DNA were recovered, consistent with a lysis rate of ∼11%.Citation67 However, when trypsin of a higher purity was used, less than 2% of cells were lysed.Citation68 Accordingly, trypsin impurities were identified to be responsible for the increased cell damage observed. More recent studies utilized a highly purified trypsin with an activity of 15,000 U/mg and resulted in cell lysis rates of less than 0.1%.Citation46 These data demonstrate that mammalian cells could be treated with trypsin without inducing lysis if highly pure trypsin is used, and they provide the background for preparing SANTAVAC formulations consisting of pure cell surface targets.
Another aspect to consider regarding the preparation of SANTAVAC vaccines is the contamination of antigens by trypsin itself. It is possible that the collected antigens could be contaminated with the trypsin used in their generation. Trypsin at a working concentration of ∼300 μg/mL has previously been widely used to cleave cell surface material.Citation56,69 A recent study showed that the treatment of 5 millions of cancer cells with 1 mL of trypsin solution yielded only 10 to 20 μg of cell surface glycoprotein fragments,Citation46,56 representing ∼1% of the total protein content of a cell. These results and others indicate that the trypsin concentration used to cleave cell surface antigens significantly exceeds that of the cleaved cell surface antigens. Correspondingly, because trypsin contains numerous impurities, including other types of proteases, differently degraded forms of trypsin, and trypsin autolysis products,Citation70 the identification of cell surface antigens derived from trypsin treatment requires complex analytical methods.Citation56,70–75
However, the need for antigen identification is more relevant to the characterization of individual antigens than to vaccine production. It is important that highly purified trypsin be used in the preparation of cell surface proteins for vaccines. Cells treated with highly purified trypsin resulted in a solution of cell specific peptides that were not contaminated with proteases, cytosolic proteins, or serum from the cell growth medium.Citation41 These results confirm the idea that highly purified trypsin can provide a pure sample of cell surface targets that can be used in SANTAVAC vaccines.
SANTAVAC contamination associated with cell damage
Animal cells are sensitive to fluid shearing in serum-free medium.Citation76–80 To obtain pure samples of cell surface antigens, cells are treated with protease in serum-free medium. Consequent damage to the cell membrane leads to the release of its intracellular contents. As shown in , the amount of cell surface antigens obtained from 100 cancer cells is comparable to the quantity of intracellular molecules contained within a single cell. In a study by Lau and Tchao (2007), Nara Bladder Tumor (NBT) II cells exhibited different cell damage due to fluid shear depending on the cell grown conditions (from 5% to 56%).Citation77 Therefore, a critical aspect to consider in the preparation of cell surface antigens is minimizing the destruction of cells due to fluid shearing. When protocol conditions were optimized and careful manipulation of the cells was maintained, an observed adenocarcinoma cell death rate of less than 0.1% was achieved after trypsinization.Citation46 For cell cultures that are relatively sensitive to fluid shear stress, cell growth conditions may need to be optimized and cyto-protectants applied to prevent cell damage in serum-free medium and to decrease the rate of cell death during manipulations.Citation76
Thus, the careful treatment of live cells with highly purified proteases facilitates the collection of cell surface antigens (i.e., SANTAVAC) with minimal contamination of undesired intracellular contents. summarizes the influence of protease impurities and cell death rates associated with fluid shearing on the purity of cell surface antigens collected. Recent publicationsCitation37,38,41 have further demonstrated how cell surface antigens can be obtained from HMECs after trypsinization. Other proteases may also be able to be used to cleave proteins from the cell surface, but their ability to generate SANTAVAC preparations would need to be confirmed. Proteases should generate peptides with preserved immunogenicity and length compatible with analysis using CPF. In as much as CPF relates to proteomics, and tryptic peptides form specific mass spectrometry signatures of biologic objects, trypsin is also considered a crucial protease for generating SANTAVAC preparations.
The SANTAVAC approach versus other candidates for developing UCVs
There have been several attempts to prepare UCVs, including the better-known vaccines targeting telomerase and mucin. Herein, we consider these approaches from the perspective of the SANTAVAC concept. Telomerase is highly expressed in almost all cancer types, whereas its expression in normal tissues is restricted. Telomerase activity is indispensable for tumor immortalization and growth; therefore, the catalytic and rate-limiting subunit of telomerase (hTERT) is an attractive target for UCV development.Citation81 For example, the recently concluded TeloVac trial in the UK aimed to determine if adding GV1001, a peptide vaccine representing a 16-aa hTERT sequence,Citation82 to gemcitabine and capecitabine chemotherapy regimens would extend the survival time for patients with advanced pancreatic cancer. Although the SANTAVAC concept does not consider immunization against telomerase to be a favorable approach because telomerase is an intracellular enzyme, hTERT can be displayed on the surface of tumor cells in the context of major histocompatibility complex (MHC) class I molecules.
ImMucin in another immunotherapeutic approach that is often inaccurately referred to as a UCV. ImMucin targets mucin 1, a molecule present in 90% of all cancers. ImMucin is a 21-mer synthetic vaccine composed of the entire signal peptide domain of the cell surface-associated mucin 1.Citation83 Overexpression of mucin 1 is associated with many cancers.Citation84 For this reason, mucin 1 is considered a target for cancer therapyCitation85 and a candidate for developing a UCV.Citation86
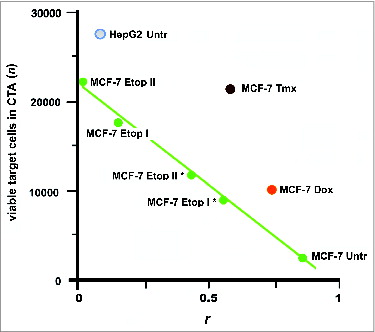
Even though the TeloVac and ImMucin were designed to target different types of cancers and they can be considered as UCVs. However, their efficacy would be diminished if the cancer returned after vaccination. One study found that the specific cell surface antigens of cancer cells were substantially modified under the selective pressures of drug treatment.Citation47 These induced changes may be sufficient to allow the cancer cells to escape from the immune response ().
Figure 6. Escape of cancer cells from the immune response in CТA as a result of cell surface profile changes induced by the selective pressure of drug treatment. These changes can lead to a loss of vaccine efficacy based on cancer cell antigens. Points are presented for MCF-7 and HepG2 cells that were untreated (MCF-7 Untr and HepG2) or MCF-7 cells treated as follows: IC96 doses of doxorubicin (MCF-7 Dox) or tamoxifen (MCF-7 Tmx); a single dose of IC96 etoposide (MCF-7 Etop I) or IC50 etoposide (MCF-7 Etop I*); and two separate doses of IC96 etoposide (MCF-7 Etop II) or IC50 etoposide (MCF-7 Etop II*). Linear approximations for MCF-7 Untr and MCF-7 Etop I, II, I*, and II* (green line) are shown. The average number of viable cells in 3 wells is presented. Correlation values (coefficient r) were calculated for MCF-7 and HepG2 cell surface profiles used to generate antigens for eliciting immune response and the surface profile of target MCF-7 cells used in same CTA. Adapted from.Citation47
Conclusion and Perspectives
The data described in this report indicate that SANTAVAC is a highly effective vaccine approach in vitro. The approach appears to be devoid of the evident shortcomings associated with other approaches and represents a highly promising antigenic composition for developing UCVs. By using SANTAVAC preparations, safe and efficacious UCVs can be developed from cell surface targets collected from primary HMEC cultures stimulated to grow in the presence of cancer cells. The nature and composition of the cell surface targets can be confirmed by using CPF. Future studies will be required, aimed at testing the vaccine efficacy using SANTAVAC. These studies should include the application of animal models and the selection of adjuvant and vaccination schedules. Previous animal and human studies have already demonstrated the capability of in vitro-induced specific cytotoxic cells to mediate in vivo protection against tumor challenge.Citation87,88 These reports and CТA data described in this paper suggest that highly effective UCVs can be developed by using the SANTAVAC approach.
Disclosure of Potential Conflicts of Interest
PGL declares that he has Eurasian, Japanese, Korean, and Chinese patents, as well as a pending European patent, related to the cancer vaccine preparation method described in this paper.
Funding
This work was funded by ZAO BioBohemia (Moscow, Russia).
References
- Lollini P-L, Cavallo F, Nanni P, Forni G. Vaccines for tumour prevention. Nat Rev Cancer 2006; 6:204-16; PMID:16498443; http://dx.doi.org/10.1038/nrc1815
- Schlom J. Therapeutic cancer vaccines: current status and moving forward. J Natl Cancer Inst 2012; 104:599-613; PMID:22395641; http://dx.doi.org/10.1093/jnci/djs033
- Boehm T, Folkman J, Browder T, O’Reilly MS. Antiangiogenic therapy of experimental cancer does not induce acquired drug resistance. Nature 1997; 390:404-7; PMID:9389480; http://dx.doi.org/10.1038/37126
- Bussolino F, Arese M, Audero E, Giraudo E, Marchiò S, Mitola S, Primo L, Serini G. Aspects of Tumor Angiogenesis. In: Preziosi L, editor. Cancer Modelling and Simulation. London: Chapman and Hall/CRC; 2003. page 1-22.
- Folkman J. Tumor angiogenesis: therapeutic implications. N Engl J Med 1971; 285:1182-6; PMID:4938153; http://dx.doi.org/10.1056/NEJM197108122850711
- Folkman J. What is the evidence that tumors are angiogenesis dependent? J Natl Cancer Inst 1990; 82:4-6; PMID:1688381; http://dx.doi.org/10.1093/jnci/82.1.4
- Pluda JM. Tumor-associated angiogenesis: mechanisms, clinical implications, and therapeutic strategies. Semin Oncol 1997; 24:203-18; PMID:9129690
- Carmeliet P, Jain RK. Angiogenesis in cancer and other diseases. Nature 2000; 407:249-57; PMID:11001068; http://dx.doi.org/10.1038/35025220
- Copier J, Dalgleish A. Overview of tumor cell-based vaccines. Int Rev Immunol 2006; 25:297-319; PMID:17169778; http://dx.doi.org/10.1080/08830180600992472
- Old LJ. Cancer vaccines: an overview. Cancer Immun a J Acad Cancer Immunol 2008; 8 Suppl 1:1.
- Chiang CLL, Benencia F, Coukos G. Whole tumor antigen vaccines. Semin Immunol 2010; 22:132-43.
- Wei YQ, Wang QR, Zhao X, Yang L, Tian L, Lu Y, Kang B, Lu CJ, Huang MJ, Lou YY, et al. Immunotherapy of tumors with xenogeneic endothelial cells as a vaccine. Nat Med 2000; 6:1160-6; PMID:11017149; http://dx.doi.org/10.1038/80506
- Corsini E, Gelati M, Calatozzolo C, Alessandri G, Frigerio S, De Francesco M, Poiesi C, Parati E, Croci D, Boiardi A, et al. Immunotherapy with bovine aortic endothelial cells in subcutaneous and intracerebral glioma models in rats: Effects on survival time, tumor growth, and tumor neovascularization. Cancer Immunol Immunother 2004; 53:955-62; PMID:15449042; http://dx.doi.org/10.1007/s00262-004-0529-5
- Okaji Y, Tsuno NH, Kitayama J, Saito S, Takahashi T, Kawai K, Yazawa K, Asakage M, Hori N, Watanabe T, et al. Vaccination with autologous endothelium inhibits angiogenesis and metastasis of colon cancer through autoimmunity. Cancer Sci 2004; 95:85-90; PMID:14720332; http://dx.doi.org/10.1111/j.1349-7006.2004.tb03175.x
- Chen X-Y, Zhang W, Zhang W, Wu S, Bi F, Su Y-J, Tan X-Y, Liu J-N, Zhang J. Vaccination with viable human umbilical vein endothelial cells prevents metastatic tumors by attack on tumor vasculature with both cellular and humoral immunity. Clin Cancer Res 2006; 12:5834-40; PMID:17020991; http://dx.doi.org/10.1158/1078-0432.CCR-06-1105
- Okaji Y, Tsuno NH, Saito S, Yoneyama S, Tanaka M, Nagawa H, Takahashi K. Vaccines targeting tumour angiogenesis-a novel strategy for cancer immunotherapy. Eur J Surg Oncol 2006; 32:363-70.
- Scappaticci FA, Nolan GP. Induction of anti-tumor immunity in mice using a syngeneic endothelial cell vaccine. Anticancer Res 2003; 23:1165-72; PMID:12820367
- Ludewig B, Ochsenbein AF, Odermatt B, Paulin D, Hengartner H, Zinkernagel RM. Immunotherapy with dendritic cells directed against tumor antigens shared with normal host cells results in severe autoimmune disease. J Exp Med 2000; 191:795-804; PMID:10704461; http://dx.doi.org/10.1084/jem.191.5.795
- Dudley ME, Wunderlich JR, Robbins PF, Yang JC, Hwu P, Schwartzentruber DJ, Topalian SL, Sherry R, Restifo NP, Hubicki AM, et al. Cancer regression and autoimmunity in patients after clonal repopulation with antitumor lymphocytes. Science 2002; 298:850-4; PMID:12242449
- Phan GQ, Yang JC, Sherry RM, Hwu P, Topalian SL, Schwartzentruber DJ, Restifo NP, Haworth LR, Seipp CA, Freezer LJ, et al. Cancer regression and autoimmunity induced by cytotoxic T lymphocyte-associated antigen 4 blockade in patients with metastatic melanoma. Proc Natl Acad Sci U S A 2003; 100:8372-7; PMID:12826605
- Maker A V, Phan GQ, Attia P, Yang JC, Sherry RM, Topalian SL, Kammula US, Royal RE, Haworth LR, Levy C, et al. Tumor regression and autoimmunity in patients treated with cytotoxic T lymphocyte-associated antigen 4 blockade and interleukin 2: a phase I/II study. Ann Surg Oncol 2005; 12:1005-16; PMID:16283570
- Attia P, Phan GQ, Maker A V, Robinson MR, Quezado MM, Yang JC, Sherry RM, Topalian SL, Kammula US, Royal RE, et al. Autoimmunity correlates with tumor regression in patients with metastatic melanoma treated with anti-cytotoxic T-lymphocyte antigen-4. J Clin Oncol 2005; 23:6043-53; PMID:16087944
- Finn OJ. Cancer vaccines: between the idea and the reality. Nat Rev Immunol 2003; 3:630-41; PMID:12974478; http://dx.doi.org/10.1038/nri1150
- Hart MN, Sadewasser KL, Cancilla PA, DeBault LE. Experimental autoimmune type of vasculitis resulting from activation of mouse lymphocytes to cultured endothelium. Lab Invest 1983; 48:419-27; PMID:6834786
- Matsuda M. Experimental glomerular tissue injury induced by immunization with cultured endothelial cell plasma membrane. Acta Pathol Jpn 1988; 38:823-39; PMID:3055806
- Aird WC. Phenotypic heterogeneity of the endothelium: I. Ssructure, function, and mechanisms. Circ Res 2007; 100:158-73; PMID:17272818; http://dx.doi.org/10.1161/01.RES.0000255691.76142.4a
- Aird WC. Phenotypic heterogeneity of the endothelium: II. representative vascular beds. Circ Res 2007; 100:174-90; PMID:17272819; http://dx.doi.org/10.1161/01.RES.0000255690.03436.ae
- St Croix B, Rago C, Velculescu V, Traverso G, Romans KE, Montgomery E, Lal A, Riggins GJ, Lengauer C, Vogelstein B, et al. Genes expressed in human tumor endothelium. Science 2000; 289:1197-202; PMID:10947988; http://dx.doi.org/10.1126/science.289.5482.1197
- Khodarev NN, Yu J, Labay E, Darga T, Brown CK, Mauceri HJ, Yassari R, Gupta N, Weichselbaum RR. Tumour-endothelium interactions in co-culture: coordinated changes of gene expression profiles and phenotypic properties of endothelial cells. J Cell Sci 2003; 116:1013-22; PMID:12584245; http://dx.doi.org/10.1242/jcs.00281
- Bhati R, Patterson C, Livasy CA, Fan C, Ketelsen D, Hu Z, Reynolds E, Tanner C, Moore DT, Gabrielli F, et al. Molecular characterization of human breast tumor vascular cells. Am J Pathol 2008; 172:1381-90; PMID:18403594; http://dx.doi.org/10.2353/ajpath.2008.070988
- Hellebrekers DMEI, Castermans K, Viré E, Dings RPM, Hoebers NTH, Mayo KH, Oude Egbrink MGA, Molema G, Fuks F, van Engeland M, et al. Epigenetic regulation of tumor endothelial cell anergy: silencing of intercellular adhesion molecule-1 by histone modifications. Cancer Res 2006; 66:10770-7; PMID:17108113; http://dx.doi.org/10.1158/0008-5472.CAN-06-1609
- Hellebrekers DMEI, Jair K-W, Viré E, Eguchi S, Hoebers NTH, Fraga MF, Esteller M, Fuks F, Baylin SB, van Engeland M, et al. Angiostatic activity of DNA methyltransferase inhibitors. Mol Cancer Ther 2006; 5:467-75; PMID:16505122; http://dx.doi.org/10.1158/1535-7163.MCT-05-0417
- Hellebrekers DMEI, Melotte V, Viré E, Langenkamp E, Molema G, Fuks F, Herman JG, Van Criekinge W, Griffioen AW, van Engeland M. Identification of epigenetically silenced genes in tumor endothelial cells. Cancer Res 2007; 67:4138-48; PMID:17483324; http://dx.doi.org/10.1158/0008-5472.CAN-06-3032
- Unger RE, Oltrogge JB, von Briesen H, Engelhardt B, Woelki U, Schlote W, Lorenz R, Bratzke H, Kirkpatrick CJ. Isolation and molecular characterization of brain microvascular endothelial cells from human brain tumors. In Vitro Cell Dev Biol Anim 2002; 38:273-81; PMID:12418924; http://dx.doi.org/10.1290/1071-2690(2002)038<0273:IAMCOB>2.0.CO;2
- Bussolati B, Deambrosis I, Russo S, Deregibus MC, Camussi G. Altered angiogenesis and survival in human tumor-derived endothelial cells. FASEB J 2003; 17:1159-61; PMID:12709414
- Maciag T, Cerundolo J, Ilsley S, Kelley PR, Forand R. An endothelial cell growth factor from bovine hypothalamus: identification and partial characterization. Proc Natl Acad Sci U S A 1979; 76:5674-8; PMID:293671; http://dx.doi.org/10.1073/pnas.76.11.5674
- Lokhov PG, Balashova EE. Tumor-induced endothelial cell surface heterogeneity directly affects endothelial cell escape from a cell-mediated immune response in vitro. Hum Vaccin Immunother 2013; 9:198-209; PMID:23442592; http://dx.doi.org/10.4161/hv.22828
- Balashova EE, Lokhov PG. Proteolytically-cleaved fragments of cell surface proteins stimulate a cytotoxic immune response against tumor-activated endothelial cells in vitro. J Cancer Sci Ther 2010; 2:126-131; http://dx.doi.org/10.4172/1948-5956.1000037
- Lokhov PG, Balashova EE. Universal cancer vaccine: an update on the design of cancer vaccines generated from endothelial cells. Hum Vaccin Immunother 2013; 9:1549-52; PMID:23571178; http://dx.doi.org/10.4161/hv.24300
- Lokhov PG, Balashova EE. Cellular cancer vaccines: an update on the development of vaccines generated from cell surface antigens. J Cancer 2010; 1:230-41; PMID:21151581; http://dx.doi.org/10.7150/jca.1.230
- Lokhov P, Balashova E, Dashtiev M. Cell proteomic footprint. Rapid Commun Mass Spectrom 2009; 23:680-2; PMID:19184978; http://dx.doi.org/10.1002/rcm.3928
- Thompson PL, Dessureault S. Tumor cell Vaccines. In: Shurin MR, Smolkin YS, editors. Immune-Mediated Diseases From Theory to Therapy. New York: Springer; 2007. page 345-55.
- De Gruijl TD, Van Den Eertwegh AJM, Pinedo HM, Scheper RJ. Whole-cell cancer vaccination: From autologous to allogeneic tumor- and dendritic cell-based vaccines. Cancer Immunology, Immunotherapy. 2008; 57:1569-77.
- Cheever MA, Allison JP, Ferris AS, Finn OJ, Hastings BM, Hecht TT, Mellman I, Prindiville SA, Viner JL, Weiner LM, et al. The prioritization of cancer antigens: a national cancer institute pilot project for the acceleration of translational research. Clin Cancer Res 2009; 15:5323-37; PMID:19723653; http://dx.doi.org/10.1158/1078-0432.CCR-09-0737
- Lang JM, Andrei AC, McNeel DG. Prioritization of cancer antigens: keeping the target in sight. Expert Rev Vaccines 2009; 8:1657-61; PMID:19943761; http://dx.doi.org/10.1586/erv.09.134
- Balashova EE, Lokhov PG. Proteolytically-cleaved fragments of cell-surface proteins from live tumor cells stimulate anti-tumor immune response in vitro. J Carcinog Mutagen 2010; 1:103; http://dx.doi.org/10.4172/2157-2518.1000103
- Balashova EE, Dashtiev MI, Lokhov PG. Proteomic Footprinting of Drug-Treated Cancer Cells as a Measure of Cellular Vaccine Efficacy for the Prevention of Cancer Recurrence. Mol Cell Proteomics 2012; 11:M111.014480-M111.014480; PMID:22074704; http://dx.doi.org/10.1074/mcp.M111.014480
- Kumar S, West DC, Ager A. Heterogeneity in endothelial cells from large vessels and microvessels. Differentiation 1987; 36:57-70; PMID:2451631; http://dx.doi.org/10.1111/j.1432-0436.1987.tb00181.x
- Lang I, Pabst MA, Hiden U, Blaschitz A, Dohr G, Hahn T, Desoye G. Heterogeneity of microvascular endothelial cells isolated from human term placenta and macrovascular umbilical vein endothelial cells. Eur J Cell Biol 2003; 82:163-73; PMID:12751902; http://dx.doi.org/10.1078/0171-9335-00306
- Shreeniwas R, Ogawa S, Cozzolino F, Torcia G, Braunstein N, Butura C, Brett J, Lieberman HB, Furie MB, Joseph-Silverstein J. Macrovascular and microvascular endothelium during long-term hypoxia: alterations in cell growth, monolayer permeability, and cell surface coagulant properties. J Cell Physiol 1991; 146:8-17; PMID:1990021; http://dx.doi.org/10.1002/jcp.1041460103
- Hewett PW. Identification of tumour-induced changes in endothelial cell surface protein expression: an in vitro model. Int J Biochem Cell Biol 2001; 33:325-35; PMID:11312103; http://dx.doi.org/10.1016/S1357-2725(01)00020-6
- Swerlick RA, Lee KH, Wick TM, Lawley TJ. Human dermal microvascular endothelial but not human umbilical vein endothelial cells express CD36 in vivo and in vitro. J Immunol 1992; 148:78-83; PMID:1370173
- Swerlick RA, Lee KH, Li LJ, Sepp NT, Caughman SW, Lawley TJ. Regulation of vascular cell adhesion molecule 1 on human dermal microvascular endothelial cells. J Immunol 1992; 149:698-705; PMID:1378077
- Lee KH, Lawley TJ, Xu YL, Swerlick RA. VCAM-1-, ELAM-1-, and ICAM-1-independent adhesion of melanoma cells to cultured human dermal microvascular endothelial cells. J Invest Dermatol 1992; 98:79-85; PMID:1370233; http://dx.doi.org/10.1111/1523-1747.ep12495643
- Takeichi N, Economou GC, Boone CW. Accelerated regeneration of trypsin-treated surface antigens of simian virus 40-transformed BALB/3T3 cells induced by X-irradiation. Cancer Res 1976; 36:1258-62; PMID:177205
- Glick MC, Kimhi Y, Littauer UZ. Glycopeptides from surface membranes of neuroblastoma cells. Proc Natl Acad Sci U S A 1973; 70:1682-7; PMID:4515927; http://dx.doi.org/10.1073/pnas.70.6.1682
- Anglhileri LJ, Dermietzel LJ. Cell coat in tumor cells–effects of trypsin and EDTA: a biochemical and morphological study. Oncology 1976; 33:17-23; PMID:185564; http://dx.doi.org/10.1159/000225095
- Baumann H, Doyle D. Effect of trypsin on the cell surface proteins of hepatoma tissue culture cells. Characterization of a carbohydrate-rich glycopeptide released from a calcium binding membrane glycoprotein. J Biol Chem 1979; 254:3935-46; PMID:438168
- Weiss L. The effects of trypsin on the size, viability and dry mass of sarcoma 37 cells. Exp Cell Res 1958; 14:80-3; PMID:13512305; http://dx.doi.org/10.1016/0014-4827(58)90214-3
- Barnard PJ, Weiss L, Ratcliffe T. Changes in the surface properties of embryonic chick neural retina cells after dissociation. Exp Cell Res 1969; 54:293-301; PMID:4975920; http://dx.doi.org/10.1016/0014-4827(69)90205-5
- Kraemer PM. Regeneration of sialic acid on the surface of Chinese hamster cells in culture. I. General characteristics of the replacement process. J Cell Physiol 1966; 68:85-90; PMID:5967192; http://dx.doi.org/10.1002/jcp.1040680112
- Kraemer PM. Sialic acid of mammalian cell lines. J Cell Physiol 1966; 67:23-34; PMID:5327858; http://dx.doi.org/10.1002/jcp.1040670104
- Kemp RB, Jones BM, Cunningham I, James MC. Quantitative investigation on the effect of puromycin on the aggregation of trypsin- and versene-dissociated chick fibroblast cells. J Cell Sci 1967; 2:323-40; PMID:4293085
- Pitelka DR, Kerkof PR, Gagne HT, Smith S, Abraham S. Characteristics of cells dissociated from mouse mammary glands. I. Method of separation and morphology of parenchymal cells from lactating glands.
- De Luca C. The use of trypsin for the determination of cellular viability. Exp Cell Res 1965; 40:186-8; PMID:5891332; http://dx.doi.org/10.1016/0014-4827(65)90312-5
- Kraemer PM. Regeneration of sialic acid on the surface of Chinese hamster cells in culture. II. Incorporation of radioactivity from glucosamine-1-14C. J Cell Physiol 1967; 69:199-207; PMID:6033950; http://dx.doi.org/10.1002/jcp.1040690210
- Allen A, Snow C. The effect of trypsin or ethylenediaminetetraacetate on the surface of cells in tissue culture. Biochem J 1970; 117:32 P; PMID:4986872
- Snow C, Allen A. The release of radioactive nucleic acids and mucoproteins by trypsin and ethylenediaminetetra-acetate treatment of baby-hamster cells in tissue culture. Biochem J 1970; 119:707-14; PMID:4992781
- Buck CA, Glick MC, Warren L. A comparative study of glycoproteins from the surface of control and Rous sarcoma virus transformed hamster cells. Biochemistry 1970; 9:4567-76; PMID:4319754; http://dx.doi.org/10.1021/bi00825a016
- Vestling MM, Murphy CM, Fenselau C. Recognition of trypsin autolysis products by high-performance liquid chromatography and mass spectrometry. Anal Chem 1990; 62:2391-4; PMID:2291484; http://dx.doi.org/10.1021/ac00220a025
- Shin BK, Wang H, Yim AM, Le Naour F, Brichory F, Jang JH, Zhao R, Puravs E, Tra J, Michael CW, et al. Global profiling of the cell surface proteome of cancer cells uncovers an abundance of proteins with chaperone function. J Biol Chem 2003; 278:7607-16; PMID:12493773; http://dx.doi.org/10.1074/jbc.M210455200
- Jang JH, Hanash S. Profiling of the cell surface proteome. Proteomics 2003; 3:1947-54; PMID:14625857; http://dx.doi.org/10.1002/pmic.200300563
- Rodríguez-Ortega MJ, Norais N, Bensi G, Liberatori S, Capo S, Mora M, Scarselli M, Doro F, Ferrari G, Garaguso I, et al. Characterization and identification of vaccine candidate proteins through analysis of the group A Streptococcus surface proteome. Nat Biotechnol 2006; 24:191-7; http://dx.doi.org/10.1038/nbt1179
- Lund R, Leth-Larsen R, Jensen ON, Ditzel HJ. Efficient isolation and quantitative proteomic analysis of cancer cell plasma membrane proteins for identification of metastasis-associated cell surface markers. J Proteome Res 2009; 8:3078-90; PMID:19341246; http://dx.doi.org/10.1021/pr801091k
- Garcia J, Faca V, Jarzembowski J, Zhang Q, Park J, Hanash S. Comprehensive profiling of the cell surface proteome of Sy5Y neuroblastoma cells yields a subset of proteins associated with tumor differentiation. J Proteome Res 2009; 8:3791-6; PMID:19505085; http://dx.doi.org/10.1021/pr800964v
- Van Der Pol L, Tramper J. Shear sensitivity of animal cells from a culture-medium perspective. Trends Biotechnol.1998; 16:323-8; PMID:9720320; http://dx.doi.org/10.1016/S0167-7799(98)01209-8
- Lau JY, Tchao R. Stressed polystyrene causes increased membrane sensitivity of adherent cells to fluid shear force: technical note. Eur Cell Mater 2007; 14:40-43; discussion 43-44; PMID:17828704
- McQueen A, Bailey JE. Influence of serum level, cell line, flow type and viscosity on flow-induced lysis of suspended mammalian cells. Biotechnol Lett 1989; 11:531-536; http://dx.doi.org/10.1007/BF01040030
- McQueen A, Meilhoc E, Bailey JE. Flow effects on the viability and lysis of suspended mammalian cells. Biotechnol Lett 1987; 9:831-6; http://dx.doi.org/10.1007/BF01026191
- Tchao R. Fluid shear force and turbulence-induced cell death in plastic tissue culture flasks. In Vitro Toxicol 1996; 9:93-100.
- Kyte JA. Cancer vaccination with telomerase peptide GV1001. Expert Opin Investig Drugs 2009; 18:687-94; PMID:19388882; http://dx.doi.org/10.1517/13543780902897631
- Shaw VE, Naisbitt DJ, Costello E, Greenhalf W, Park BK, Neoptolemos JP, Middleton GW. Current status of GV1001 and other telomerase vaccination strategies in the treatment of cancer. Expert Rev Vaccines 2010; 9:1007-16; PMID:20822343; http://dx.doi.org/10.1586/erv.10.92
- Gendler SJ, Lancaster CA, Taylor-Papadimitriou J, Duhig T, Peat N, Burchell J, Pemberton L, Lalani EN, Wilson D. Molecular cloning and expression of human tumor-associated polymorphic epithelial mucin. J Biol Chem 1990; 265:15286-93; PMID:1697589
- Gendler SJ. MUC1, The renaissance molecule. J Mammary Gland Biol Neoplasia 2001; 6:339-53; PMID:11547902; http://dx.doi.org/10.1023/A:1011379725811
- Singh R, Bandyopadhyay D. MUC1: a target molecule for cancer therapy. Cancer Biol Ther 2007; 6:481-6; PMID:18027437; http://dx.doi.org/10.4161/cbt.6.4.4201
- McCarthy N. Running a MUC1. Nat Rev Cancer 2012; 12:317-317; PMID:22495322
- Paczesny S, Shi H, Saito H, Mannoni P, Fay J, Banchereau J, Palucka AK. Measuring melanoma-specific cytotoxic T lymphocytes elicited by dendritic cell vaccines with a tumor inhibition assay in vitro. J Immunother 2005; 28:148-57; PMID:15725959; http://dx.doi.org/10.1097/01.cji.0000154247.97254.ef
- Ossevoort MA, Feltkamp MC, van Veen KJ, Melief CJ, Kast WM. Dendritic cells as carriers for a cytotoxic T-lymphocyte epitope-based peptide vaccine in protection against a human papillomavirus type 16-induced tumor. J Immunother Emphasis Tumor Immunol 1995; 18:86-94; PMID:8574470; http://dx.doi.org/10.1097/00002371-199508000-00002
- Kern PA, Knedler A, Eckel RH. Isolation and culture of microvascular endothelium from human adipose tissue. J Clin Invest 1983; 71:1822-9; PMID:6306056; http://dx.doi.org/10.1172/JCI110937
- Correa de Sampaio P, Auslaender D, Krubasik D, Failla AV, Skepper JN, Murphy G, English WR. A heterogeneous in vitro three dimensional model of tumour-stroma interactions regulating sprouting angiogenesis. PLoS One 2012; 7; PMID:22363483; http://dx.doi.org/10.1371/journal.pone.0030753
