Abstract
Quality control of vaccine strains is directly associated with the safety and efficacy of inactivated whole bacterial vaccines. The assessment of genetic stability is one of the essential elements to guarantee the quality of vaccine strains. The multiple-valence inactivated leptospiral vaccine, comprising the main circulating serogroups, has played an important role in the control of Leptospira infection in China. In the present study, to assess the genetic stability of vaccine strains and develop novel quality control tests that enhance and extend the existing procedures, 7 Chinese leptospiral vaccine strains were characterized during in vivo and in vitro passages by multilocus sequence typing (MLST) and pulsed-field gel electrophoresis (PFGE) analysis. The seven vaccine strains were found to have distinct sequence types (STs) and PFGE profiles. Further analysis showed that the ST and PFGE pattern of each vaccine strain, after in vivo or serial in vitro passages (up to 20 passages), were identical to those of the initial strain, demonstrating that these strains were genetically stable and homogeneous. Taken together, PFGE and MLST provide a reproducible and reliable means for confirming the identity and genetic stability of vaccine seeds, suggesting that these approaches can be used to evaluate the quality of leptospiral vaccine strains.
Leptospirosis is a global zoonotic disease caused by pathogenic species of the Leptospira genus, which are most commonly found in tropical and subtropical countries.Citation1-4 At least 200 animal host species have been identified for pathogenic Leptospira spp. Leptospirosis can be maintained in chronic carrier mammalian hosts (e.g., rats and dogs), and it is typically transmitted to humans by direct or indirect contact with urine or body fluids from the reservoir hosts.Citation2,4 Although this condition manifests relatively mild symptoms in humans, some severe cases presenting with pulmonary hemorrhage, jaundice, and meningitis have been reported.Citation5,6 According to World Health Organization (WHO) estimates, the number of human leptospirosis cases averages over 500,000 annually, and the mortality rate can reach up to 25%.Citation7 Most cases of leptospirosis occur in developing countries; however, globalization and international travel have led to an apparent increase in its incidence in industrialized countries.Citation4 Therefore, leptospirosis has the potential to become even more prevalent and has recently been recognized as a re-emerging infectious disease.
In China, leptospirosis is a nationally monitored infectious disease. The first human leptospirosis case in mainland China can be traced to the 1930s. To date, more than 2.5 million cases and over 20,000 deaths due to leptospirosis have been reported in China.Citation8,9 An inactivated, whole leptospiral vaccine composed of prevalent serovars was successfully developed in the 1950s, and it has played a critical role in controlling Leptospira infections in China.Citation8-10 In 2007, the leptospiral vaccine was included in the Chinese Expanded Program on Immunization (EPI), and it is recommended for use in high-risk populations from leptospirosis-endemic regions.Citation11
Currently, the multiple-valence, inactivated leptospiral vaccine used in the EPI contains Icterohaemorragiae, Grippotyphosa, Autumnalis, Canicola, Pomona, Australis, and Hebdomadis,Citation12 which are the main circulating L. interrogans serogroups in China. Quality control of vaccine strains is considered an important process in the production of vaccine, and it is directly associated with the quality of the final product. According to Chinese Pharmacopoeia (2010 Edition),Citation12 analyses of leptospiral vaccine strains include physicochemical and microbiological tests such as morphological and cultural characteristics, serovars, virulence, immunity, and antigenicity. To preserve the virulence of the leptospiral strains, it is also recommended that the strain should be passaged in guinea pigs after 3–6 passages in vitro.Citation12 Bacterial adaptation to the animal host and in vitro culture may lead to changes in the bacterial genome.Citation13 For example, it was determined that tandem duplications varied between Bacille Calmette-Guérin (BCG) vaccine sub-strains after in vitro passages for vaccine production.Citation14,15 Although the role of tandem duplications in attenuation and/or altered immunogenicity of the BCG vaccine is yet unknown, it is thought that knowledge of their existence will facilitate quality control of the BCG vaccine lot.Citation14,16 Therefore, to ensure appropriate quality control, the aforementioned criteria should be supplemented with an evaluation of genetic homogeneity in vaccine seeds. In this context, analysis of genetic stability may be an essential element to guarantee the quality of a vaccine strain.
Recently, advances in science and technology have resulted in the development of several techniques for investigating the molecular epidemiology of Leptospira, such as pulsed-field gel electrophoresis (PFGE) fingerprinting. PFGE is currently considered the gold standard in epidemiological analysis of pathogenic organisms, and it has been applied to Leptospira studies since 1990s.Citation17–19 In addition, multilocus sequence typing (MLST), the most widespread genotyping methodology for bacterial pathogens, is used to describe the molecular epidemiology of leptospirosis in some countries or regions.Citation20,21 These approaches have potential to confirm the identity and genetic stability of bacterial strains over multiple vaccine seed passages. Therefore, the aims of the present study were to assess the genetic stability of leptospiral vaccine strains by using PFGE and MLST. Furthermore, we evaluated the ability of PFGE and MLST methodologies to be used as tools to facilitate the quality control of vaccine strains.
The seven leptospiral vaccine strains used in this study were from the Wuhan Institute of Biological Products Co., Ltd (). To assess the genetic homogeneity of each strain, the vaccine strains after animal passage (to preserve virulence) were also included. Vaccine strains passaged before animal inoculation were arbitrarily labeled as passage −1, while strains after passage in animals were designated as passage 1. These strains were identified according to standard tests from Chinese Pharmacopoeia (2010 Edition). To further determine the genetic stability of the 7 vaccine strains, 20 serial passages of each strain were performed in vitro. All leptospiral strains were grown at 28°C on modified TEPCKNN medium (10 mM Na2HPO4, 4 mM KH2PO4, pH 7.2) (NIFDC, Beijing) supplemented with 10% rabbit serum. DNA was extracted using a Wizard Genomic DNA Purification Kit (Promega, Southampton, UK) following the manufacturer's instructions. MLST was performed on 7 housekeeping genes (including glmU, pntA, sucA, tpiA, pfkB, mreA, and caiB) as previously described.Citation20 PCR amplification of MLST target genes was performed in a volume of 50 μl and contained a final concentration of 1.5–3.5 mM MgCl2, 0.2 mM dNTP, 1 unit Taq DNA polymerase (Takara, Dalian, China), 0.25 μM of each primer, and 20–50 ng of Leptospira DNA. Amplification was performed under the following thermocycler conditions: one cycle of 95°C for 2 min, 30 cycles each of 95°C for 10 s; 46°C for 15 s; and 72°C for 30 s, followed by a final period of 72°C for 7 min. All amplified fragments were visualized on 1.5% agarose gels by Goldview staining, and the amplicons were sequenced by Taihe Biotechnology Co., LTD. The sequence types (STs) were determined from the resulting allelic profiles and compared to an established internet database (http://leptospira.mlst.net/). PFGE analysis was performed as previously describedCitation18 with minor modifications. Briefly, after the DNA plugs were treated with 10 U of NotI (Takara, Dalian, China), the cleaved DNA fragments were separated by electrophoresis in a 1% agarose gel using a Chef Mapper (Bio-Rad, Hercules, CA, USA) with pulse times of 5–65 s for 20 h. The band patterns were analyzed with BioNumerics software (Applied Maths, Kortrijk, Belgium). The clustering method used was the unweighted pair group with arithmetic averages (UPGMA).
Table 1. Information for 7 leptospiral vaccine strains used in this study
The seven loci selected in MLST were successfully amplified from all vaccine strains and then sequenced. The ST of housekeeping gene from each isolate was queried against the Leptospira sequence-type database, and 7 STs were detected. Each vaccine strain harbored a different ST (). Furthermore, the genetic stability of each vaccine strain was investigated by passaging the strains in animals, in vivo, and in culture, in vitro, and then comparing the STs obtained at each passage. The results showed that the ST of each strain had not changed after in vivo and in vitro passages (up to 20 passages) ().
Figure 1. Selected PFGE patterns and MLST types from 7 leptospiral vaccine strains during in vivo and in vitro passages. The unweighted pair group method with arithmetic averages (UPGMA) was used as the clustering method for PFGE profiles. Number of passages: −1 represented vaccine strains before inoculation in animals; 1, 10 and 20 represented the first, tenth, and twentieth passage of each strain in culture, respectively.
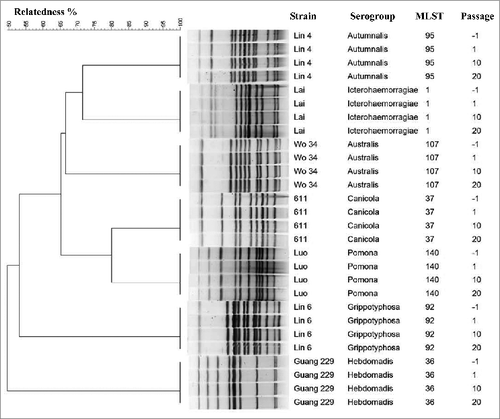
PFGE analysis showed that the 7 vaccine strains fell into distinct PFGE profiles (), indicating that these 7 serovars can be differentiated by Not I restriction analysis.Citation18,19 Consistent with the MLST analysis, we found that PFGE profiles of the 7 vaccine stains, after serial passages, were 100% identical to the initial strains, based on phylogenetic analysis with UPGMA (). Moreover, the PFGE fingerprint of each serovar had not changed after 3 replicates were performed at each passage, suggesting that reproducibility of PFGE may represent a powerful technique for identifying Leptospira strains.
Since vaccination is one of the most effective measures to control Leptospira infection, the use of a high quality vaccine is essential for the success of leptospiral vaccination programs. Therefore, to ensure the vaccine developed is of adequate quality, it is important to strengthen quality control methodologies for testing vaccine strains. In the present study, the nucleotide sequencing-based MLST and PFGE techniques were used to characterize leptospiral vaccine strains at the bacterial chromosomal level to evaluate genetic homogeneity. Although these vaccine seeds have been used in China for leptospiral vaccine production since 1950s, the genetic stability of each vaccine seeds has not been verified. To our knowledge, this is the first report demonstrating the genomic integrity and stability of vaccine seeds used for Chinese leptospiral vaccine production.
MLST analysis for Leptospira spp has the advantage of direct traceability and association with housekeeping gene polymorphisms because this analysis is sequence based. Housekeeping genes are constitutively expressed, are essential for the maintenance of basic cellular functions, and evolve more rapidly than rRNA genes.Citation22 Therefore, analysis of multiple housekeeping genes by MLST may offer information about the genetic stability of bacterial strains under different conditions. In this study, we found that each housekeeping gene, from 7 different vaccine strains, did not acquire any mutations during multiple in vivo and in vitro passages. These results suggested that housekeeping genes of the leptospiral vaccine strains studied were genetically stable and homogeneous.
Many studies have demonstrated that PFGE permits a rapid and detailed comparative analysis of chromosomes in bacterial strains by exploiting restriction endonucleases that cut the chromosome infrequently. The resulting restriction patterns are characteristic of a particular vaccine strain, and this information can be used to investigate the physical structure of the bacterial chromosome.Citation19,23 Reproducible PFGE profiles were found in 7 vaccine strains after in vitro and in vivo passage, further reflecting the genetic stability of leptospiral vaccine strains at the whole genome level. Previous studies have shown that repetitions of the same serovar consistently produced reproducible patterns, and the PFGE patterns of different strains of the same serovar are closely related. This corresponds well to serologic classification, suggesting that this method has the capability of rapidly identifying the serovar of most clinical isolates.Citation17,19 Furthermore, PFGE also has the potential to determine the taxonomic status of isolates in question due to mislabeling or contamination, as Cicerono et al. reported a case with strain 5621, the serovar mozdok reference strain, and K1, formerly described as the serovar dania reference strain, but was recognized to be a mozdok-like strain.Citation24 Taken together, the generation of genomic restriction patterns by PFGE may provide a comparatively reliable means of confirming the identity and genetic stability of leptospiral vaccine strains.
Different from other bacterial pathogens, the virulence of pathogenic Leptospira spp. is gradually lost after several passages in culture media.Citation25 Until now, lyophilization had also not been developed successfully.Citation26 Considering that the virulence of vaccine strains is associated with the efficacy of the vaccine product, leptospiral vaccine seeds must be used to inoculate animal models to preserve virulence.Citation12 However, current existing procedures for quality control of leptospiral vaccine strains are mainly based on phenotypic and biological characterization.Citation12 These methods are insufficient to determine the identity of vaccine strains, especially since they are poor indicators of strain identity at whole genome level and cannot be used to evaluate genetic stability. The data presented in this study show that molecular and genetic techniques such as PFGE and MLST could be used not only for investigating the chromosome structure and genetic stability of leptospiral vaccine strains but also for controlling the consistency of inactivated vaccines. These approaches are rapid and reproducible, as they conform well to standardization, thereby suggesting that they should be applied to maintain quality control of leptospiral vaccines. Currently, in China, there are some bacterial vaccine manufacturers who produce a single variety of vaccine on a small scale, and with variable quality. Almost all of the vaccine-producing bacteria are consistently provided by the Chinese national regulatory authorities.Citation12 Although these vaccine seeds have been used to produce vaccines (such as the pertussis or BCG vaccines) for many years by different manufactures, to date, the genetic consistency among vaccine seed strains has not been verified. Therefore, the molecular approaches described in the present study have the potential to be adopted for use in national regulation of vaccine quality for many bacterial vaccines produced in China.
Recently, a pathogenomic approach was used to infer leptospiral virulence genes by whole genome comparison of the culture-attenuated L. interrogans serovar Lai with its virulent parent. A total of 45 non-synonymous, single nucleotide polymorphisms (SNPs) in 35 protein-coding genes were discovered.Citation27 Consistently accumulating mutations confers higher fitness, and it was assumed that some SNPs should occur in the bacterial genome during the in vivo and in vitro passages in this study. However, no nucleotide changes were found using PFGE and MLST analysis. Although the exact cause is unknown, a possible explanation for these results is that the assumed nucleotide changes do not occur in the housekeeping genes analyzed by MLST or affect the Not I restriction sites within the bacterial chromosome. In this sense, whole-genome sequencing of leptospiral vaccine strains would be the best way to confirm genomic stability in these vaccine strains. The cost, time, and standardization requirements for this analysis preclude its use on a routine basis. However, if the sequencing data on each strain after passage is made available, it will provide adequate information to further assess the sensitivities of the methods described in the study. Collectively, the molecular tools described in the study should be applied to quality control practices and combined with existing tests for a more comprehensive evaluation of the biological quality and genetic stability of leptospiral vaccine strains.
In summary, the 7 leptospiral vaccine strains examined proved to be homogeneous and genetically stable after MLST and PFGE analyses, even after multiple in vivo and in vitro passages. PFGE and MLST provide a reproducible and reliable means for molecular characterization of Leptospira spp, suggesting that these are essential assays for evaluating the quality and stability of leptospiral vaccine strains. Furthermore, the proposed methods have potential for use in the quality control programs for other bacterial vaccines, especially for assessing the genetic consistency of vaccine seed strains between different manufacturers, when the strains originate from a common source.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by National Major Scientific and Technological Special Project for “Major new drug development” (No. 2013ZX09304101) from the Ministry of Science and Technology, China and The National natural science Funds, China (No. 81471968), and NIFDC Professional Leader Fund (NO.2013×4).
References
- Plank R, Dean D. Overview of the epidemiology, microbiology, and pathogenesis of Leptospira spp. in humans. Microbes Infect 2000; 2:1265-76; PMID:11008116; http://dx.doi.org/10.1016/S1286-4579(00)01280-6.
- Ko AI, Goarant C, Picardeau M. Leptospira: the dawn of the molecular genetics era for an emerging zoonotic pathogen. Nat Rev Microbiol 2009; 7:736-47; PMID:19756012; http://dx.doi.org/10.1038/nrmicro2208.
- World Health Organization. Leptospirosis worldwide, 1999. Wkly Epidemiol Rec 1999; 74:237-42; PMID:10437435.
- Bharti AR, Nally JE, Ricaldi JN, Matthias MA, Diaz MM, Lovett MA, Levett PN, Gilman RH, Willig MR, Gotuzzo E, et al. Leptospirosis: a zoonotic disease of global importance. Lancet Infect Dis 2003; 3:757-71; PMID:14652202; http://dx.doi.org/10.1016/S1473-3099(03)00830-2.
- Seijo A, Coto H, San Juan J, Videla J, Deodato B, Cernigoi B, Messina OG, Collia O, de Bassadoni D, Schtirbu R, et al. Lethal leptospiral pulmonary hemorrhage: an emerging disease in Buenos Aires, Argentina. Emerg Infect Dis 2002; 8:1004-5; PMID:12194784; http://dx.doi.org/10.3201/eid0809.010499.
- Thammakumpee K, Silpapojakul K, Borrirak B. Leptospirosis and its pulmonary complications. Respirology 2005; 10:656-9; PMID:16268921; http://dx.doi.org/10.1111/j.1440-1843.2005.00764.x.
- World Health Organization. Report of the Second Meeting of the Leptospirosis Burden Epidemiology Reference Group. 2010.
- Hu W, Lin X, Yan J. Leptospira and leptospirosis in China. Curr Opin Infect Dis 2014; 27:432-6; PMID:25061933; http://dx.doi.org/10.1097/QCO.0000000000000097.
- Shi MH, Jiang X Leptospirosis in the past fifty years in China. Chin J Epidemiol 2000; 21: 228-30.
- Zhang C, Wang H, Yan J. Leptospirosis prevalence in Chinese populations in the last two decades. Microbes Infect 2012; 14:317-23; PMID:22155621; http://dx.doi.org/10.1016/j.micinf.2011.11.007.
- Liang X, Wu Z. Implementation of EPI for 30 years to protect hundreds of millions of people's health. Chinese J Preventive Med 2008; 42:4-6; PMID:18512318.
- National Pharmacopoeia Committee.The Chinese Pharmacopoeia (2010 version) 3rd Part. Beijing: Taylor & Francis, 2010.
- Blot M. Transposable elements and adaptation of host bacteria. Genetica 1994; 93:5-12; PMID:7813917; http://dx.doi.org/10.1007/BF01435235.
- Brosch R, Gordon SV, Buchrieser C, Pym AS, Garnier T, Cole ST. Comparative genomics uncovers large tandem chromosomal duplications in Mycobacterium bovis BCG Pasteur. Yeast 2000; 17:111-23; PMID:10900457; http://dx.doi.org/10.1002/1097-0061(20000630)17:2%3c111::AID-YEA17%3e3.0.CO;2-G.
- Brosch R, Gordon SV, Garnier T, Eiglmeier K, Frigui W, Valenti P, Dos Santos S, Duthoy S, Lacroix C, Garcia-Pelayo C, et al. Genome plasticity of BCG and impact on vaccine efficacy. Proc Natl Acad Sci U S A 2007; 104:5596-601; PMID:17372194; http://dx.doi.org/10.1073/pnas.0700869104.
- World Health Organization. Report. Consultation on the characterization of BCG vaccines. Geneva, Switzerland: Taylor & Francis; 8–9 December 2004.
- Galloway RL, Levett PN. Application and validation of PFGE for serovar identification of Leptospira clinical isolates. PLoS Negl Trop Dis 2010; 4; e824; PMID:20856859.
- Galloway RL, Levett PN. Evaluation of a modified pulsed-field gel electrophoresis approach for the identification of Leptospira serovars. Am J Trop Med Hyg 2008; 78: 628-32; PMID:18385360.
- Herrmann JL, Bellenger E, Perolat P, Baranton G, Saint Girons I. Pulsed-field gel electrophoresis of NotI digests of leptospiral DNA: a new rapid method of serovar identification. J Clin Microbiol 1992; 30:1696-702; PMID:1629323.
- Boonsilp S, Thaipadungpanit J, Amornchai P, Wuthiekanun V, Bailey MS, Holden MT, Zhang C, Jiang X, Koizumi N, Taylor K, et al. A single multilocus sequence typing (MLST) scheme for seven pathogenic Leptospira species. PLoS Negl Trop Dis 2013; 7:e1954; PMID:23359622; http://dx.doi.org/10.1371/journal.pntd.0001954.
- Thaipadungpanit J, Wuthiekanun V, Chierakul W, Smythe LD, Petkanchanapong W, Limpaiboon R, Apiwatanaporn A, Slack AT, Suputtamongkol Y, White NJ, et al. A dominant clone of Leptospira interrogans associated with an outbreak of human leptospirosis in Thailand. PLoS Negl Trop Dis 2007; 1:e56; PMID:17989782; http://dx.doi.org/10.1371/journal.pntd.0000056.
- Zhu J, He F, Hu S, Yu J. On the nature of human housekeeping genes. Trends Genet 2008; 24:481-4; PMID:18786740; http://dx.doi.org/10.1016/j.tig.2008.08.004.
- Herrmann JL, Baril C, Bellenger E, Perolat P, Baranton G, Saint Girons I. Genome conservation in isolates of Leptospira interrogans. J Bacteriol 1991; 173:7582-8; PMID:1938954.
- Ciceroni L, Ciarrocchi S, Ciervo A, Petrucca A, Pinto A, Calderaro A, Viani I, Galati L, Dettori G, Chezzi C. Differentiation of leptospires of the serogroup Pomona by monoclonal antibodies, pulsed-field gel electrophoresis and arbitrarily primed polymerase chain reaction. Res Microbiol 2002; 153:37-44; PMID:11881897; http://dx.doi.org/10.1016/S0923-2508(01)01284-0.
- Johnson RC, Harris VG. Differentiation of pathogenic and saprophytic letospires. I. Growth at low temperatures. J Bacteriol 1967; 94:27-31; PMID:6027998.
- Myers DM, Varela-Diaz VM, Siniuk AA. Long-term survival of Leptospira in a biphasic culture medium containing charcoal. Appl Microbiol 1973; 25:514-6; PMID:4735486.
- Lehmann JS, Fouts DE, Haft DH, Cannella AP, Ricaldi JN, Brinkac L, Harkins D, Durkin S, Sanka R, Sutton G, et al. Pathogenomic inference of virulence-associated genes in Leptospira interrogans. PLoS Negl Trop Dis 2013; 7:e2468; PMID:24098822; http://dx.doi.org/10.1371/journal.pntd.0002468.
