Abstract
Cross-reactive peptides on HIV-1 and FIV p24 protein sequences were studied using peripheral blood mononuclear cells (PBMC) from untreated HIV-1-infected long-term survivors (LTS; >10 y of infection without antiretroviral therapy, ART), short-term HIV-1 infected subjects not on ART, and ART-treated HIV-1 infected subjects. IFNγ-ELISpot and CFSE-proliferation analyses were performed with PBMC using overlapping HIV-1 and FIV p24 peptides. Over half of the HIV-1 infected subjects tested (22/31 or 71%) responded to one or more FIV p24 peptide pools by either IFNγ or T-cell proliferation analysis. PBMC and T cells from infected subjects in all 3 HIV+ groups predominantly recognized one FIV p24 peptide pool (Fp14) by IFNγ production and one additional FIV p24 peptide pool (Fp9) by T-cell proliferation analysis. Furthermore, evaluation of overlapping SIV p24 peptide sequences identified conserved epitope(s) on the Fp14/Hp15-counterpart of SIV, Sp14, but none on Fp9-counterpart of SIV, Sp9. The responses to these FIV peptide pools were highly reproducible and persisted throughout 2–4 y of monitoring. Intracellular staining analysis for cytotoxins and phenotyping for CD107a determined that peptide epitopes from Fp9 and Fp14 pools induced cytotoxic T lymphocyte-associated molecules including perforin, granzyme B, granzyme A, and/or expression of CD107a. Selected FIV and corresponding SIV epitopes recognized by HIV-1 infected patients indicate that these protein sequences are evolutionarily conserved on both SIV and HIV-1 (e.g., Hp15:Fp14:Sp14). These studies demonstrate that comparative immunogenicity analysis of HIV-1, FIV, and SIV can identify evolutionarily-conserved T cell-associated lentiviral epitopes, which could be used as a vaccine for prophylaxis or immunotherapy.
Abbreviations:
- aa, amino acid
- AIDS, acquired immune deficiency syndrome
- ART, anti-retroviral therapy
- CFSE, Carboxyfluorescein succinimidyl ester
- CMI, cell mediated immunity
- CTL, cytotoxic T cell
- FIV, feline immunodeficiency virus
- GrzA, granzyme A
- GrzB, granzyme B
- HIV, human immunodeficiency virus
- HLA, human leukocyte antigen
- HERV, human endogenous retrovirus
- ICS, intracellular staining
- LANL, Los Alamos National Laboratory
- LTS, Long term survivors
- Nab, broadly neutralizing antibody
- PHA, phytohaemagglutinin
- SFU, spot forming units
- SIV, simian immunodeficiency virus
- ST, short term survivors
Introduction
The Centers for Disease Control and Prevention reported a decline in newly HIV-1 infected individuals from 2008 to 2010 but a slow steady rise in people living with HIV/AIDS in the United States.Citation1,2 The increase in survival correlates with the use of antiretroviral therapy (ART).Citation3 Nevertheless, the incidence of HIV infection continues to increase, and an effective HIV-1 vaccine is critical to curtail this pandemic.Citation4 Over 240 phase I-III clinical HIV-1 vaccine trials have been completed or are ongoing, but only 3 vaccine candidates have progressed to phase III clinical trials (IAVI database, http://www.iavireport.org/Trials-Database/Pages/default.aspx). Two phase III trials using antibody-based HIV-1 envelope (Env) vaccines failed in high-risk groups.Citation5,6 The phase III trial, RV144, based on both antibody- and cellular-based vaccine approaches, conferred a modest protection rate of 31.2% overall, but was only 3.7% effective in the high-risk group.Citation7 Protection was positively correlated with the production of IgG antibodies to the HIV Env V1V2 region and low amounts of IgA antibody binding to Env. Broadly neutralizing antibodies (NAbs) against HIV-1 were not detected in vaccinated individuals.Citation8 Most notably, polyfunctional CD4+ T cells and CD4+ cytototoxic T lymphocyte (CTL)-like T cells were detected in vaccinated individuals that were specifically activated by HIV V2 epitopes.Citation9
Numerous studies using T cells from HIV-1 positive (HIV+) individuals have identified CD8+ cell and CD4+ T-helper (TH) cell-associated epitopes on HIV-1 proteins (Los Alamos National Laboratory (LANL) database, http://hiv-web.lanl.gov/content/immunology/maps/maps.html).Citation10,11 In addition, HLA allotypes responsive to T-cell epitopes have correlated to a delay in AIDS progression and/or lower HIV-1 viral set point in HIV+ long-term survivors and elite controllers when compared to HIV+ rapid progressors.Citation12,13 Nevertheless, it is currently unclear as to which epitopes detected at what stage of HIV infection may be useful as vaccine immunogens or in immunotherapy.
The first inactivated dual-subtype whole-lentivirus vaccine against feline immunodeficiency virus (FIV) was commercially released in the US in 2002.Citation14 FIV is a lentivirus that causes an immunodeficiency syndrome in domestic cats with clinical features similar to HIV/AIDS in people. FIV resembles both the HIV and simian immunodeficiency virus (SIV) in genomic structure, morphology, and biophysical characteristics.Citation15,16 However, FIV is the only lentivirus for which there is a commercial vaccine available in the United States.Citation14 Prototype and commercial dual-subtype (A and D) FIV vaccines conferred combined protection rates of 100% against homologous subtype-A tier-1 FIVPet, 89% against heterologous subtype-B tier-3 FIVFC1, 61% against recombinant subtype-A/B tier-2 FIVBang, and 62% against recombinant subtype-F'/C tier-3 FIVNZ1.Citation17 Both the prototype and commercial vaccines induced moderate levels of homologous and fewer heterologous subtype-specific FIV NAbs, but induced strong T-cell responses against both homologous and heterologous subtype FIV strains.Citation17,18 Using the FIV model, previous studies demonstrated that an experimental HIV-1 p24 vaccine conferred cross-protection against low-dose FIV challenge which indicated that potential vaccine epitopes for FIV protection likely reside on viral p24.Citation19
Host responses to HIV-1 and SIV p24 antigens are associated with control of virus load.Citation20-24 Therefore, to identify potential vaccine epitopes, PBMC and T cells from healthy HIV-1 infected (HIV+) subjects on or off antiretroviral therapy (ART) were evaluated for immune responses to overlapping FIV, HIV-1, and SIV p24 peptide pools. In these studies, the p24 sequences were screened for CD8+ and CD4+ T-cell epitopes that are conserved between species and recognized by HIV-1+ subjects () for future use as vaccine immunogens.
Table 1. Description of HIV+ population: Demographic information and immune status
Results
Determining conserved cell-mediated immune (CMI) peptides based on IFNγ responses
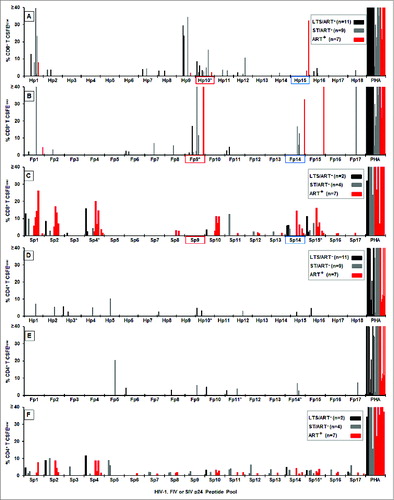
The PBMC from the 31 HIV+ subjects developed robust IFNγ responses to the full length HIV-1 p24 peptide pools (, 144 total responses), whereas minimal to no responses were observed with the PBMC from HIV-negative (HIV−) control subjects (range of 0–15 spot forming unit (SFU)). The highest responder frequencies were observed to human p24 peptide pool 3 (Hp3) (18 of 31; 58%) followed by Hp2, Hp10, and Hp15 (11 of 31; 35%). A lower number of subjects responded to Hp7 and Hp14 (10 of 31; 32%). In addition, PBMC from the HIV+ subjects produced IFNγ responses of low magnitudes and frequencies to all FIV p24 peptide pools (, 53 total responses) except for feline p24 peptide pools (Fp)13 and Fp14 which correspond to HIV sequences within Hp14 and Hp15 peptide pools. The highest responder frequencies were observed to Fp14 (11 of 31; 35%) with lower responder frequency to Fp13 (5 of 31; 16%). These findings suggested that these FIV pools contain potential cross-reactive epitopes to HIV since cats immunized with FIV vaccine also reacted to Fp14 (data not shown). In addition, overlapping SIV p24 peptide pool (Sp) analysis identified a moderate number of responses to the corresponding SIV peptide pool Sp14 (5 of 15; 33%) (), the counterpart to Fp14 and Hp15. Interestingly, the high response to Fp14 correlated to muted responses in both HIV and SIV. These results suggest that cross-reactive epitope(s) conserved between species may reside on Hp15, Fp14, and Sp14. Even though the response is diminished, reactivity of these conserved epitopes by HIV+ subjects suggest that cross-reactive epitope(s) conserved between species may reside on Hp15, Fp14, and Sp14.
Figure 1. IFNγ responses to HIV, FIV and SIV p24 peptide pools. The IFNγ ELISpot responses to overlapping peptide pools of HIV p24 (Hp1-Hp18, n = 31 ; A), FIV p24 (Fp1-Fp17, n = 31 ; B), and SIV p24 (Sp1-Sp17, n = 15 ; C) are depicted as spot forming units (SFU) per 106 PBMC. The HIV+ subjects (panel-A insert for A and B; panel-C insert) consisted of long-term survivors (LTS) without antiretroviral therapy (ART) (LTS; black bar); recently diagnosed subjects (<1 year) with short-term infection without ART (ST; gray bar); and those receiving ART with various duration of infection (ART+, red bar). Each bar represents a positive response by an individual subject with a threshold of >50 SFU per 106 PBMC. Asterisk after the peptide pool(s) represent those with highest frequency of responders. The FIV p24 pool with the highest and the most frequent responses and its counterpart HIV-1 and SIV pools are highlighted in blue.
Conserved CMI peptides based on T-cell proliferation responses
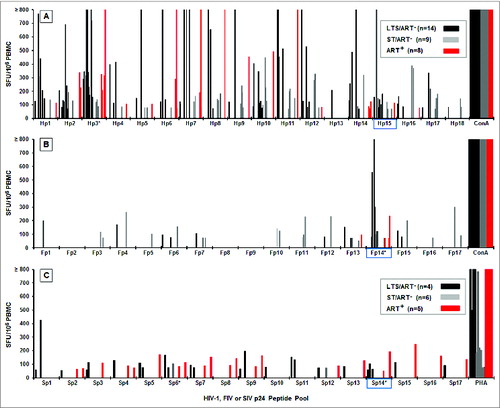
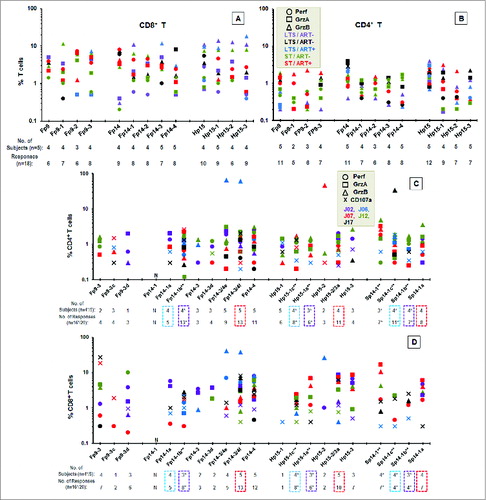
The CD8+ T cells of the HIV+ subjects () proliferated more frequently and at higher magnitudes than the CD4+ T cells () to all HIV p24 peptide pools (36 CD8+ T-cell responses vs. 12 CD4+ T-cell responses). The highest frequency of CD8+ T-cell proliferation responses to HIV p24 was against pools Hp1 (6 of 27; 22%) and Hp10 (8 of 27; 30%) followed by Hp9 (4 of 27; 15%) (). The same analysis performed on T cells from healthy HIV− subjects indicated that their CD4+ and CD8+ T cells did not significantly recognize HIV p24 pools except for the Hp15 pool with a frequency of 42%. All results are shown after subtraction of the average result of the responders for each peptide pool or peptide. CD8+ T cells from 42% (5 of 12) of the HIV− subjects had a substantial response to the Hp15 peptide pool. The number of HIV+ responders to Hp15 is low due to a high average result (28% CFSElow) of HIV− responders to Hp15 that were subtracted from the HIV+ responses (). Except for the Hp15 pool (the counterpart of the Fp14 pool), the Fp9 pool, the Hp15–3 peptide in Hp15 pool, and the Fp9–3 peptide in Fp9 pool with 25–51% of the value before subtraction, none of the other peptide pools or peptides induced substantial CD8+ or CD4+ T-cell proliferation in HIV− subjects (0–20% of the value before subtraction).
Twelve HIV+ subjects had CD8+ T-cell proliferation responses to FIV p24 pools () compared to only 5 subjects with CD4+ T-cell proliferation responses to the same peptide pools (). Remarkably, substantial CD8+ T-cell proliferation responses were detected in HIV+ subjects to Fp9 (8 of 27; 30%) and to Fp14 (4 of 27; 15%) (). A lower responder frequency at a lower magnitude was detected in HIV− subjects to Fp9 with a minimal response to Fp14. When comparing SIV p24 pools, the Fp14-counterpart Sp14 pool, but not the Fp9-counterpart Sp9 pool, was recognized by CD8+ T cells from the HIV+ cohorts (). In addition, CD8+ T cells from HIV+ subjects recognized multiple SIV peptide pools at a frequency of 38–54% (Sp1, Sp2, Sp4, Sp10, Sp14, Sp15) (). However, Sp2, Sp4, Sp10, Sp14, and Sp15 were also recognized by CD8+ T cells from HIV− subjects at a low frequency of 20–30%. Notably, both the Sp1 pool and its counterpart Hp1 pool were recognized by the CD8+ T cells from HIV+ subjects ().
Among the FIV peptide pools, the Fp9 pool induced strong CD8+ T-cell proliferation responses but few IFNγ responses, while the Fp14 pool induced both IFNγ and CD8+ T-cell proliferation responses (). As a result, subsequent studies focused on the Fp14 and Fp9 peptide pools and their HIV counterparts, Hp15 and Hp10, respectively.
The persistence of IFNγ responses to Fp14 and CD8+ T-cell proliferation to Fp9
PBMC from 8 of 10 (80%) HIV+ subjects who initially responded to the Fp14 pool had a declining but persistent IFNγ response for the duration of the 2-yr study period while 3 of 7 tested continued to respond in the 4th yr (, left graph). Although there was no statistical difference between the IFNγ responses during the 1st yr and the 2nd yr of the 7 HIV+ subjects monitored (t1 and t2, p = 0.39), a statistically significant decrease in IFNγ response was detected when the levels were compared between the 2nd yr and 4th yr (t2 and t3, p = 0.035) and between the initial time point and the 4th yr (t1 and t3, p = 0.014). Similarly, IFNγ responses in the PBMC in 6 of 8 (75%) initial responders to Hp15, the counterpart for Fp14, were sustained in response to Hp15 through the 2nd yr but showed a substantial declining trend in the magnitude of IFNγ responses in by the 4th yr (, right graph).
Figure 3. The persistence of IFNγ and T-cell proliferation responses to selected peptide pools. The IFNγ (A) and CD8+ T cell proliferation (B) responses of HIV+ subjects who responded at the first sample collection (t1), 2 y later (t2), and 4 y later (t3) are shown for peptide pool Fp14 (IFNγ), Hp15 (IFNγ), Hp10 (proliferation), and Fp9 (proliferation). The p-value indicates statistical differences between t1 and t2, t2 and t3, and t1 and t3. The threshold for IFNγ and proliferation responses are >50 SFU and >1% CFSElow, respectively.
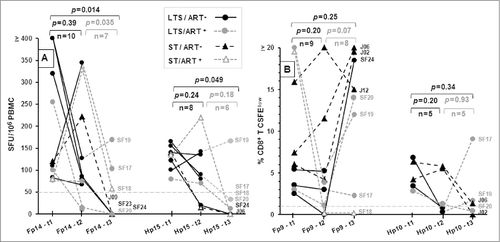
CD8+ T cells from 7 of 9 (78%) initial Fp9 responders retained T-cell proliferation responses to Fp9 during the 2-yr monitoring period (, left graph). Two of the 7 subjects responding to the Fp9 pool (SF17, SF19) were treated with ART during or shortly after the 2nd yr time point () leaving 4 subjects on ART and 4 subjects not on ART by yr 4. Three of 4 subjects on ART (SF17, SF19, SF20) maintained stable or increased levels of proliferation responses to Fp9, while one subject (SF18) continued to remain non-responsive. This finding suggests that the magnitude of response to Fp9 can improve in subjects (SF19, SF20) undergoing ART and may be related to either recovery of CD4+ lymphocytes or a reduction in viral load. In comparison, only 3 of 5 (60%) initial responders retained activity against Hp10, the counterpart of Fp9, through yr 2 (, right graph). This response declined in magnitude by the 4th yr. A large majority of these responders had either a major decrease or a loss of response to the Hp10 peptide pool, while the response to pools Fp9, Fp14, and Hp15 persisted for at least 2 y and some for as long as 4 y The Fp9 and Hp10 pools have very little sequence similarity (), and the response to Fp9, but not Hp10, was retained over time (). Therefore, subsequent studies focused on evaluating the Fp9, Fp14, and Hp15 peptide pools. Due to the high frequency and magnitude of the response, Fp9 was selected for epitope mapping.
Table 2. Fp9/Fp9–3, Fp14/Fp14–3, and their counterpart aa sequences and CMI responses
Identifying the p24 epitope(s) that induce CMI responses
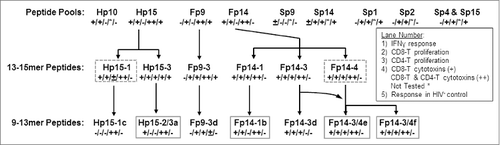
The HIV Hp15 pool has 3 well-established CD8+ CTL epitopes described in LANL database that are present within the Hp15–1a, Hp15–1c, and Hp15–2/3a peptides (). These CTL epitopes have high sequence similarity to FIV Fp14–1b, Fp14–1a, and Fp14–3/4f respectively (). Furthermore, SIV Sp14–1b and Sp14–1a have sequence similarity to their direct counterparts Hp15–1a/Fp14–1b and Hp15–2/3a/Fp14–3/4f. In addition, these peptide epitopes show moderate to high conservation of the Hp15/Fp14/Sp14 sequence between individual subtypes of species-specific lentiviruses (). Three to 4 overlapping 13–15mer peptides constitute each of the peptide pools Fp9 (Fp9–1, Fp9–2, Fp9–3), Fp14 (Fp14–1, Fp14–2, Fp14–3, Fp14–4), and Hp15 (Hp15–1, Hp15–2, Hp15–3). Fp9–3 and Hp10–3 have an aa sequence similarity of 29% and identity of 12% with 4 single aa differences due to gaps (). This low degree of sequence similarity and identity further supports the concept that epitope(s) on Fp9 are most likely not in the same location as those on Hp10. The analysis of individual 13–15mer peptides in the Fp9 pool indicates that the CD8+ T cells of the Fp9 responders proliferate predominantly in response to Fp9–3 (6 of 7) and to a lesser extent to Fp9–2 (3 of 7) (). Based on this result, the 15mer Fp9–3 peptide was further evaluated to map specific proliferative epitope(s) using shorter (9mer) overlapping peptides.
Figure 4. IFNγ and CD8+ T-cell proliferation responses to Fp9, Fp14, and Hp15 peptide epitopes. Peptide pool Fp14 consists of 4 overlapping 13–15mer peptides spanning the amino- to carboxyl-terminus (Fp14–1, Fp14–2, Fp14–3, Fp14–4), while pools Hp15 (Hp15–1, Hp15–2, Hp15–3) and Fp9 (Fp9–1, Fp9–2, Fp9–3) each consist of 3 overlapping 13–15mer peptides. IFNγ responses to individual Fp14 peptides (A; n=9 ), Hp15 peptides (B; n=9 ) and CD8+ T-cell proliferation responses to individual Fp9 peptides (C; n=7 ) are shown along with responses to their corresponding peptide pools. Only responders to pools Fp14 (A), Hp15 (B), and Fp9 (C) were tested. The responders consisted of long-term survivors (LTS) without ART (LTS; black bar); recently diagnosed subjects (<1 year) with short-term infection without ART (ST; gray bar); and those on ART with various duration of infection (ART+, red bar). Each bar with (*) in panels A and B are from the same 3 LTS.
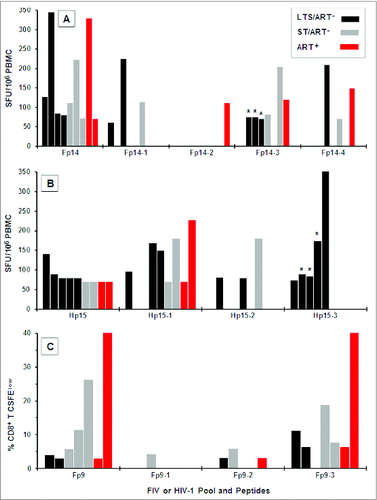
Table 3. Sequence conservation in EC p24 epitopes of Hp15/Fp14/Sp14 pools
When compared to Fp9 and Fp10 pools, Fp14 and Hp15 pools have a higher aa sequence similarity (65%) and identity (35%) with one aa difference due to a gap (). Based on aa sequence alignment analysis, the approximate counterpart for Hp15–1 and Hp15–2 peptides are Fp14–1 and Fp14–2 peptides respectively, whereas the Hp15–3 peptide contains regions that overlap both Fp14–3 and Fp14–4 peptides. Smaller regions have more similarity between Fp14–1 and Hp15–1 peptides () and between Fp14–4 and Hp15–3 peptides (). PBMC from Fp14 responders had substantial IFNγ responses to peptide Fp14–3 (6 of 9 responders) followed by peptides Fp14–1 and Fp14–4 (both 3 of 9) (). The majority of Hp15 responders had substantial IFNγ responses to Hp15–1 (7 of 9) and fewer responses to Hp15–3 (5 of 9) (). Thus, Fp14–3 and Hp15–1 contain epitopes that induce significant IFNγ responses. These peptides are not counterpart FIV and HIV peptides based on aa sequence analysis and therefore do not share common epitope(s). However, PBMC from 3 LTS responded to both Fp14–3 and its counterpart Hp15–3, indicating a conserved CMI epitope within these peptides (,B; bars with*).
When specific epitope analyses of Fp9 and Fp14 regions were performed, 2 9mer peptides (Fp9–3c and Fp9–3d) of the Fp9 region, differing by a single aa in carboxyl-end or amino-end, provided the highest frequency of a CD8+ T-cell proliferation response (5/6 of 9) () but at a low magnitude (<13 % CFSElow) (). Proliferation responses to Fp9–3c and Fp9–3d peptides were much lower than the levels of proliferation observed with the Fp9 pool (). Furthermore, this result is in stark contrast to the high frequency of responders (3 of 6, 50%) and the higher levels (average magnitude of 14% CFSElow) of CD8+ T-cell proliferation to the 15mer Fp9–3 peptide (). Thus, the 15mer, but not the 9mer Fp9–3 peptides, appears to contain the epitope that induces the bulk of the proliferation responses of both CD8+ and CD4+ T cells ().
Figure 5. IFNγ and T-cell proliferation responses to 9–13mers of FIV peptides Fp9–3, Fp14–3, and Fp14–4. Six 9–12mer peptides for the Fp9 pool (A1, B1, C1) and 7 9–13mer peptides for the Fp14 pool (A2, B2, C2) described in were tested for their ability to induce CD8+ T-cell proliferation (A), CD4+ T-cell proliferation (B), and IFNγ production (C) and compared to results with the Fp9 and Fp14 peptide pools. Thirteen–15mer peptides Fp9–3, Fp14–3, and Fp14–4; and mitogen (PHA) were included as controls. A total of 9 HIV+ responders to the Fp9 pool (A1, B1, C1) and 10 HIV+ responders to the Fp14 pool (A2, B2, C2) were tested up to 4 y after the beginning of the study. Two to 4 of the responders were lost to follow up. Their proliferation and IFNγ results to the 13–15mer peptides Fp9–3, Fp14–3, and Fp14–4, and are denoted with an (*) for each individual missing.
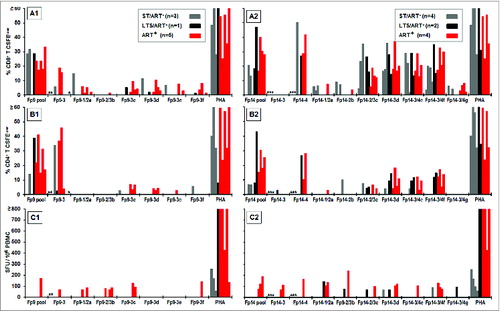
Table 4. 9–13mer T-cell epitope mapping of Fp9 and Fp14 sequences using responders to Fp9 or Fp14
Similarly specific epitope analysis of the Fp14–3 region with an overlap with the Fp14–2 and Fp14–4 regions determined that a higher frequency of HIV+ subjects respond to epitopes in Fp14–3 (Fp14–3d) and Fp14–4 (Fp14–3/4f, overlapping both Fp14–3 and Fp14–4) more than in Fp14–2, based on both CD8+ T-cell proliferation and IFNγ responses (). Since the shorter sequence of 10mer Fp14–3/4f also resides in the larger 13mer Fp14–3d sequence (), it is still possible that both contain the same epitope (i.e., Fp14–3/4f). This observation is supported by in silico analysis using the HLA algorithm where the predicted HLA A2 supertype is common among all 4 responders for both Fp14–3d and Fp14–3/4f (). Thus, Fp14–3/4f contains the major epitope residing in Fp14–3 and Fp14–4 that is identified by the HLA A2 supertype.
Table 5. HLA supertypes of the responders to key short and long peptides
Characterization of CTL-associated activity induced by Fp9, Fp14, and Hp15 peptides
In intracellular staining (ICS) analysis for cytotoxins, both CD8+ and CD4+ T cells expressed granzyme B (GrzB) most consistently in response to all 3 peptide pools tested (Fp9, Fp14, Hp15) (). 40% (2 of 5) HIV+ subjects had CD8+ T cell GrzB expression to individual peptide pool Fp9, whereas 80% (4 of 5) responded to Fp14, and 80% (4 of 5) to Hp15 (). Notably, GrzB was expressed by both CD4+ and CD8+ T cells from the same individuals although the overall expression magnitude of GzB, but not the frequency, was lower in the CD4+ T cells (). A few of the subjects who did not have GrzB responses did demonstrate either GrzA or perforin expression (). When individual 13–15mer peptides from each pool were examined, CD8+ T cells from all 5 HIV+ subjects responded to Fp14–3, Fp14–4, and Hp15–1, whereas up to 4 HIV+ subjects responded to Fp9–3 with the production of one or more cytotoxins. Hence, the majority of the Fp9, Fp14, and Hp15 peptides induced expression of one or more cytotoxins.
Figure 6. Stimulation of cytotoxins by CTL epitopes on Fp9, Fp14, and Hp15 peptide pools, and their individual peptides. The ICS analyses for perforin (Perf, ○), granzyme A (GrzA, □), and granzyme B (GrzB, Δ) are shown for CD8+ T cells (A, D) and CD4+ T cells (B, C) from 5 HIV+ responders with or without ART. Two LTS/ART−, one LTS/ART+, one ST/ART+, and one ST/ART− were first evaluated (A, B). Peptides tested included Fp9, Fp14, and/or Hp15 peptide pools or large 13–15mer peptides (A, B). Responses from 5 HIV+ responders of short-term HIV-infected subjects not on ART (ST/ART−) were tested using small 9–13mer peptides within Fp9–3, Fp14–1, Fp14–3, Fp14–4, and Sp14–1 (C, D). Note that Sp14 pool is a single 13mer peptide Sp14–1. Counterpart peptides are shown with blue-dashed box for Fp14–1a/Hp15–1c/Sp14–1c, purple-dashed box for Fp14–1b/Hp15–1a/Sp14–1b, and red-dashed box for Fp14–3/4f/Hp15–2/3a/Sp14–1a. The peptides tested with one less subject are denoted by (**), while their number of subjects and number of responses are denoted by (*). Each separate symbol color represents one subject, and each color-coded subject is shown with his/her infection status. The threshold for T cells expressing cytotoxin is set at >0.1% CD4+ or CD8+ T cells.
The short (9–13mer) peptides of Fp9–3, Fp14–3, and Fp14–4 from previous IFNγ and proliferation studies as well as a few additional short peptides () were further tested with T cells from short-term HIV-infected subjects not on ART (ST/ART−) for production of cytotoxins and expression of CD107a (), which are commonly expressed by CTLs. These short 9–13mer epitopes were produced based on the CTL algorithm of NetCTL 1.2 and HLA algorithm of NetMHC 3.4. The shortest peptides that predicted the highest CD8+ T-cell responder frequency were 9mer peptides Fp9–3c (RMQCRAWYL) and Fp9–3d (ARMQCRAWY), 10mer peptide Fp14–3/4f (KLYLKQSLSI), and 13mer peptide Fp14–3d (AEVKLYLKQSLSI) (). Peptides Fp9–3, Fp9–3c and Fp9–3d induced very little cytotoxin expression in CD4+ T cells (). Among these peptides, Fp9–3 and Fp9–3d induced more cytotoxins in CD8+ T cells from a slightly larger number of ST/ART− subjects (). In comparison, Fp14–4 and Fp14–3/4f peptides followed by Fp14–1b and Fp14–3/4e induced a high frequency of cytotoxin expression in both CD4+ and CD8+ T cells from a majority of ST/ART− subjects (,D). Remarkably, the Fp14–4 and Fp14–3/4f peptides had a higher number of cytotoxin responses and a higher frequency of responders for both CD4+ and CD8+ T cells than their counterparts on HIV (Hp15–3; Hp15–2/3a) and SIV (Sp14–1; Sp14–1a) as well as when compared to those induced by Fp9–3 and its shorter peptides.
When NetCTL and NetMHC predictions were compared to the responders' HLA class-I supertype(s), 4 responders to peptide Fp9–3c had the predicted responder HLA supertype A2 (). Three of them also had an additional HLA supertype (A1, A3, B27, or B62) predicted to have a strong binding affinity to Fp9–3c. The same analysis performed on Fp9–3d determined that 3 of 4 responders possessed supertype B44 while one responder had HLA supertype A1. Both supertypes are predicted to have a strong binding affinity to Fp9–3d. Similarly, Fp14–3/4f showed supertype A2 as the common HLA supertype correlating with all 4 responders. Moreover, 3 more subjects had additional HLA supertypes (A1, B27, or B58) with strong predicted binding affinity for Fp14–3/4f (). Hence, the ICS and the combined NetCTL/NetMHC analyses support the presence of CD8+ T-cell epitopes on Fp9–3, Fp14–3, and Fp14–4 peptides.
Direct effect of peptides Fp9–3, Hp15–1, and Hp15–3 on in vitro HIV-1 infection
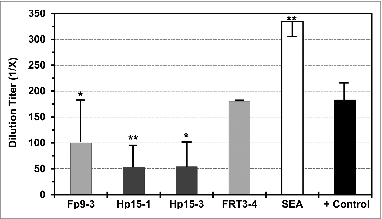
Peptides Fp9–3 and Hp15–3 induced proliferation responses in PMBC and T cells of HIV− healthy subjects. This observation raised a concern on whether such T-cell proliferation can non-specifically enhance HIV-1 infection. Remarkably, these peptides and peptide Hp15–1 did not enhance HIV-1 infection but instead significantly suppressed the in vitro HIV-1 infection of uninfected PBMC upon inoculation with HIV-1LAV(). As expected, enhancement of HIV infection was observed in the T-cell mitogen stimulated control, whereas the negative control consisting of non-reactive FIV reverse transcriptase peptide FRT3–4 (see Methods) had no effect.
Figure 7. Direct effect of Fp9–3, Hp15–1, and Hp15–3 peptides on in vitro HIV-1 infection. The direct effect of FIV peptides Fp9–3 and FRT3–4 (light gray bar), HIV-1 peptides Hp15–1 and Hp15–3 (dark gray bar), and virus positive control (+ control, black bar) on HIV-1 infection of PBMC from healthy uninfected subjects is shown as average end-point dilution titer with standard deviation. Staphylococcal enterotoxin A control (SEA, white bar) is included as a T-cell mitogen control to serve as positive control for FIV enhancement. FIV reverse transcriptase peptide FRT3–4 previously reported to be non-reactive to T cells from HIV− subjects is included as a negative control.Citation48 Statistically significant difference between the average result of each peptide and the average virus control are shown with (*) for P < 0.02 and (**) for p < 0.001.
Discussion
Vaccination that selectively stimulates the immune system through both neutralizing antibody and polyfunctional T cells and avoiding areas that result in viral enhancement is critical to producing a safe and effective HIV vaccine. Although the focus of vaccination has largely centered around using entire regions of the virus to modulate the immune response, inadvertent selection of epitopes that enhance viral infection constitutes significant risk. Clearly, it has been well documented in both HIV and FIV that enhancement of infection can occur via antibody, complement and potentially T cell activation and upregulation of surface markers resulting from antigenic stimulation.Citation25,26 In addition, previous immunizations using non-targeted sections of FIV p24 and envelope have also resulted in enhancement of infection or at the very least, failure to induce protective immunity.Citation26-29 In this study, we have identified selected areas of p24 of both FIV and HIV that stimulate polyfunctional T cells and suppress viral replication, thus making them prime candidates for inclusion in an HIV vaccine.
Based on IFNγ ELISpot and CFSE-proliferation analysis, the PBMC and T cells from HIV+ subjects () identified at least 2 cell-mediated immune (CMI) peptide epitopes in the FIV p24 pools Fp9 and Fp14 that could serve as potential T-cell immunogens (). These peptides were effective in the majority of subjects that had a corresponding in silico predicted HLA and did not induce substantial responses in CD4+ T cells from HIV− subjects. Peptide pool Fp14 induced robust IFNγ production with the highest frequency of responders (32%) and moderate CD8+ T-cell proliferation responses ( and ). In contrast, peptide pool Fp9 induced robust CD8+ T-cell proliferation with the highest frequency of responders (26%) and no IFNγ responses. Most notably, the immune activity induced by the Fp9 and Fp14 peptide pools was detected on repeated patient testing and persisted for up to 4 y ().
Unexpectedly, SIV p24 pools induced more CD4+ and CD8+ T-cell proliferation responses than the corresponding HIV p24 pools in HIV+ subjects (). Moreover, the attenuated CD4+ T-cell proliferation responses to the SIV pools often mirrored the CD8+ T-cell proliferation responses since CD4+ T-cell proliferation responses were observed only from those who also had CD8+ T-cell proliferation responses to the same peptides (). In some cases, these CD4+ T cells could be more sensitive to HIV-1 infection (see below). However, possibly the CD4+ T cells that are cross-reactive to these peptides persist in HIV+ subjects and can perhaps be stimulated by these conserved epitopes to enhance the CD8+ T-cell responses against HIV.
In addition to high aa sequence similarity (), the peptide pools Hp15 and Fp14 induced IFNγ responses, notably, only in PBMC from HIV+ subjects (). Furthermore, Fp14–3 and its HIV counterpart, Hp15–3, had a high frequency of IFNγ responders. When small 9–13mer peptides from the Fp14 region were evaluated (), 2 overlapping peptides within Fp14–3 and Fp14–4 (Fp14–3d, Fp14–3/4f) showed high CD8+ and CD4+ T-cell proliferation responses, low IFNγ responses in HIV+ subjects () and elicited no response in HIV− subjects (average of <10 SFU). Thus, the epitope(s) present in peptide pools Hp15 and Fp14 are specific and likely to be evolutionarily conserved.
Peptide analysis of the Fp9 region gave a high frequency of responders measured by CD8+ T-cell proliferation to peptides Fp9–3c and Fp9–3d but higher CD8+ T-cell proliferation responses to the 15mer peptide Fp9–3 (, ). One concern regarding the Fp9–3 peptide is its ability to elicit non-specific CD8+ and CD4+ T-cell proliferation (enhanced immunogenicity); this enhanced immunogenicity can serve as an activation signal and could enhance HIV-1 infection.Citation30,31 However, Fp9–3 peptide is not a classical enhancer as compared with other T-cell mitogens (PHA and concanvalin A) because it does not induce IFNγ in T cells from either HIV+ or HIV− subjects (). IFNγ production in some cases could enhance HIV-1 infection.Citation32,33 A percentage of CD8+ T cells from HIV− subjects proliferated in response to the Fp9 pool and the 15mer Fp9–3 peptide but not to Fp9–3c and Fp9–3d 9mer peptides (data not shown). These less enhancatory peptides have a strong algorithmic prediction for CD8+ T-cell activity with the most common HLA supertypes A2 and B44 ().Citation34 Therefore, the less enhancatory Fp9–3c and Fp9–3d peptides are likely better candidates as vaccine immunogens.
In this report, the CTL epitopes Hp15–1c, Hp15–1a, and Hp15–2/3a were further evaluated for cytotoxin expression along with their counterpart in FIV Fp14–1a, Fp14–1b, and Fp14–3/4f, respectively (). In these studies, the HIV peptides and their FIV counterparts, Hp15–1c/Fp14–1a (blue box), Hp15–1a/Fp14–1b (purple box), and Hp15–2/3a/Fp14–3/4f (red box) had high cytotoxin and CD107a expression. The respective SIV counterparts Sp14–1c, Sp14–1b, and Sp14–1a had a slightly lower number of responses than either their FIV or HIV-1 counterpart ().
The CTL epitope within the Hp15–1 peptide described in LANL database is predicted to bind strongly to HLA supertype B44.Citation35 The B44 supertype is associated with a lower incidence of HIV disease progression in study subjects in South Africa, Botswana, and Zimbabwe.Citation36,37 The inducers of CD8+ T-cell proliferation in HIV− subjects are epitopes within Hp15–3 and, to a lesser extent Hp15–2, but not in the Hp15–1 peptide (). Thus, 2 known CTL epitopes within Hp15–1c and Hp15–1a elicited substantial levels of cytotoxin and CD107a expression but not as high as the well characterized epitope Hp15–2/3a. Hp15–2/3a induced expression in both CD4+ and CD8+ T cells and did not stimulate T cells from HIV− subjects. As a result, it should be a better candidate for use as a vaccine immunogen.
Among the 3 peptides in Fp9 pool, the 15mer Fp9–3 had the highest frequency of responders expressing one or more cytotoxins by ICS analysis (). Based on NetCTL analysis, this peptide can mediate CD8+ T-cell activity by expressing peptide-specific cytotoxin(s) and using multiple HLA supertypes (A2, B7, B8, B27, B62). This finding makes it a strong candidate as a HIV-1 vaccine immunogen (). Similarly, the epitopes in Fp14–4 and lesser extent in Fp14–3 () had the highest frequency of responders expressing cytotoxin(s) with high binding affinity for supertypes A2 and/or B44 (). Since the HLA B44 supertype is associated with either control of HIV infection and/or slow progression to AIDSCitation38-40 and the HLA A2 supertype is associated with low HIV transmissionCitation41-43, targeting these supertypes are likely beneficial in the development of an effective vaccine. In addition, a recent study correlated HLA A2 alleles with vaccine efficacy in the RV144 HIV vaccine trial and highlighted the importance of HLA allotypes in developing an effective HIV vaccine.Citation44 The percent of patients expressing these supertypes in the US is significant (A2, 27–50%; B44, 17–27% based on ethnicity) with similar frequencies of A2 supertype observed in sub-Saharan Africa and Thailand.Citation45 Study subjects demonstrated an overall prevalence of 48% of A2 and higher than estimated prevalence of B44 at 44% (Table S1). Notably, the current observations indicate that the cross-reactive peptides Fp9–3, Fp9–3c, Fp9–3d, Fp14–3d, and Fp14–3/4f induce CMI responses in HIV+ subjects and are predicted to bind with highly prevalent HLA supertypes A2 and/or B44 ().Citation34,45,46
Interestingly, a large number of HIV− subjects also responded to HIV and to a lesser degree FIV and SIV peptides. Although a reason for the response in the uninfected population was not specifically determined, it is important to note that peptide design was targeted heavily toward common HLA types (A2, B44) as described. As such, although patients were HIV− and supposed not exposed to antigens, those HIV− subjects who responded to the HIV peptides and to Fp9–3c do represent an HLA subset with a greater number of predicted HLA supertypes representing MHC restricted peptide targeting (Table S1). In the case of HIV− subjects who responded Fp14, there was an overrepresentation of additional supertypes, specifically A03 (7/7 responders), that were not predicted as targeted supertypes. This may represent additional responding supertypes that may be included and not predicted by the algorithm. Alternatively, reaction to these sequences in HIV− individuals may be due to exposure of naïve individuals to molecular HIV mimics such as human endogenous retrovirus (HERV). HERV sequences within the genome of naïve subjects have been shown to elicit a T cell specific response to HIV-1/2 as well as SIV.Citation47 Although a non-specific response in uninfected individuals may indicate broad stimulation of the immune system, this is not necessarily detrimental, as immune recognition by naïve individuals is required for an effective vaccine. It is possible for such a phenomenon to be beneficial however, through priming the immune response or harmful by upregulating cell surface markers of inflammation and enhancing HIV infection. Interestingly, further analysis of this phenomenon using in vitro viral enhancement/suppression assay revealed a viral suppressive effect of the identified peptides Fp9–3 and Hp15–3 () suggesting a beneficial effect of immune stimulation by these candidate peptides.
Previously, we described evolutionarily conserved CD8+ T-cell epitopes on the FIV reverse transcriptase.Citation48 In the current study, we have identified cross-reactive p24 epitopes that are found in both HIV and FIV peptide sequences. These results support the existence of an evolutionary lineage among essential proteins of inter-species lentiviruses. Being conserved, these sequences are most likely essential for viral fitness, and thus less likely to mutate.Citation13
In summary, by evaluating IFNγ production, CFSE proliferation, and cytotoxin production in both HIV+ and HIV− subjects (), we can conclude that the large 13–15mer peptides, HIV Hp15–1 and FIV Fp14–4, and small 9–10mers Hp15–2/3a, Fp14–1b, Fp14–3/4e, and Fp14–3/4f induce robust CMI responses without enhancement of infection. Inclusion of epitopes that generate polyfunctional activity (a combination of IFNγ, T-cell proliferation and/or cytotoxin responses), is considered an important component in HIV vaccine design in that increased numbers of polyfunctional T cells are present in HLA-B27 supertype patients (those demonstrating a higher natural resistance to HIV infection) and are more prevalent in HIV nonprogressors.Citation9,49 Since the Fp9 and Fp14 epitopes possess polyfunctional activity, they also merit consideration as potential immunogens for inclusion in an effective HIV-1 vaccine. Selectively targeting these conserved sequences and monitoring non-enhancing, T-cell specific responses allow the identification of conserved FIV, HIV, and SIV immunogenic peptides that could be included in an HIV vaccine for prophylaxis and immunotherapy.
Figure 8. Summary of functional epitope analyses. Results from 3 functional analyses are summarized according to frequency of HIV+ (lanes 1–4) or HIV− (lane 5) responders and are shown as (+) for frequency of >25 % responders, (±) for 19–25% responders, (−) for <19% responders, or as (*) for not available. Each lane, abridged in the insert, shows the ability of the peptide pool or peptide to stimulate an IFNγ response (lane 1), CD8+ T-cell proliferation (lane 2), and CD4+ T-cell proliferation (lane 3). Lane 4 denotes the ability to induce cytotoxin(s) in only CD8+ T cells (+) or in both CD8+ and CD4+ T cells (++) by 4 or more HIV+ subjects when at least 9 subjects tested. In lane 5, the positive result (+) indicates substantial frequency of proliferation responses from HIV− subjects and may indicate a safety concern; whereas a negative result (−) indicates no substantial HIV− response to the peptide pool or peptide. Positive response of HIV− control had CD8+ T-cell proliferation response in 30–42% of HIV− subjects. The large 13–15mer peptides with the best CMI responses without stimulation in HIV− subjects are shown with dashed boxes, while the best small peptides are shown with solid boxes. Therefore, Hp15–1, Hp15–2/3a, Fp14–4, Fp14–1b, Fp14–3/4e, and Fp14–3/4f peptides appear to contain the best potential epitopes to target for use as HIV immunogens.
Materials and Methods
Study population
The blood samples of HIV+ subjects were obtained from the University of California at San Francisco (UCSF) and the University of Florida Center for HIV/AIDS Research, Education and Service (UF CARES) in Jacksonville using the protocol approved by the Institutional Review Board at UF. The HIV+ subjects consisted of 14 long-term survivors (LTS) without antiretroviral therapy (ART) (LTS), 10 recently diagnosed subjects (<1 year) with short-term infection without ART (ST); and eleven HIV+ subjects receiving ART with various duration of infection (ART+). Age, gender, and race as well as the viral and immune status of the HIV+ subjects used in the current study are outlined in . All patients were considered to be in clinical latency. The blood samples were processed within 48 hours of collection. T-cell phenotyping and HIV-1 load were performed by the clinical laboratories at the UCSF Medical Center and UF CARES. The samples from 22 healthy HIV seronegative (HIV−) subjects were obtained from LifeSouth Community Blood Centers (Gainesville, FL) or from UF. HLA typing was performed by the clinical laboratories at the UF Shands Medical Center. Supertypes were determined based on previous reports.Citation50
ELISpot assays
Human enzyme-linked immunosorbent spot assays (ELISpot) (R&D Systems, Cat# XEL285) which measure IFNγ production were performed.Citation51 The positive threshold for human IFNγ responses was >50 spot forming units (SFU)/10Citation6 cells. The final value for each subject was derived after subtracting the result of each HIV+ subject with the media control followed by subtraction with the average response of the HIV− subjects from each peptide pool which was rarely more than 10 SFU.
Flow cytometry (FACS) for measuring CFSE-proliferation and intracellular cytokine staining (ICS)
Carboxyfluorescein diacetate succinimide ester (CFSE)-proliferation analysis was performed according to the manufacturer's protocol (Invitrogen) and processed as previously describedCitation52 using the following modification: 2.5–5.0×10Citation5 CFSE-labeled PBMC stimulated for 4–5 d (37°C, 5% CO2) with 15–30 μg of peptides in culture media (AIM V medium, 25 μg/mL gentamycin, and 10% heat-inactivated fetal bovine serum). The ICS analysis was performed as previously described.Citation53,54
The antibodies used for the proliferation analysis consisted of anti-CD4 allophycocyanin (APC) anti-CD3 APC-H7, and anti-CD8 Pacific Blue, and those for ICS were anti-CD3 APC-H7, anti-CD4 BD Horizon V450, anti-CD8 FITC, anti-granzyme B (GrzB) Alexa 700, anti-granzyme A (GrzA) PE, CD107a Alexa 700 (BD Biosciences, Cat# 555349, 560176, 558207, 560345, 555366, 560213, 558904, 561340), and anti-perforin PerCP (Abcam, Cat# ab86319). Both analyses were performed with BD LSRII and FACSDiva™ Software (BD Biosciences), using a positive threshold of >1 % CFSElow for CFSE-proliferation except for ICS studies with threshold of >0 .1% T cells expressing cytotoxin. The final value for each subject was derived after subtracting the result of each HIV+ subject with the media control followed by subtraction with the average response of the media-control subtracted HIV− subjects from each peptide pool.
Peptide design and Human leukocyte antigen (HLA) binding analyses
Peptides were derived from p24 sequences of HIV-1 UCD-1 (subtype B), SIV MM251 and FIV FC-1 (subtype B). Thirteen–15mer peptides with an 11 amino acid overlap were used for initial screening followed by targeted 9mers using previously responding subjects (). The affinity of peptide binding to HLA was determined by NetMHC version 3.2 for HLA class-I (http://www.cbs.dtu.dk/services/NetMHC/), NetMHCII version 2.2 for HLA class-II (http://www.cbs.dtu.dk/services/NetMHCII/), and NetCTL version 1.2 for CTL-associated epitopes (http://www.cbs.dtu.dk/services/NetCTL/). The LANL database for CD8+ and CD4+ epitopes are based on the HIV-1 HXB2 sequence and identifies the epitope-interacting HLA allele(s).
Viral Enhancement/Suppression Assay
Few of the T-cell peptides were tested in vitro for HIV-1 enhancement or suppression of viral infection. This assay is a 48-well modification of the 25-cmCitation2 flask method described for detecting HIV-1 enhancing activities of IFNγ and HIV/FIV suppressive activities of IFNα and/or IFNβ.Citation32 Briefly, 1.0–1.5×10Citation6 unstimulated PBMC from HIV− donor was cultured in a final volume of 1-mL/well with 7 μg/mL of peptides Hp15–1, Hp15–3, Fp9–3, or negative control peptide FRT3–4. FRT3–4 is an FIV reverse transcriptase peptide previously reported to be reactive with T cells from HIV+ subjects but do not react with T cells from HIV− subjects.Citation48 The infection enhancing control was cultured with 0.4 μg/mL of Staphylococcal enterotoxin A (SEA). Eight hours after the culturing with peptides or mitogen control (SEA), varying dilutions of HIV-1LAV were added to the wells. On Day 3 (72 hours) the cells were spun down and re-cultured in fresh culture media with PHA-stimulated autologous PBMC (1.0–1.5×10Citation6) using the same amount of corresponding peptide or media for media and mitogen controls. On Days 6, 9, and 12 of culture, 0.5 mL of the culture fluid was collected and fresh media added to keep a consistent volume. The harvest culture samples were analyzed to determine HIV-1 titer by RT assay.Citation17 Results are shown as end-point dilution titer of samples from 2 to 3 studies using PBMC from different HIV− donors.
Statistical analysis
Statistically significant differences between the results from 2 time points were calculated using a paired Student t-test with a 2-tailed distribution (SigmaPlot version 11.0) and were considered statistically significant when p<0.05.
Disclosure of Potential Conflicts of Interest
JKY is the inventor of record on a patent held by the University of Florida and may be entitled to royalties from companies developing commercial products related to the research described in this paper.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Supplemental_Table_S1.docx
Download MS Word (16.3 KB)Acknowledgments
The authors thank Melissa Volz, Shweta Kaushik, Melissa S. Abreu, and Sue H. Fujimura for their technical assistance, and Kaylynn Peter and Kathy Thoma for administrative assistance in organizing study subjects.
Funding
This work was supported by NIH R01-AI65276 (JKY and JAL), R01-AI30904 (JKY), and JKY Miscellaneous Donors Fund. This work was in part supported by the NIH/NCRR Clinical and Translational Science Award UL1 RR029890 (MHR) and TL1 TR00066 (SRR).
References
- Center for Disease Control. HIV/AIDS Surveillance Report 2010, Supplemental Report, Estimated HIV incidence in the United States, 2007-2010. 2010; (Vol. 17, Number 4). http://www.cdc.gov/hiv/pdf/statistics_hssr_vol_17_no_4.pdf. Accessed May 30, 2014
- Centers for Disease Control and Prevention. HIV Surveillance Report, 2011; 2013; vol. 23. Rates of diagnoses of HIV infection among adults and adolescents, by area of residence, 2011 - United States and 6 dependent areas. Published February 2013. http://www.cdc.gov/hiv/topics/surveillance/resources/reports/. Accessed May 30, 2014.
- Hankins, C.. Overview of the current state of the epidemic. Current HIV/AIDS reports 2013; 10, 113-123.
- O'Connell RJ, Kim JH, Corey L, Michael NL. Human immunodeficiency virus vaccine trials. Cold Spring Harb Perspect Med 2 2012; a007351
- Flynn NM, Forthal DN, Harro CD, Judson FN, Mayer KH, Para MF, rgp, H.I.V. Vaccine Study Group. Placebo-controlled phase 3 trial of a recombinant glycoprotein 120 vaccine to prevent HIV-1 infection. J Infect Dis 2005; 191, 654-65; PMID:15688278; http://dx.doi.org/10.1086/428404
- Pitisuttithum P, Gilbert P, Gurwith M, Heyward W, Martin M, van Griensven F, Hu D, Tappero JW, Choopanya K, Bangkok Vaccine Evaluation, G.. Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. J Infect Dis 2006; 194, 1661-71; PMID:17109337; http://dx.doi.org/10.1086/508748
- Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med 2009; 361, 2209-20; PMID:19843557; http://dx.doi.org/10.1056/NEJMoa0908492
- Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med 2012; 366, 1275-86; PMID:22475592; http://dx.doi.org/10.1056/NEJMoa1113425
- de Souza MS, Ratto-Kim S, Chuenarom W, Schuetz A, Chantakulkij S, Nuntapinit B, Valencia-Micolta A, Thelian D, Nitayaphan S, Pitisuttithum P, et al. The Thai phase III trial (RV144) vaccine regimen induces T cell responses that preferentially target epitopes within the V2 region of HIV-1 envelope. J Immunol 2012; 188, 5166-76; http://dx.doi.org/10.4049/jimmunol.1102756
- Cao J, McNevin J, Holte S, Fink L, Corey L, McElrath MJ. Comprehensive analysis of human immunodeficiency virus type 1 (HIV-1)-specific gamma interferon-secreting CD8+ T cells in primary HIV-1 infection. J Virol 2003; 77, 6867-78; PMID:12768006; http://dx.doi.org/10.1128/JVI.77.12.6867-6878.2003
- Kaufmann DE, Bailey PM, Sidney J, Wagner B, Norris PJ, Johnston MN, Cosimi LA, Addo MM, Lichterfeld M, Altfeld M, et al. Comprehensive analysis of human immunodeficiency virus type 1-specific CD4 responses reveals marked immunodominance of gag and nef and the presence of broadly recognized peptides. J Virol 2004; 78, 4463-77; PMID:15078927; http://dx.doi.org/10.1128/JVI.78.9.4463-4477.2004
- Pontesilli O, Klein MR, Kerkhof-Garde SR, Pakker NG, de Wolf F, Schuitemaker H, Miedema F. Kinetics of immune functions and virus replication during HIV-1 infection. Immunol Lett 1997; 57, 125-30; PMID:9232438; http://dx.doi.org/10.1016/S0165-2478(97)00047-3
- Sanou MP, De Groot AS, Murphey-Corb M, Levy JA, Yamamoto JK. HIV-1 Vaccine Trials: Evolving Concepts and Designs. open AIDS J 2012; 6, 274-88; PMID:23289052; http://dx.doi.org/10.2174/1874613601206010274
- Uhl EW, Heaton-Jones TG, Pu R, Yamamoto JK. FIV vaccine development and its importance to veterinary and human medicine: a review FIV vaccine 2002 update and review. Vet Immunol Immunopathol 2002; 90, 113-32; PMID:12459160; http://dx.doi.org/10.1016/S0165-2427(02)00227-1
- Elder JH, Lin YC, Fink E, Grant CK. Feline immunodeficiency virus (FIV) as a model for study of lentivirus infections: parallels with HIV. Curr HIV Res 2010; 8, 73-80; PMID:20210782; http://dx.doi.org/10.2174/157016210790416389
- Kenyon JC, Lever AM. The molecular biology of feline immunodeficiency virus (FIV). Viruses 2011; 3, 2192-213; PMID:22163340; http://dx.doi.org/10.3390/v3112192
- Coleman JK, Pu R, Martin MM, Noon-Song EN, Zwijnenberg R, Yamamoto JK. Feline immunodeficiency virus (FIV) vaccine efficacy and FIV neutralizing antibodies. Vaccine 2014; 32, 746-54; PMID:23800540; http://dx.doi.org/10.1016/j.vaccine.2013.05.024
- Uhl EW, Martin M, Coleman JK, Yamamoto JK. Advances in FIV vaccine technology. Vet Immunol Immunopathol 2008; 123, 65-80; PMID:18295907; http://dx.doi.org/10.1016/j.vetimm.2008.01.030
- Coleman JK, Pu R, Martin M, Sato E, Yamamoto JK. HIV-1 p24 vaccine protects cats against feline immunodeficiency virus infection. Aids 2005; 19, 1457-66; PMID:16135898; http://dx.doi.org/10.1097/01.aids.0000183627.81922.be
- Pontesilli O, Carotenuto P, Kerkhof-Garde SR, Roos MT, Keet IP, Coutinho RA, Goudsmit J, Miedema F. Lymphoproliferative response to HIV type 1 p24 in long-term survivors of HIV type 1 infection is predictive of persistent AIDS-free infection. AIDS Res Hum Retroviruses 1999; 15, 973-81; PMID:10445809; http://dx.doi.org/10.1089/088922299310485
- Rosenberg ES, Billingsley JM, Caliendo AM, Boswell SL, Sax PE, Kalams SA, Walker BD. Vigorous HIV-1-specific CD4+ T cell responses associated with control of viremia. Science 1997; 278, 1447-50; PMID:9367954; http://dx.doi.org/10.1126/science.278.5342.1447
- Gillespie GM, Kaul R, Dong T, Yang HB, Rostron T, Bwayo JJ, Kiama P, Peto T, Plummer FA, McMichael AJ, et al. Cross-reactive cytotoxic T lymphocytes against a HIV-1 p24 epitope in slow progressors with B*57. Aids 2002; 16, 961-72; PMID:11953462; http://dx.doi.org/10.1097/00002030-200205030-00002
- Zhang Y, Huber M, Euler-Konig I, Sussmuth R, Jung G, Jassoy C. Analysis of the proliferative responses to peptides in individuals with vigorous Gag protein-specific proliferation. Immunol Lett 2001; 79, 93-6; PMID:11595294; http://dx.doi.org/10.1016/S0165-2478(01)00270-X
- Mothe B, Llano A, Ibarrondo J, Zamarreno J, Schiaulini M, Miranda C, Ruiz-Riol M, Berger CT, Herrero MJ, Palou E, et al. CTL responses of high functional avidity and broad variant cross-reactivity are associated with HIV control. PloS one 2012; 7, e29717; PMID:22238642; http://dx.doi.org/10.1371/journal.pone.0029717
- Shmelkov E, Nadas A, Cardozo T. Could vaccination with AIDSVAX immunogens have resulted in antibody-dependent enhancement of HIV infection in human subjects? Hum Vacc Immunother. 2014; Ten:10, 3013-16
- Hosie MJ, Osborne R, Reid G, Neil JC, Jarrett O. Enhancement after feline immunodeficiency virus vaccination. Vet Immunol Immunopathol. 1992; 35:191-7; PMID:1337397; http://dx.doi.org/10.1016/0165-2427(92)90131-9
- Flynn JN, Cannon CA, Neil JC, Jarrett O. Vaccination with a Feline Immunodeficiency Virus Multiepitopic Peptide Induces Cell-Mediated and Humoral Immune Responses in Cats, but Does Not Confer Protection. J Virol. 1997; 71:10 7586-92. J Virol. 1997 Dec; 71(12):9640-9; PMID:9371628
- Richardson J1, Moraillon A, Baud S, Cuisinier AM, Sonigo P, Pancino G. Enhancement of feline immunodeficiency virus (FIV) infection after DNA vaccination with the FIV envelope. J Virol 1997; Dec;71(12):9640-9; PMID:9371628
- Siebelink KH1, Tijhaar E, Huisman RC, Huisman W, de Ronde A, Darby IH, Francis MJ, Rimmelzwaan GF, Osterhaus AD. Enhancement of feline immunodeficiency virus infection after immunization with envelope glycoprotein subunit vaccines. J Virol 1995; 1995 Jun;69(6):3704-11
- Spina CA, Prince HE, Richman DD. Preferential replication of HIV-1 in the CD45RO memory cell subset of primary CD4 lymphocytes in vitro. J Clin Investig 1997; 99, 1774-85; PMID:9120023; http://dx.doi.org/10.1172/JCI119342
- Stevenson M, Stanwick TL, Dempsey MP, Lamonica CA. HIV-1 replication is controlled at the level of T cell activation and proviral integration. EMBO J 1990; 9, 1551-60; PMID:2184033
- Yamamoto JK, Barre-Sinoussi F, Bolton V, Pedersen NC, Gardner MB. Human alpha- and beta-interferon but not gamma- suppress the in vitro replication of LAV, HTLV-III, and ARV-2. J Interferon Res 1986; 6, 143-52. -152; PMID:2425014; http://dx.doi.org/10.1089/jir.1986.6.143
- Roff SR, Noon-Song EN, Yamamoto JK. The Significance of Interferon-gamma in HIV-1 Pathogenesis, Therapy, and Prophylaxis. Front Immunol 2014; 4, 498; PMID:24454311; http://dx.doi.org/10.3389/fimmu.2013.00498
- Marsh SG, Parham P, Barber LD 2000. The HLA Class I and Class II Loci. In Marsh S.G., Parham P., Barber L.D. (eds), The HLA Facts Book. London: Academy Press; p. 93-272
- Kiepiela P, Ngumbela K, Thobakgale C, Ramduth D, Honeyborne I, Moodley E, Reddy S, de Pierres C, Mncube Z, Mkhwanazi N, et al. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med 2007; 13, 46-53; PMID:17173051; http://dx.doi.org/10.1038/nm1520
- Leslie A, Matthews PC, Listgarten J, Carlson JM, Kadie C, Ndung'u T, Brander C, Coovadia H, Walker BD, Heckerman D, et al. Additive contribution of HLA class I alleles in the immune control of HIV-1 infection. J Virol 2010; 84, 9879-88; PMID:20660184; http://dx.doi.org/10.1128/JVI.00320-10
- Carlson JM, Listgarten J, Pfeifer N, Tan V, Kadie C, Walker BD, Ndung'u T, Shapiro R, Frater J, Brumme ZL, et al. Widespread impact of HLA restriction on immune control and escape pathways of HIV-1. J Virol 2012; 86, 5230-43; PMID:22379086; http://dx.doi.org/10.1128/JVI.06728-11
- Tang J, Cormier E, Gilmour J, Price MA, Prentice HA, Song W, Kamali A, Karita E, Lakhi S, Sanders EJ, et al. Human leukocyte antigen variants B*44 and B*57 are consistently favorable during two distinct phases of primary HIV-1 infection in sub-Saharan Africans with several viral subtypes. J Virol 2011; 85, 8894-902; PMID:21715491; http://dx.doi.org/10.1128/JVI.00439-11
- Zhang X, Huang X, Xia W, Li W, Zhang T, Wu H, Xu X, Yan H. HLA-B*44 is associated with a lower viral set point and slow CD4 decline in a cohort of Chinese homosexual men acutely infected with HIV-1. Clin Vaccine Immunol 2013; 20, 1048-54; PMID:23677320; http://dx.doi.org/10.1128/CVI.00015-13
- Goulder PJ, Walker BD. HIV and HLA class I: an evolving relationship. Immunity 2012; 37, 426-40; PMID:22999948; http://dx.doi.org/10.1016/j.immuni.2012.09.005
- MacDonald KS, Embree JE, Nagelkerke NJ, Castillo J, Ramhadin S, Njenga S, Oyug J, Ndinya-Achola J, Barber BH, Bwayo JJ, et al. The HLA A2/6802 supertype is associated with reduced risk of perinatal human immunodeficiency virus type 1 transmission. J Infect Dis 2001; 183, 503-6; PMID:11133384; http://dx.doi.org/10.1086/318092
- MacDonald KS, Matukas L, Embree JE, Fowke K, Kimani J, Nagelkerke NJ, Oyugi J, Kiama P, Kaul R, Luscher MA, et al. Human leucocyte antigen supertypes and immune susceptibility to HIV-1, implications for vaccine design. Immunol Lett 2001; 79, 151-7; PMID:11595302; http://dx.doi.org/10.1016/S0165-2478(01)00277-2
- Liu C, Carrington M, Kaslow RA, Gao X, Rinaldo CR, Jacobson LP, Margolick JB, Phair J, O'Brien SJ, Detels R. Association of polymorphisms in human leukocyte antigen class I and transporter associated with antigen processing genes with resistance to human immunodeficiency virus type 1 infection. J Infect Dis 2003; 187, 1404-10; PMID:12717621; http://dx.doi.org/10.1086/374394
- Gartland AJ, Li S, McNevin J, Tomaras GD, Gottardo R, Janes H, Fong Y, Morris D, Geraghty DE, Kijak GH, et al. Analysis of HLA A*02 Association with Vaccine Efficacy in the RV144 HIV-1 Vaccine Trial. J Virol 2014; 88, 8242-55; PMID:24829343; http://dx.doi.org/10.1128/JVI.01164-14
- Gonzalez-Galarza FF, Christmas S, Middleton D, Jones AR. Allele frequency net: a database and online repository for immune gene frequencies in worldwide populations. Nucleic Acid Res 2011; 39, D913-D919; http://dx.doi.org/10.1093/nar/gkq1128
- Allele Frequency Net Database, http://www.allelefrequencies.net/ Accessed October 2, 2014
- Jones RB, Garrison KE, Mujib S, Mihajlovic V, Aidarus N, Hunter DV, Martin E, John VM, Zhan W, Faruk NF, et al. HERV-K-specific T cells eliminate diverse HIV-1/2 and SIV primary isolates. J Clin Invest 2012; 122:4473-4489; PMID:23143309; http://dx.doi.org/10.1172/JCI64560
- Sanou MP, Roff SR, Mennella A, Sleasman JW, Rathore MH, Yamamoto JK, Levy JA. Evolutionarily conserved epitopes on human immunodeficiency virus type 1 (HIV-1) and feline immunodeficiency virus reverse transcriptases detected by HIV-1-infected subjects. J Virol 2013; 87, 10004-15; PMID:23824804; http://dx.doi.org/10.1128/JVI.00359-13
- Almeida JR, Price DA, Papagno L, Arkoub ZA, Sauce D, Bornstein E, Asher TE, Samri A, Schnuriger A, Theodorou I, et al. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J Exp Med 2007; 204, 2473-85; PMID:17893201; http://dx.doi.org/10.1084/jem.20070784
- Sidney J, Peters B, Frahm N, Brander C, Setter A. HLA class I supertypes: a revised and updated classification. BMC Immunol 2008; 9:1; PMID:18211710; http://dx.doi.org/10.1186/1471-2172-9-1
- Abbott JR, Pu R, Coleman JK, Yamamoto JK. Utilization of feline ELISPOT for mapping vaccine epitopes. Methods in Molecular Biology 2012; 792, 47-63; PMID:21956500; http://dx.doi.org/10.1007/978-1-61779-325-7_4
- Lichterfeld M, Kaufmann DE, Yu XG, Mui SK, Addo MM, Johnston MN, Cohen D, Robbins GK, Pae E, Alter G, et al. Loss of HIV-1-specific CD8+ T cell proliferation after acute HIV-1 infection and restoration by vaccine-induced HIV-1-specific CD4+ T cells. J Exp Med 2004; 200, 701-12; PMID:15381726; http://dx.doi.org/10.1084/jem.20041270
- Horton H, Thomas EP, Stucky JA, Frank I, Moodie Z, Huang Y, Chiu YL, McElrath MJ, De Rosa SC. Optimization and validation of an 8-color intracellular cytokine staining (ICS) assay to quantify antigen-specific T cells induced by vaccination. J Immunol Methods 2007; 323, 39-54; PMID:17451739; http://dx.doi.org/10.1016/j.jim.2007.03.002
- Pattacini L, Mize GJ, Graham JB, Fluharty TR, Graham TM, Lingnau K, Wizel B, Perdiguero B, Esteban M, Pantaleo G, et al. A novel HIV vaccine adjuvanted by IC31 induces robust and persistent humoral and cellular immunity. PloS one 2012; 7, e42163; PMID:22848738; http://dx.doi.org/10.1371/journal.pone.0042163