Abstract
The outbreak of human infections of a novel avian influenza virus A (H7N9) prompted the development of the vaccines against this virus. Like all types of influenza vaccines, H7N9 vaccine must be tested for its potency prior to being used in humans. However, the unavailability of international reference reagents for the potency determination of H7N9 vaccines substantially hinders the progress in vaccine development. To facilitate clinical development, we enlisted 5 participants in a collaborative study to develop critical reagents used in Single Radial Immunodiffusion (SRID), the currently acceptable assay for potency determination of influenza vaccine. Specifically, the hemagglutinin (HA) content of one vaccine bulk for influenza A (H7N9), herein designated as Primary Liquid Standard (PLS), was determined by SDS-PAGE. In addition, the freeze-dried antigen references derived from PLS were prepared to enhance the stability for long term storage. The final HA content of lyophilized antigen references were calibrated against PLS by SRID assay in a collaborative study. Importantly, application of these national reference standards to potency analyses greatly facilitated the development of H7N9 vaccines in China.
Introduction
Human infections with avian-origin influenza virus A (H7N9) were first identified in the spring of 2013 in China. As of 28 February 2014, a total of 375 laboratory-confirmed cases of infection were reported, including more than one hundred deaths.Citation1 The H7N9 outbreak is significantly different from the previous infection with other H7 subtypes of virus in human, in which mild respiratory disease commonly accompanied by conjunctivitis were presented, with only a few deaths.Citation2-6 At this time, the exact mechanisms underlying the high pathogenicity of H7N9 in humans remain to be fully understood. However, recent reports have shed some light on the virological and immunological characteristics of H7N9 and related infections under detailed investigation. Firstly, 6 internal genes of H7N9 virus were found to be closely related to those of the avian influenza A/brambling/Beijing/16/2012 (H9N2) strain while hemagglutinin (HA) and neuraminidase (NA) genes were traced to A/duck/Zhejiang/12/2011 and A/wild bird/Korea/A14/2011 (H7N9) virus, respectively.Citation6,7 Secondly, the surveillance on the H7N9 is more difficult than that of highly pathogenic avian influenza virus A, such as H5N1, as the latter cause the massive die-off in the affected poultries, making it easier to be observed. In contrast, H7N9 viruses are low pathogenic avian influenza viruses, with infection of poultry or wild bird resulting in mild or asymptomatic diseases; therefore, the virus may continuously transmit in animal reservoirs and subject to silent mutations. Recent study indicate that H7N9 has acquired genetic characteristics known to be adaptive for transmission in humans as well as ferrets, which is the most relevant animal model for human influenza infection.Citation8 Specifically, there are several mutations in the H7N9 virus associated with adaptation to human; for example, the HA protein has the Q226L mutation which could enhance viral binding to mammalian receptors in the human upper airway while the E627K mutation in PB2 facilitates replication of the virus in human.Citation6,9,Citation10 Thirdly, some H7N9 virus apparently has evolved to be resistant to current antiviral drugs.Citation11 All these findings suggest that H7N9 has retained the fitness to continuously circulate in avian and human hosts and stands out as one of avian virus with the greatest pandemic potential.
Vaccination remains the most effective way to prevent and contain influenza. There is an urgent need to develop H7N9 influenza vaccine for the protection of humans. It has been known that the influenza virus HA is the most important surface antigen of the virus and constitutes the main component of the vaccines. The potency or antigenicity of influenza vaccines must be measured prior to its use in clinical development and eventual mass immunization of humans. Currently, Single Radial Immunodiffusion (SRID) is the routine method recommended by the WHO for the determination of HA content of flu vaccines. In SRID, the HA antigen and corresponding anti-serum form the precipitation ring in the agarose gel, with the potency of vaccines determined by calibrating samples against the antigen references.Citation12 If these 2 crucial reagents were not available (antigens and antiserum), the vaccine development could be hindered. In this paper, we report the development of the national antigen reference standards for H7N9 vaccines by enlisting 5 participants for full characterization of the reference standard sin according to the protocol of WHO Essential Regulatory Laboratories (ERL).Citation13 The successful development of these reagents would greatly facilitate clinical trials of H7N9 vaccine in China.
Results
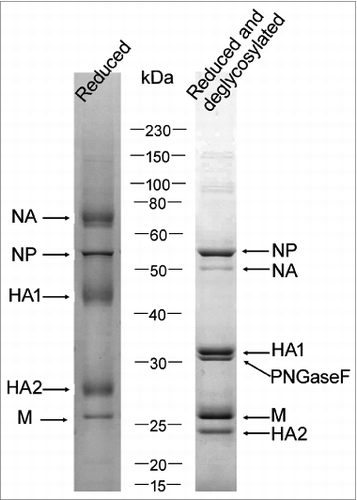
SDS-PAGE analysis of H7N9 bulk
The HA protein exists as 2 subunits, HA1 and HA2 with molecular weights of around 41 and 28kDa, respectively, in reduced conditions. In our previous reportCitation14 and during this study, we found that split samples from various subtype/types (H1N1, H3N2, B) as well as H7N9 bulk samples were stable for at least 6 months in SDS-PAGE profiles (data not shown). Unlike H1N1 and H3N2 virus, the HA1 and HA2 molecules of H7N9 can be easily separated from other viral proteins in reduced conditions. However, these 2 bands are not sharp, making it difficult to precisely quantify these 2 HA subunits. Clearly, after deglycosylation treatment with PNGase F, the HA1 and HA2 bands became much sharper as is the case for both H5N1 and H1N1.Citation14,15 Although the PNGase F band is close to HA1, the 2 bands were well separated (), making it easier for quantification (below).
Determination of HA contents of H7N9 vaccine bulk (PLS): phase I collaborative study
The HA contents of H7N9 was calculated based on the ratio of the HA to the total protein as described.Citation14,15 Three participants with prior experiences in 2009 H1N1 National Reference Standards validation were enlisted to take part in this collaborative study. Specifically, H7N9 vaccine bulk (batch No. A7-1306-001) was sent to all labs for total protein and SDS-PAGE analysis. As presented in , the average total protein contents from all participating labs demonstrated good consistency, with the lowest of 349.60 μg/ml and highest of 370.33 μg/ml; the average HA percentages from different labs ranged from 35.88% to 37.28%, with coefficient of variation being less than 3%. Therefore, this H7N9 bulk (PLS) was assigned to 132 μg/ml HA based on the averaged values from all participants. The PLS was used as primary standard to calibrate the lyophilized H7N9 antigen references.
Table 1. Collaborative study to determine the HA content of H7N9 vaccine bulk by SDS-PAGE
Preparation of H7 antiserum for vaccine quantification
The immunizing rHA was analyzed by SDS-PAGE and confirmed by mass spectrum, showing that the purity is 91.5% and the apparent molecular weight is 66 kDa, the same as proved by vendor (data not shown). The antiserum was collected from the sheep which received 4 immunizations as described in Materials and methods. The titers of the antiserum tested in indirect ELISA against recombinant HA protein were over 1:1 000 000, suggesting that the recombinant proteins are highly immunogenic. In double immunodiffusion test, the titer of antisera was 1:32 against the H7N9 viral antigens. The obvious difference of titers from 2 methods is due to the nature of the 2 different assays. In indirect ELISA, the rHA was used as the coating antigen and the specific antibodies were detected and amplified by the HRP-labeled secondary antibody as well as the following substrate color development. The ELISA is a much sensitive assay compared to double immunodiffusion test, in which antigen-antibody precipitation is visualized by Coomassie blue staining. However, the data generated from double immunodiffusion was comparable to those obtained using antisera for seasonal influenza vaccine analyses using SRID. In SRID of this study, approximately 10 μl antiserum was sufficient in a 1 ml gel in which clear precipitation of antigen-antibody complex could be observed when the antigen concentrations ranges from 10 to 40 μg/ml HA antigen (data not shown).
HA content of the lyophilized H7N9 antigen reference standards: phase II Collaborative study
Having determined the HA contents of the H7N9 reference antigens in liquid form, we next sought to determine the contents of lyophilized antigens as such antigen preparations are needed for long-term storage to ensure the continual supply of these reagents in vaccine production and clinical studies. To this end, a total of 15 vials of samples were weighed before and after filling in order to check the filling variation. The mean net weight was 0.5004 g with a coefficient of variation of 0.94%, which met the requirements of China Pharmacopoeia, 2010, Vol III (less than 1%). The residual moisture content of freeze-dried H7N9 antigen was 2.30% as measured by Karl Fischer method, conforming to the requirements of China Pharmacopoeia, 2010, Vol III (less than 3%). Finally, the consistency of the lyophilized H7N9 antigen was tested by SRID, in which 18 vials samples were taken randomly and assayed 3 times on the same day using the PLS as standard. As shown in , the average HA content from different vials was found to be within 54.17 to 59.50 μg/vial with a small value in coefficient of variation (2.99%).
Table 2. HA contents of 18 vials freeze-dried H7N9 antigen reference
Five participants then carried out SRID tests using H7N9 PLS as standard and H7 antiserum to measure the HA content of lyophilized references. As shown in , the results from different labs are very consistent. Specifically, the lowest average HA content is 52.69 μg/vial and the highest is 57.51 μg/vial, with SD being less than 3.49. Based on the averaged value from 5 labs, a final 55 μg/vial of HA content was assigned to the national antigen reference standards.
Table 3. Collaborative study to determine the HA content of freeze-dried H7N9 antigen reference by SRID
The suitability of antigen references for products derived from other strain
As two vaccine strains for H7N9 vaccine were provided by NIBSC, i.e., NIBRG-268 (A/Anhui/1/2013(H7N9) like strain) and NIBRG-267 (A/Shanghai/2/2013(H7N9) like strain), we next wished to confirm the suitability of our references (from NIBRG-268) to quantify HA contents of products derived from NIBRG-267 strain. To this end, HA contents of 4 lots vaccine bulks (NIBRG-267 strains, 2 lots of whole virus and 2 lots of split vaccines) were tested by SRID as described in this paper, with results being compared to those obtained from SDS-PAGE determination. As presented in , the HA contents of 4 vaccine bulks determined by the SRID methods are similar to those from the SDS-PAGE assay; the recovery rates varied from 95.11% to 113.67%, falling within the range validated previously, in which the agreement in results between these 2 assays is between 88–122% for different subtypes of vaccine samples.Citation14 These data indicated that the lyophilized H7N9 reference antigen (from NIBRG-268) could be used to quantify HA contents of vaccine produced by NIBRG-267 strain, in agreement with the observation that there is no amino acid differences in the HA sequences between 2 donor strains.
Table 4. Comparison of 2 methods for HA measurement of vaccine bulk derived from strain NRBRG-267
Discussions
As one of the 2 important surface proteins of influenza virus, HA is commonly recognized as the major component of influenza vaccines; quantification of HA prior to human immunization is required. The quantification of HA is routinely conducted using SRID. The two crucial reagents, HA antigen references and corresponding antiserum used in SRID are provided annually by WHO collaborative centers to manufacturers and national control laboratories for release of vaccines. The HA antigen reference was prepared and calibrated according to protocols of ERL.Citation13 The preparation of antiserum is a time-consuming process since it involves the virus propagation and purification, followed by truncating and purifying the HA, The truncated and purified HA preparations were subsequently used to inject animals for the generation of high-titer antisera. Usually, it takes 3˜6 months to complete the whole process and to provide these 2 reagents for seasonal influenza vaccines production and quality control. This would not be an ideal situation should a new virus strain emerge or an influenza pandemic happen as the lack of these reagents in the early phase of a pandemic could delay the process of vaccine development. In this study, we use the rHA to immunize sheep and obtained higher titer of antiserum. This strategy enabled us to immunize the sheep before antigens from vaccine preparations becomes available. It is of note that the availability of antigens for immunization is often the first hurdle in vaccine production, particularly should a flu pandemic happen. Another advantage with rHA is that the immunogen is free of other viral proteins and is better for specific antiserum generation. The similar studies have been previously reported by scientists from CBER, FDA.Citation16,17
There have been many reports in recent years on the development of alternative methods for the potency analyses of the vaccine. Garcia et al. reported the applicability of 2D-HPLC for the characterization of vaccine proteins. This method combines the on-line coupling of size exclusion HPLC to reversed-phase HPLC to effectively separate 3 subtypes of HA proteins in the trivalent vaccines based on the different elution profiles.Citation18 However, this method still needs antigen reference to quantitate HA content. Creskey et al. report a method for vaccine analysis for simultaneous and absolute quantification of HA and NA levels. Specifically, enzymatically digested vaccine proteins were analyzed by LC-MS, allowing identification and quantification of HA and NA antigens as well as other virus or host proteins within one run. This method has the potential to increase the accuracy determination of the reference antigen standards and to verify label claims for HA content in formulated vaccines.Citation19 Chun et al. prepared the universal antibody against conserved epitope on HA2 subunit and proved that it can react with all subtypes of influenza HA proteins, and the Slot-Blot assay was established basing on this universal antibody.Citation20,21 All these efforts provide the promising tools to overcome the difficulties due to the unavailability of references in SRID method, although the extensive validations are still needed before these techniques are adopted internationally.
During the 2009 H1N1 flu pandemic, we successfully established the alternative method to determine the HA content, in which the samples were treated by deglycosylation and subject to SDS-PAGE separation. In the pilot study, we used the gradient gel to separate the samples to retain the smaller peptides as much as possible. However, no smaller molecules were detectable on the gel due to the high purity of samples; therefore, 10% gel was used to achieve better separation resolution. The HA content was calculated based on the ratio of HA vs. total proteins. This method was used by multiple manufacturers and facilitated the vaccine formulation in 2009 pandemic influenza vaccines development earlier before the SRID reagents arrival.Citation14,22,Citation23 More importantly, the vaccines released by the alternative method have been proved to be effective and safe in subsequent clinical trials.Citation24 The global community has been on alert for the H7N9 avian influenza virus and its infections of humans. Two vaccines strains (NIBRG-267 and NIBRG-268) have been promptly prepared by NIBSC as seeds to produce vaccines. As mentioned earlier, both antigen and anti-serum references are needed in SRID for vaccine potency determination. Based on our prior experiences in the H1N1 flu pandemic, we prepared the influenza A (H7N9) antigen references with additional targeted improvements, particularly by a collaborative study enrolling 5 participants. These reference antigens have been shown to be suitable for potency determination of H7N9 vaccines in SRID and have greatly facilitated vaccines development in 5 H7N9 vaccine manufacturers in China. The lyophilized antigen references are very stable for at least 3 years when stored at −20°C based our previous data, nevertheless, the HA content is continuously monitored by titration against references stored in liquid nitrogen. In this study, the preliminary data proved that the reagents are suitable for both whole virion and split vaccines. While several clinical trials have shown that SRID data correlate well with sero-conversion and vaccine efficacy.Citation25-27 Our future plan is to collect sera of patients enrolled in the clinical trials on these H7N9 vaccines and investigate the correlation of sero-conversion to potency of the vaccines determined by SRID using our reagents. Interested investigators may contact the authors for the antigen and antibody reagents in their exploratory research.
Materials and Methods
Influenza virus A (H7N9) vaccine bulk
H7N9 vaccine bulk (batch No. A7-1306-001) was kindly provided by Sinovac Biotech Co., Ltd., Beijing, China. The vaccine strain is NIBRG-268 (A/Anhui/1/2013(H7N9) like strain) obtained from National Institute for Biological Standards and Control (NIBSC), Potters Bar, UK. The viruses were propagated in chicken embryos, inactivated by formaldehyde, and then purified by centrifugation and chromatography. This bulk, designed as Primary Liquid Standard (PLS), was subsequently used as starting materials for the preparation of the reference materials.
Protein content measurement
Total protein content of the PLS was measured by Lowry method according to Chinese Pharmacopoeia, 2010, Vol III. For collaborative study purpose, each laboratory measured the contents in 3 independent tests.
Deglycosylation treatment
Prior to gel electrophoresis analyses, the PLS samples were treated with PNGase F (New England Biolabs) as described before.Citation14 In brief, samples were diluted to approximately 400 μg/ml of total protein and then denatured by mixing 36 μl of sample with 4 μl denature buffer prior to being boiled for 10 minutes. The samples were then placed on ice, followed by addition of 5 μl reaction buffer provided by the vender, 5 ul 10% NP40 and 1 μl PNGase F according to the manual's instruction. The samples were incubated at 37°C for approximately 16 hours. For collaborative study purpose, 6 independent treatments of the PLS samples were carried out in each participating laboratory.
SDS-PAGE analysis of the vaccine bulk
SDS-PAGE analysis was performed as described by Harvey et al. with slight modification as described elsewhere.Citation14,15 In brief, vaccine samples, after deglycosylation treatment, were mixed with sample buffer, then heated to 95°C for 5 min. Approximately 4 μg of protein samples were loaded on 10% Bis-Tris gels (Life Technologies) and run at 120 V using MOPS running buffer (Life Technologies) until the tracking dye reached the end of the gel. The gel was stained with Coomassie blue. The bands on the gel were confirmed by Q-TOF identification (Beijing Protein Innovation).
Densitometry quantification was carried out using scanner and Quantity One software (Bio-Rad). The HA content of PLS was calculated based on the ratio between HA (HA1 and HA2) and the total protein content which was determined by Lowry assay.Citation13
Preparation of lyophilized national antigen references
Vaccine sample derived from H7N9 bulk (also named as PLS) was diluted in PBS buffer containing sucrose at final concentration of 1% (w/v); these samples were then aliquot into 0.5 ml in each vial and subject to freeze-drying.Citation28 The filling process was monitored by measuring the net weight of 1 in every 50 vials of samples.
Preparation of H7 antiserum for SRID test
The H7 antiserum was prepared by immunizing sheep with recombinant HA protein derived from strain A/Shanghai/1/2013(H7N9).Citation29 In brief, HA proteins expressed in eukaryotic cells were purchased from Sino Biological Inc., Beijing, China and were used to immunize sheep. A DNA sequence encoding the Influenza A virus (A/Shanghai/1/2013(H7N9)) hemagglutinin (Met1-Val524) with a C-terminal polyhistidine tag was inserted into pCMV3 plasmid which is similar to PC DNA 3.1. The target proteins were expressed in HEK293 cells and purified by affinity chromatography. The rHA was analyzed by SDS-PAGE to check integrity and purity, with the identity of the HA band on the gel verified by mass spectrometry.100 μg of HA with Freund's Complete Adjuvant (Sigma-Aldrich) was given to the sheep, followed by a second injection of 50 μg of HA with Freund's Incomplete Adjuvant 2 weeks later. The animals were given 2 more injections of 50 μg with Freund's Incomplete Adjuvant with an interval of one week. Five weeks after the initial immunization, all bleeds were collected. The titer of antiserum was determined by indirect ELISA and double immune-diffusion.
Indirect ELISA was carried out by coating 1 μg/ml of recombinant HA protein on 96-well plates. After washing and blocking, serially diluted antisera were added, followed by incubation at 37°C for 1 h. Subsequently, peroxidase-conjugated anti-goat immunoglobulin was added for an additional incubation at 30°C for 1 h before TMB substrate was used for colorimetric development.
H7N9 vaccine bulk was used in the double immune-diffusion. In brief, approximately 0.8 μg HA was loaded in the middle well on the agarose gel, serially diluted antisera were loaded in peripheral wells. After diffusion for 18 h at 25°C, the agarose gel was dried and stained with Coomassie blue to detect the antigen-antibody precipitation on the gel. The data obtained for this H7N9 serum as well as other standard antiserum can facilitate the optimizing the working concentration in SRID test.
Collaborative study of lyophilized H7N9national reference standards
The study was conducted in accordance to the WHO ERL protocol for the calibration of influenza antigen.Citation13 SRID tests were carried out in a procedure as described by Wood et al.Citation12 In brief, 10 μl antiserum was added to 1 ml agarose gel and PLS was used as antigen standard to calibrate the HA content of national reference standards. Five labs (NIFDC and 4 manufacturers) participated in this study. The SRID assays were performed 6 times by 2 persons in each lab. All data were collected to determine the final potency of H7N9 reference antigen.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Sinovac Biotech Co. Ltd. for providing the influenza (H7N9) vaccine bulks for this study. We appreciated all the participators in the collaborative study to establish this national reference standard.
Funding
This work is supported by the Ministry of Science and Technology, PR China (KJYJ-2013-01-06 to J.W; 2013DFA31680 to C.L.)(Canadian Regulatory Strategy for Biotechnology: XL).
References
- World Health Organization. 28 February 2014, Summary of surveillance and investigation findings. http://www.who.int/influenza/human_animal_interface/influenza_h7n9/140225_H7N9RA_for_web_20140306FM.pdf?ua=1
- Tweed SA, Skowronski DM, David ST, Larder A, Petric M, Lees W, Li Y, Katz J, Krajden M, Tellier R, et al. Human illness from avian influenza H7N3, British Columbia. Emerg Infect Dis 2004; 10:2196-9; PMID:15663860; http://dx.doi.org/10.3201/eid1012.040961
- Centers for Disease Control and Prevention (CDC). Notes from the field: Highly pathogenic avian influenza A (H7N3) virus infection in two poultry workers–Jalisco, Mexico, July 2012. MMWR Morb Mortal Wkly Rep 2012; 61:726-7; PMID:22971746
- Belser JA, Bridges CB, Katz JM, Tumpey TM. Past, present, and possible future human infection with influenza virus A subtype H7. Emerg Infect Dis 2009; 15:859-65; PMID:19523282; http://dx.doi.org/10.3201/eid1506.090072
- Fouchier RA, Schneeberger PM, Rozendaal FW, Broekman JM, Kemink SA, Munster V, Kuiken T, Rimmelzwaan GF, Schutten M, Van Doornum GJ, et al. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc Natl Acad Sci U S A 2004; 101:1356-61; PMID:14745020; http://dx.doi.org/10.1073/pnas.0308352100
- Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med 2013; 368:1888-97; PMID:23577628; http://dx.doi.org/10.1056/NEJMoa1304459
- Chen Y, Liang W, Yang S, Wu N, Gao H, Sheng J, Yao H, Wo J, Fang Q, Cui D, et al. Human infections with the emerging avian influenza A H7N9 virus from wet market poultry: clinical analysis and characterisation of viral genome. Lancet 2013; 381:1916-25; PMID:23623390
- Hu Y, Lu S, Song Z, Wang W, Hao P, Li J, Zhang X, Yen HL, Shi B, Li T, et al. Association between adverse clinical outcome in human disease caused by novel influenza A H7N9 virus and sustained viral shedding and emergence of antiviral resistance. Lancet 2013; 381:2273-9; PMID:23726392
- Liu D, Shi W, Shi Y, Wang D, Xiao H, Li W, Bi Y, Wu Y, Li X, Yan J, et al. Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: phylogenetic, structural, and coalescent analyses. Lancet 2013; 381:1926-32; PMID:23643111; http://dx.doi.org/10.1016/S0140-6736(13)60938-1
- Liu Q, Lu L, Sun Z, Chen GW, Wen Y, Jiang S. Genomic signature and protein sequence analysis of a novel influenza A (H7N9) virus that causes an outbreak in humans in China. Microbes Infect 2013; 15:432-9; PMID:23628410; http://dx.doi.org/10.1016/j.micinf.2013.04.004
- Hai R, Schmolke M, Leyva-Grado VH, Thangavel RR, Margine I, Jaffe EL, Krammer F, Solórzano A, García-Sastre A, Palese P, et al. Influenza A(H7N9) virus gains neuraminidase inhibitor resistance without loss of in vivo virulence or transmissibility. Nat Commun 2013; 4:2854; PMID:24326875; http://dx.doi.org/10.1038/ncomms3854
- Wood JM, Schild GC, Newman RW, Seagroatt V. An improved single-radial-immunodiffusion technique for the assay of influenza haemagglutinin antigen: application for potency determinations of inactivated whole virus and subunit vaccines. J Biol Stand 1977; 5:237-47; PMID:408355; http://dx.doi.org/10.1016/S0092-1157(77)80008-5
- WHO. Generic protocol for the calibration of seasonal/pandemic influenza antigen working reagents by WHO Essential Regulatory Laboratories. 2012
- Li C, Shao M, Cui X, Song Y, Li J, Yuan L, Fang H, Liang Z, Cyr TD, Li F, et al. Application of deglycosylation and electrophoresis to the quantification of influenza viral hemagglutinins facilitating the production of 2009 pandemic influenza (H1N1) vaccines at multiple manufacturing sites in China. Biologicals 2010; 38:284-9; PMID:20074976; http://dx.doi.org/10.1016/j.biologicals.2009.12.004
- Harvey R, Wheeler JX, Wallis CL, Robertson JS, Engelhardt OG. Quantitation of haemagglutinin in H5N1 influenza viruses reveals low haemagglutinin content of vaccine virus NIBRG-14 (H5N1). Vaccine 2008; 26:6550-4; PMID:18840494; http://dx.doi.org/10.1016/j.vaccine.2008.09.050
- Schmeisser F, Vodeiko GM, Lugovtsev VY, Stout RR, Weir JP. An alternative method for preparation of pandemic influenza strain-specific antibody for vaccine potency determination. Vaccine 2010; 28:2442-9; PMID:20074687; http://dx.doi.org/10.1016/j.vaccine.2009.12.079
- Khurana S, Larkin C, Verma S, Joshi MB, Fontana J, Steven AC, King LR, Manischewitz J, McCormick W, Gupta RK, et al. Recombinant HA1 produced in E. coli forms functional oligomers and generates strain-specific SRID potency antibodies for pandemic influenza vaccines. Vaccine 2011; 29:5657-6518.
- Garcia-Canas V, Lorbetskie B, Bertrand D, Cyr TD, Girard M. Selective and quantitative detection of influenza virus proteins in commercial vaccines using two-dimensional high-performance liquid chromatography and fluorescence detection. Anal Chem 2007; 79:3164-72; PMID:21704111; http://dx.doi.org/10.1016/j.vaccine.2011.06.014
- Creskey MC, Li C, Wang J, Girard M, Lorbetskie B, Gravel C, Farnsworth A, Li X, Smith DG, Cyr TD. Simultaneous quantification of the viral antigens hemagglutinin and neuraminidase in influenza vaccines by LC-MSE. Vaccine 2012; 30:4762-70; PMID:22643214; http://dx.doi.org/10.1016/j.vaccine.2012.05.036
- Chun S, Li C, Van Domselaar G, Wang J, Farnsworth A, Cui X, Rode H, Cyr TD, He R, Li X. Universal antibodies and their applications to the quantitative determination of virtually all subtypes of the influenza A viral hemagglutinins. Vaccine 2008; 26:6068-76; PMID:19007587; http://dx.doi.org/10.1016/j.vaccine.2008.09.015
- Li C, Jaentschke B, Song Y, Wang J, Cyr TD, Van Domselaar G, He R, Li X. A simple slot blot for the detection of virtually all subtypes of the influenza A viral hemagglutinins using universal antibodies targeting the fusion peptide. Nat Protoc 2010; 5:14-9; PMID:20010723; http://dx.doi.org/10.1038/nprot.2009.200
- Hardy S, Eichelberger M, Griffiths E, Weir JP, Wood D, Alfonso C. Confronting the next pandemic–workshop on lessons learned from potency testing of pandemic (H1N1) 2009 influenza vaccines and considerations for future potency tests, Ottawa, Canada, July 27–29, 2010. Influenza Other Respir Viruses 2011; 5:438-42; PMID:21668676; http://dx.doi.org/10.1111/j.1750-2659.2011.00250.x
- Elmgren L, Li X, Wilson C, Ball R, Wang J, Cichutek K, Pfleiderer M, Kato A, Cavaleri M, Southern J, et al. A global regulatory science agenda for vaccines. Vaccine 2013; 31 Suppl 2:B163-75; PMID:23598478
- Wu J, Xu F, Lu L, Lu M, Miao L, Gao T, Ji W, Suo L, Liu D, Ma R, et al. Safety and effectiveness of a 2009 H1N1 vaccine in Beijing. N Engl J Med 2010; 363:2416-23; PMID:21158658; http://dx.doi.org/10.1056/NEJMoa1006736
- La Montagne JR, Noble GR, Quinnan GV, Curlin GT, Blackwelder WC, Smith JI, Ennis FA, Bozeman FM. Summary of clinical trials of inactivated influenza vaccine – 1978. Rev Infect Dis 1983; 5(4):723-36; PMID:6353529; http://dx.doi.org/10.1093/clinids/5.4.723
- Cate TR, Couch RB, Parker D, Baxter B. Reactogenicity, immunogenicity, and antibody persistence in adults given inactivated influenza virus vaccines – 1978. Rev Infect Dis 1983; 5(4):737-47; PMID:6622888; http://dx.doi.org/10.1093/clinids/5.4.737
- Hashem AM, Gravel C, Farnsworth A, Zou W, Lemieux M, Xu K, Li C, Wang J, Goneau MF, Merziotis M, et al. A novel synthetic receptor-based immunoassay for influenza vaccine quantification. PLoS One 2013; 8(2):e55428; PMID:23424631; http://dx.doi.org/10.1371/journal.pone.0055428
- Campbell PJ. International biological standards and reference preparations. I. Preparation and presentation of materials to serve as standards and reference preparations. J Biol Stand 1974; 2:249-58; PMID:4459394; http://dx.doi.org/10.1016/0092-1157(74)90033-X
- Xu K, Shao M, Liu S, Cai F, Gao Q, Li C, Wang J. Rapid preparation of antiserum against influenza virus(H7N9) hemagglutinin for single radial immunodiffusion assay. Chin J Microbiol Immunol 2014; 24:146-8