Abstract
Over the past few decades, climate warming has caused profound changes in our living environment, and human diseases, including infectious diseases, have also been influenced by these changes. However, it remains unclear if a warm-wet climate can influence the infectivity of influenza and result in influenza pandemics. This study focused on observations of how the hydrothermal environment influences the infectivity of the influenza virus and the resulting immunoreactions of the infected mice. We used a manual climatic box to establish the following 3 environments with different temperatures and humidity: normal environment (T: 24 ± 1°C, RH: 50% ± 4%), wet environment (T: 24 ± 1°C, RH: 95% ± 4%) and warm-wet environment (T: 33 ± 1 °C, RH: 95% ± 4%), and the mice were fed and maintained in these 3 different environments. After 14 days, half of the mice were infected with H1N1 (A/FM1/1/47, a lung adapted strain of the flu virus specific for the mouse lung) virus for 4 d After establishing the animal model, we observed the microstructure of the lung tissue, the Th1/Th2 T cell subsets, the Th17/Treg balance, the expression of cytokines in the peripheral blood serum and the expression of the immune recognition RLH signal pathway. The results showed that mice in different environments have different reaction. Results showed that after infection, the proportion of Th1/Th2 and Th17/Treg cells in the spleen was significantly increased, and these proportions were increased the most in the infected group kept in wet-hot conditions. After infection, the mRNA levels and protein expression of the RLH (RIG-1-like helicases) signal pathway components were up-regulated while the uninfected animals in the 3 diverse environments showed no significant change. The infected mice kept in the wet and warm-wet environments showed a slight elevation in the expression of RLH pathway components compared to infected mice maintained in the normal environment. Our study suggested that the warm-wet environment may have interfered with the immune response and balance. The mice kept in the warm-wet environment displayed immune tolerance when they were exposed to the influenza virus, and the body was not able to effectively clear the virus, leading to a persistent infection. A warm-wet climate may thus be a factor that contributes to influenza pandemics, people should focus on the warm-wet climate coming and advance prepare to vaccine manufacture.
Keywords:
Introduction
Influenza pandemics can severely damage people's health and socioeconomic development.Citation1 The most recent outbreak of influenza was in 2009 when infections with H1N1 caused a great panic around the world.Citation2 Currently, influenza vaccinations, M2 protein inhibitors and neuraminidase inhibitors are the main therapeutics to prevent and treat influenza.Citation3-5 Vaccines are one of the most effective medical interventions for influenza pandemic, but due to the influenza virus evolve frequently, vaccines will failure to influenza pandemic prevention. Until recently, there was no effective method to accurately forecast epidemics, and people had no therapeutic regimens to prevent outbreaks.Citation6
The climate and humans are closely related. Over the past 20 years, scientists have focused on the associations between climate change and infectious diseases. They suggest that climate change will continue to limit or influence the transmission of some pathogens and give other diseases opportunities to expand. Scientists have already demonstrated that warm temperatures can lower host immunity; for example, the activity of phagocytic cells was depressed at higher temperatures.Citation7 In addition, warm-wet weather has been shown to increase the incidence of Legionnaires' disease in the Netherlands.Citation8 Novel climates also can influence the diseases of plants, such as the fungal diseases of the Oil Palm.Citation9 Southern China has a typical warm-wet climate, and it has been long-term regarded as an “influenza epicenter” for the genesis and emergence of pandemic viruses Citation10; this fact seems to indicate that warm-wet climates are favorable for influenza epidemics.
However, it remains unknown whether a warm-wet climate can actually influence the development of an influenza epidemic. To better elucidate the relationship between a warm-wet climate and influenza virus infections and to establish whether the climate can cause influenza pandemic, our study used manual climatic boxes to establish 3 different temperature and humidity conditions to simulate climate change. The mice were maintained in these environments. After acclimation for 1 week, the mice were infected with the H1N1 (A/FM1/1/47) virus through nasal inhalation, and the symptoms, pathologic changes and immunologic functions of the mice were compared to evaluate the effect of climate on influenza infections.
Results
Histological changes of the lung tissue
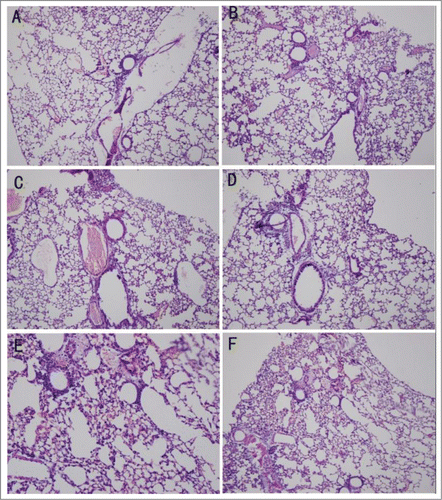
After establishing the model, the histological changes were observed in control animals. When observed by light microscopy, the organization of the pulmonary alveoli and bronchioles were chaotic and twisty, the pulmonary alveoli were enlargement, the interstices were broadened, and lymphocytes had infiltrated the pulmonary mesenchyme ().
Figure 1. Pathological changes detected by HE staining. (A) Uninfected mice in a normal environment. (B) Infected mice in a normal environment. (C) Uninfected mice in the wet environment. (D) Infected mice in the wet environment. (E) Uninfected mice in the warm-wet environment. (F) Infected mice in the warm-wet environment.
Cytokine expression
Infection with the influenza virus significantly elevated the serum levels of IL-4, IFN-γ, IL-10 and IL-17(P < 0.05) compared to the uninfected groups. The infected mice maintained in the wet or wet-hot environments showed cytokine serum levels lower than those of the infected mice kept in the normal environment. Thus, the wet-hot environment can suppress cytokine expression during the viral infection ().
Figure 2. Serum cytokine levels detected by ELISA. (A) Serum levels of IL-4 in each group. (B) Serum levels of IFN-γ in each group. (C) Serum levels of IL-10 in each group. (D) Serum levels of IL-17 in each group. The cytokine levels were elevated during infection, and the levels in the group B animals were higher than group A. In the wet and wet-hot environments, the cytokines levels were suppressed, and the levels of groups D and F were lower than those of group B. There was no significant difference between group D and group F. *P < 0.05; ns, not significant.
Detection of T cell subset balance
FCM was used to detect the proportion of the T cell subsets Th1/Th2 and Th17/Treg. These proportions increased after infection. The Th1/Th2 differentiated toward Th1, and the Th17/Treg cells differentiated toward Th17.The infected mice in the wet and wet-hot environments had lower proportions of these T cells subsets than in infected mice in the normal environment. The wet-hot environment suppressed T cells from differentiating into Th1 or Th17 cells, and the proinflammatory roles of T cells were inhibited ().
Figure 3. The T cell subset proportions were detected by FCM. (A) Th1/Th2 proportions. In the normal environment, the proportion of group B was higher than that of group A, the proportions of Th1/Th2 in groups D and F were lower than group B. (B) Th17/Treg proportions. In the normal environment, the proportion of group B was higher than that of group A, the proportions of Th17/Treg cells in groups D and F were lower than group B. There were no significant differences between group D and group F. *P < 0.05; ns, not significant.
Detection of the RLH signal pathway
The relative expression of the RLH signal pathway was elevated in the infected mice. We observed that the levels of IRF3, IRF7, RIG-1, IPS-1 and TBK-1 mRNA were elevated compared to the uninfected group. The infected mice in the wet and wet-hot environment had lower mRNA expression levels for the RLH pathway components compared to infected mice that were maintained in the normal environment. The IPS-1 and RIG-1 protein levels were elevated in all of the infected animals but to a lesser degree when the animals were kept in the wet-hot environment. This indicates that the wet-hot environment suppressed the RLH signal pathway and affected the immune response of the mice ().
Figure 4. Expression of the RLH signal pathway as detected by RT-PCR and Western blotting. (A) Relative expression of IRF3 mRNA. (B) Relative expression of IRF7 mRNA. (C) Relative expression of RIG-1 mRNA. (D) Relative expression of IPS-1 mRNA. (E) Relative expression of TBK-1 mRNA. (F) Expression IPS-1 and RIG-1 proteins. The expression of the RLH pathway in groups D and F were lower than group B. The relative expression of mRNA in the uninfected animals in the normal environment was set as 1. *P < 0.05; ns, not significant.
Discussion
Influenza continues to infect people around the world. A worldwide outbreak of swine-origin influenza A/H1N1 occurred in 2009,Citation11 and patients infected with the novel type influenza virus H7N9 have been identified throughout our country.Citation12 The recent epidemic was thought to be influenced by various factors, such as a lack of information on the viral origin and toxicity.Citation13 We also lack information on the influence of climate influenza pandemics.
Climate changes can make humans more vulnerable to infectious diseases.Citation2 There is growing evidence that food-and water-borne diseases will increase in the future.Citation14 Due to the changing climate, some areas may no longer be capable of supporting vegetation, so diseases will become irrelevant. However, new locations where diseases could be reduced may also emerge.Citation9 The influenza virus also can be influenced by the plant and animal populations. Researchers have shown that at least 22% of infections in adult patients admitted to the emergency room with respiratory complaints were caused by respiratory viral infection (RVI), and climate factors may be significant underlying contributors to the prevalence of RVI.Citation15 Climatechange also causes immune suppression and protein damage in the Nephropidae, and high temperatures may potentially affect disease severity and spread.Citation16 Increased temperature resulted in the increased activity of 2 immune-related enzymes, phenoloxidase and a lysozyme-like enzyme, in the cricket Gryllustexensis.Citation17 Taken together, this research demonstrates that climate can influence the immunity.
T cell subsets play an important role in combating influenza; for example, Th17 cells may contribute to protection against high doses of influenza.Citation18 Th1 cell-mediated adaptive immunity is an important component in the response to intracellular microbes such as influenza virus.Citation19 Th1 and Th17 cells mediate the evolution of mild A/H1N1 infections, and hypercytokinemia of Th1 and Th17 during the early host response is a signature of severe pandemic influenza.Citation20 The response of Th1 cells can enhance immunity and the ability of the host to defend against pathogens. Our studies showed that after influenza virus invasion, the proportion of Th1/Th2 and Th17/Treg cells increased. Th1 and Th17 cells induce inflammation, and the level of IFN-γ in the serum was also increased. Mice maintained in the wet-hot environment demonstrated a shift in the balance of T cell subsets toward Th2 and Treg cells; these cell types play anti-inflammatory roles. The warm-wet climate suppressed the inflammatory response and the immunity of the mice and induced immune tolerance; thus the mice were unable to clear the influenza virus and displayed a persistent infection. Persistent infections of patients exposed to a warm-wet climate may increase during an influenza pandemic.
The RLH (RIG-1-like helicases) signaling pathway is involved in immunity to the influenza virus. The activation of RIG leads to the production of type I interferon and inflammatory cytokines to achieve anti-virus activity.Citation21 The RLH signal pathway is an important pathway of immunological recognition. During the influenza virus infection, the RNA helicase retinoic acid inducible gene I (RIG-I) detects viral ribonucleoprotein complexesas one pathogen-associated molecular pattern among others by combined recognition of RNA secondary base-pairing structures and 5-triphospate termini.Citation22 RIG-I interacts with TRIM25 and is recruited to mitochondria by the scaffold protein MAVS, finally leading to the activation of the IFNβ induction pathway.Citation23 Our studies showed that after influenza virus invasion, the IRF3, IRF7, RIG-1, IPS-1 and TBK-1 mRNA levels and the RIG-1 and IPS-1 protein levels were upregulated. The up-regulation of the RLH pathway in response to influenza virus invasion induces the production of IFN to clear the virus. The warm-wet environment suppressed the mRNA and protein levels of the components of the RLH pathway, and thus the mice were unable to recognize the influenza virus infection. This deficiency in the ability of the immune system to recognize pathogens will increase susceptibility to the influenza virus.
Whether the influenza vaccines is effective depends on the epidemic strain, it may not depends on the climate changes. But the warm-wet climate might be a factor that contributes to influenza pandemic, so when the warm-wet climate coming, people should think of the possibility of influenza epidemics and advance prepare to vaccine manufacture.
Conclusion
In conclusion, the warm-wet environment suppressed the recognition of the influenza virus by the immune system and also induced immune tolerance. Thus, the mice were unable to recognize the virus and could not effectively clear the virus. A warm-wet climate may be one of the factors that contribute to influenza pandemics, people should advance prepare to vaccine manufacture in warm-wet climate.
Materials and Methods
Experimental group and model establishment
120 NIH (National Institutes of Health) mice were separate into the following 6 groups: Group A, uninfected mice maintained in a normal environment; Group B, infected mice maintained in a normal environment; Group C, uninfected mice maintained in a wet environment; Group D, infected mice maintained in a wet environment; Group E, uninfected mice maintained in a warm-wet environment; Group F, infected mice maintained in a warm-wet environment ().
Table 1. Experimental groups of this study. All the groups were feeding in the manual climatic box and half of the rats were infected by virus
All of the mice were fed in the manual climatic box located in the P2 laboratory (this laboratory was located at the Department of Microbiology and Immunology, Medical College of Jinan University) for 2 weeks. On the 13th day, half of the mice were infected with the A/FM/1/47(H1N1) virus (Institute of Tropical Diseases, Southern Medical University).
The cryopreserved FM1 virus was revived, replicated in chick embryos and the viral titer determined for the experiment. Based on preliminary experiments, we determined that the titer of the viral stock was 1:640 and the dosage was 50 μl. The intranasal administration lasted for 4 d ().
Figure 5. The timeline of model establish. For the first 2 days, the mice were kept in our P2 laboratory to acclimate to the feeding environment. On the third day, mice were introduced to the 3 different environments. On the thirteenth day, the mice were infected with the FM1 virus through nasal inhalation for 4 days; on the 18 day, the mice were executed for viral detection.
Reagents and methods
We used HE staining (HE Kit, Beyotime Institute of Biotechnology, Shanghai, China) to observe the pathologic changes in the lung tissue. The Th1/Th2 T cell subsets and the Th17/Treg balance were detected by flow cytometry (FCM). The levels of IL-4, IL-10, IL-17 and IFN-γ in the peripheral blood serum were assayed by ELISA (NeoBioscience Technology CO Ltd., Shenzhen, China), and the expression of immune recognition RLH signal pathway was detected by RT-PCR and Western blot.
The reagents for FCM, including PMA, ionomycin and monensin, were purchased from Sigma Co. Ltd., USA. The anti-mouse CD4 PE-Cy7, anti-mouse IL-4 APC, FIX&PERM Kit, anti-mouse IFN-gamma, anti-mouse CD25 Alexa Fluor, anti-mouse/rat Foxp3 APC and anti-Mouse/ Rat IL-17A PE were purchased from eBioscience Co. Ltd., USA.
The reagents for the RT-PCR and Western blotting, including the MAVS antibody (#3993) and the RIG-1 antibody (#4520) were purchased from Cell Signaling Technology Co. Ltd., USA, and the TRIZOL and reverse transcription and PCR reagents were purchased from TakaRa Co. Ltd., JAPAN.
Cytokine measurements
Peripheral blood was collected for ELISAs. The peripheral blood was centrifuged to collect the serum, and the expression of IL-4, IFN-γ, IL-10 and IL-17 was determined by ELISA, according to the manufacturer's instructions.
Measurement of T cell subset balance
The proportions of Th1/Th2 and Th17/Treg cells were measured by flow cytometry (FCM) to evaluate the balance of the T cell subsets. The lymphocytes were extracted from the spleens of mice and stimulated with PMA/ionomycin prior to staining for CD4 and CD25. After surface staining, the cellular were permeabilized by Cytofix/Cytoperm Kit. Finally, the intracellular expression of IL-4, IFN-gamma, IL-17 and Foxp3 were determined by staining with the antibodies listed above. All procedures were performed according to the instructions of the manufactures. The data were analyzed by Flowjo software.
RLH signal pathway detected by RT-PCR and Western Blot
The total RNA was extracted by TRIZOL, and reverse transcription and PCR reaction were performed according to the instructions of the manufactures. The expression of each gene was analyzed by the 2-ΔΔCt method. The total protein was extracted by RIPA lysis and quantified using a BCA kit. SDS-PAGE electrophoresis, transfer, blocking, hatch antibodies and chemiluminescence were all performed according to the manufactures' instructions.
Statistics
All data were expressed as the mean±SEM. The means of different groups were compared using a one-way analysis of variance (ANOVA). Differences were assumed to be significant at P<0.05.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the National Natural Science Foundation of China (NO. 81273616, 81403302, 81473557), the Natural Science Foundation of Guangdong province (NO. 2014A030310477, NO.S2013010013434), the Fundamental Research Funds for the Central Universities (No. 21614306) and the Science and Technology Program of Guangzhou, China (No. 2014J4100106).
References
- Iwami S, Takeuchi Y, Liu X, Nakaoka S. A geographical spread of vaccine-resistance in avian influenza epidemics. J Theor Biol 2009; 259:219-28; PMID:19361532; http://dx.doi.org/10.1016/j.jtbi.2009.03.040
- Uchida M, Tsukahara T, Kaneko M, Washizuka S, Kawa S.. How the H1N1 influenza epidemic spread among university students in Japan:Experience from Shinshu University. Am J Infect Control 2012; 40:218-20; PMID:21764179; http://dx.doi.org/10.1016/j.ajic.2011.03.012
- Macintyre CR, Heywood AE, Kovoor P, Ridda I, Seale H, Tan T, Gao Z, Katelaris AL, Siu HW, Lo V, et al. Ischemic heart disease, influenza and influenza vaccination: a prospective case control study. Heart 2013; 99:1843-8; PMID:23966030; http://dx.doi.org/10.1136/heartjnl-2013-304320
- Gu RX, Liu LA, Wei DQ. Structural and energetic analysis of drug inhibition of the influenza A M2 proton channel. Trends Pharmacol Sci 2013; 34:571-80; PMID:24011996; http://dx.doi.org/10.1016/j.tips.2013.08.003
- Mungall BA, Xu X, Klimov A. Surveillance of influenza isolates for susceptibility to neuraminidase inhibitors during the 2000–2002 influenza seasons. Virus Res 2004; 103:195-7; PMID:15163509; http://dx.doi.org/10.1016/j.virusres.2004.02.033
- Schutten M, van Baalen C, Zoeteweij P, Fraaij P. The influenza virus: disease, diagnostics, and treatment. MLO Med Lab Obs 2013; 45:38-40; PMID:24313126
- Altizer S, Ostfeld RS, Johnson PT, Kutz S, Harvell CD. Climate change and infectious diseases: from evidenceto a pre dictive framework. Science 2013; 341:514-9; PMID:23908230; http://dx.doi.org/10.1126/science.1239401
- Karagiannis I, Brandsema P, VanderSande M. Warm, wet weather associated with increased Legionnaires' disease incidence in the Netherlands. Epidemiol Infect 2009; 137:181-7; PMID:18631425; http://dx.doi.org/10.1017/S095026880800099X
- RRM Paterson, M Sariah, N Lima. How will climate change affect oil palm fungal diseases?, Crop protection 2013; 46:113-20; http://dx.doi.org/10.1016/j.cropro.2012.12.023
- Yi Guan, Gavin JD Smith. The emergence and diversification of panzooticH5N1 influenza viruses, Virus Res 2013; 178:35-43; PMID:23735533; http://dx.doi.org/10.1016/j.virusres.2013.05.012
- Moro PL, Museru OI, Broder K, Cragan J, Zheteyeva Y, Tepper N, Revzina N, Lewis P, Arana J, Barash F, et al. Safety of Influenza A (H1N1) 2009 Live Attenuated Monovalent Vaccine in Pregnant Women. Obstet Gynecol 2013; 122:1271-8; PMID:24201689; http://dx.doi.org/10.1097/AOG.0000000000000010
- Chi Y, Zhu YF, Wen T, Cui L, Ge Y, Jiao Y, Wu T, Ge A, Ji H, Xu K, et al. Cytokine and Chemokine Levels in Patients Infected With the Novel Avian Influenza A (H7N9) Virus in China. J Infect Dis 2013; 208:1962-7; PMID:23990573; http://dx.doi.org/10.1093/infdis/jit440
- Feng Y, Mao H, Xu C, Chen Y, Yan J, Gao J, Li Z, Xia S, Lu Y. Origin and Characteristics of Internal Genes Affect Infectivity of the Novel Avian-Origin Influenza A (H7N9) Virus. Plos one 2013; 8:e81136; PMID:24278391; http://dx.doi.org/10.1371/journal.pone.0081136
- Mutasem El-Fade, Sophia Ghanimeh, Rania Maroun, Ibrahim Alameddine. Climate change and temperature rise: Implications on food- andwater-borne diseases. Sci Total Environ 2012; 437:15-21; PMID:22903000; http://dx.doi.org/10.1016/j.scitotenv.2012.07.041
- Silva DR, Viana VP, Müller AM, Livi FP, Dalcin Pde T. Respiratory viral infections and effects of meteorologicalparameters and air pollution in adults with respiratorysymptoms admitted to the emergency room. I Influenza Other Respir Viruses 2014; 8:42-52; PMID:24034701; http://dx.doi.org/10.1111/irv.12158
- Hernroth B, Sköld HN, Wiklander K, Jutfelt F, Baden S. Simulated climate change causes immune suppression and protein damagein the crustacean Nephropsnorvegicus. Fish Shellfish Immunol 2012; 33:1095-101; PMID:22974540; http://dx.doi.org/10.1016/j.fsi.2012.08.011
- Adamo SA, Lovett MM.Some like it hot: the effects of climate change on reproduction, immune function and disease resistance in the cricket Gryllustexensis. J Exp Biol 2011; 214:1997-2004; PMID:21613515; http://dx.doi.org/10.1242/jeb.056531
- Strutt TM, McKinstry KK, Swain SL. Functionally diverse subsets in CD4 T cell responses against influenza. J Clin Immunol 2009; 29:145-50; PMID:19050998; http://dx.doi.org/10.1007/s10875-008-9266-4
- Fietta P, Delsante G. The effector T helper cell triade. Riv Biol 2009; 102:61-74; PMID:19718623
- Bermeio-Martin JF, Ortiz de Leiarazu R, Pumarola T, Rello J, Almansa R, Ramírez P, Martin-Loeches I, Varillas D, Gallegos MC, Serón C, et al. Th1 and Th17 hypercytokinemia as early host response signaturein severe pandemic influenza. Crit Care 2009; 13:1-11
- Wu XL, Ju DH, Chen J, Yu B, Liu KL, He JX, Dai CQ, Wu S, Chang Z, Wang YP, et al. Immunologic Mechanism of patchouli alcohol anti-H1N1 influenza virus may through regulation of the RLH signal pathway in vitro. Curr Microbiol 2013; 64:431-6; http://dx.doi.org/10.1007/s00284-013-0381-y
- Rehwinkel J, Tan CP, Goubau D, Schulz O, Pichlmair A, Bier K, Robb N, Vreede F, Barclay W, Fodor E, et al. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell 2010; 140:397-408; PMID:20144762; http://dx.doi.org/10.1016/j.cell.2010.01.020
- Liedmann S, Hrincius ER, Guy C, Anhlan D, Dierkes R, Carter R, Wu G, Staeheli P, Green DR, Wolff T, et al. Viral suppressors of the RIG-I-mediated interferon response are pre-packaged in influenza virions. Nat Commun 2014; 5:5645; PMID:25487526; http://dx.doi.org/10.1038/ncomms6645
