abstract
HLA-DR is the most commonly expressed and likely the most medically important human MHC class II, antigen presenting protein. In a normal immune response, HLA-DR binds to antigenic peptide and the HLA-DR/peptide complex binds to a T-cell receptor, thus contributing to T-cell activation and stimulation of an immune response against the antigen. When foreign antigen is not present, HLA-DR binds endogenous peptide which, under normal conditions does not stimulate an immune response. In most cases, the human peptide is CLIP, but a certain percentage of HLA-DR molecules will be present at the cell surface with other human peptides. We have recently shown that cell surface, CLIP/HLA-DR ratios are a measure of peptide heterogeneity, and in particular, changes in CLIP/HLA-DR ratios represent changes in the occupancy of HLA-DR by other, endogenous peptides. For example, treatment of cells with the HDAC inhibitor, Entinostat, leads to an upregulation of Cathepsin L1 and replacement of Cathepsin L1 senstitive peptides with HLA-DR binding, Cathepsin L1 resistant peptides, an alteration that can be at least partially assessed via assessment of CLIP/HLA-DR cell surface ratios. Here we assay for CLIP/HLA-DR ratios following treatment of immortalized B-cells with a variety of common drugs, almost all of which indicate significant changes in the CLIP/HLA-DR ratios. Furthermore, the CLIP/HLA-DR ratio changes parallel the impact of the drug panoply on cell viability, suggesting that alterations in the HLA-DR peptidome are governed by a variety of mechanisms, rather than exclusively dependent on a dedicated peptide loading process. These results raise questions about how FDA approved drugs may affect the immune response, and whether any of these drugs could be useful as vaccine adjuvants?
Introduction
The human major histocompatibility complex (MHC) class II proteins regulate the immune response to foreign antigen, anti-cancer immune responses, and autoimmunity. In the classical scenario, the MHC class II proteins expressed on the surface of professional antigen presenting cells, such as dendritic cells, bind peptide derived from extra-cellular sources, after proteolytic degradation of the antigenic protein. The resulting, protease-derived peptides bind to MHC class II proteins in the cleft formed by the α and β chains, and the complex is transported to the cell surface. When antigenic peptide is not available, endogenous peptides are substitutes. If there are no peptides in the MHC class II binding cleft, the MHC class II dimers are unstable. The most common substitute peptide is termed, class II associated invariant peptide (CLIP), and is derived from the MHC class II chaperone, invariant chain. However, a great variety of other endogenous peptides can also occupy the MHC class II binding cleft,Citation5 and the sources of these additional, endogenous peptides, and their mode of MHC class II access and binding, remain obscure.
Several proteases have been implicated in the preparation of peptide for MHC class II binding, for examples, Cathepsin L1 Citation6,14 and Cathepsin S,Citation11,12 but the study of this aspect of peptide loading has not been extensive and many questions regarding the factors affecting the MHC class II peptidome remain to be answered. For example, how do varying levels of protease, affected by various regulatory mechanisms, impact the peptides that become part of the peptidome? And, are there differences in which proteases impact the MHC class II peptidome in distinct, differentiated cell types? And, does apoptosis, which leads to a variety of changes in cell protease composition, lead to a different peptidome?
We previously reported Entinostat up-regulation of Cathepsin L1 in Raji B-cells and an accompanying replacement of Cathepsin L1 sensitive, human peptides by Cathepsin L1 resistant peptides,Citation3 which led to the question of how frequently small molecules may be expected to alter the MHC class II peptidome, as represented by HLA-DR bound peptides in immortalized B-cells? Some of the small molecules in this study represent FDA approved drugs, to get a basic appreciation of possible, unexpected peptidome alterations in patients, which in turn could unexpectedly impact their immune responses. Here we report that CLIP/HLA-DR ratios are very commonly altered by small molecules representing a variety of drugs, and that these alterations parallel drug mediated cell death. The roles of non-canonical proteases in this process are discussed.
Results
We treated Raji B-cells with a series of drugs indicated in and assayed for their effects on the CLIP/HLA-DR ratio, as detected by flow cytometry, exactly as described.Citation3 In particular, cells were stained separately with 4 antibodies, 2 isotype controls, anti-CLIP and anti-HLA-DR. The mean fluorescence values for the isotype controls were subtracted from the anti-CLIP and anti-HLA-DR mean fluorescence values, respectively. Then, the ratios of these so adjusted, anti-CLIP and HLA-DR values were obtained. The HDACIs, Entinostat and Vorinostat, have been shown to alter the CLIP/HLA-DR ratio in the Raji cells,Citation3 and these alterations are virtually, exactly reproduced in In all cases, treatment leads to a reduction in the CLIP/HLA-DR ratios, indicating an alteration of the HLA-DR peptidome upon treatment. To verify the specificity of the antibodies and the appropriateness of the above approach as an assay of CLIP/HLA-DR ratios, we repeated the anti-CLIP and anti-HLA-DR staining and calculation of ratios for the RJ subclone of Raji,Citation1 which lacks CIITA, the obligate transactivator for HLA-DRA and HLA-DRB transcription and thus lacks HLA-DR surface expression. Results indicate only background binding of the anti-CLIP and anti-HLA-DR antibodies to RJ ().
Table 1. Drugs used to treat cells and assay for alterations in the CLIP/HLA-DR ratios.
Table 2. Correlation of cell death and reduction in CLIP/HLA-DR ratios.
Figure 1. Effect of drug panoply on the CLIP/HLA-DR ratio. Y axis = [Mean fluorescence intensity (MFI) of anti-CLIP minus the MFI of the ant-CLIP isotype control] divided by [MFI of anti-HLA-DR minus the MFI of anti-HLA-DR isotype control]. X axis represents the drug panoply, with individual drugs indicated. (A) Raji; (B) Raji repeat, but note: DMSO was not done for this experiment; (C) HLA class II-negative, RJ subclone of Raji. All MFI values represent gating on live cells.
![Figure 1. Effect of drug panoply on the CLIP/HLA-DR ratio. Y axis = [Mean fluorescence intensity (MFI) of anti-CLIP minus the MFI of the ant-CLIP isotype control] divided by [MFI of anti-HLA-DR minus the MFI of anti-HLA-DR isotype control]. X axis represents the drug panoply, with individual drugs indicated. (A) Raji; (B) Raji repeat, but note: DMSO was not done for this experiment; (C) HLA class II-negative, RJ subclone of Raji. All MFI values represent gating on live cells.](/cms/asset/87b99210-ea25-4faf-861a-55e73a424e4b/khvi_a_1089370_f0001_oc.gif)
The relatively high affinity and specificity of the anti-CLIP suggested the possibility that results above could be reproduced using anti-CLIP alone. Thus, we averaged the above results from 2 independent experiments (; ) and compared the averages with the mean fluorescence values for anti-CLIP alone (), which indicates that anti-CLIP alone is able to provide the same basic indication of alteration in the HLA-DR peptidome as is the calculation of the ratio of the isotype-adjusted CLIP and HLA-DR values.
Figure 2. Assessment of CLIP alone for drug effects. (A) Average of data above; (B) Average of anti-CLIP MFI alone (without subtracting isotype control; without obtaining a ratio with anti-HLA-DR MFI). (C) MFI for anti-CLIP for the T5–1 B-cell line, with the indicated drugs. (D) MFI for anti-CLIP for the HLA class II negative 616 Citation7 subclone of T5–1.
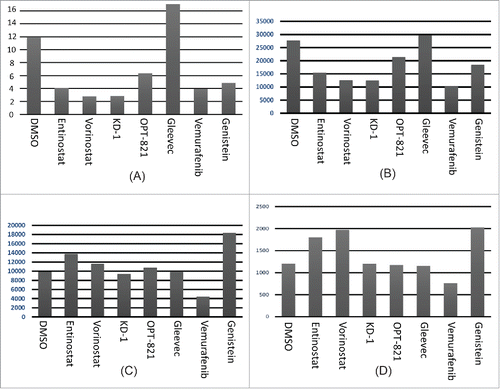
To verify the effect of the drug panoply on a second B-cell line, we assayed drug treated T5–1 B-cells with anti-CLIP followed by flow cytometry (). Overall, the drug treatments led to alterations in the HLA-DR peptidome, but there were several differences, vis a vis the DMSO control, from the Raji cells. Interestingly, the relative differences between Entinostat and Vorinostat remained the same as with Raji.
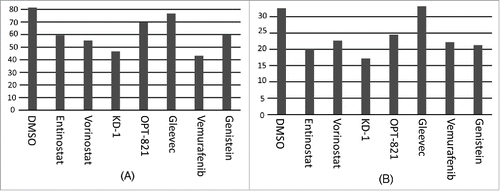
Each of the drugs tested leads to reduced cell viability. Thus, we determined the correlation between cell viability and the alteration in the HLA-DR peptidome, with results indicating that cell viability is inversely proportional to the extent of the reduction in the ratio of CLIP/HLA-DR, i.e., in the replacement of CLIP by other endogenous peptides ().
Figure 3. Percent of live cells following drug treatments. Drug treatments as indicated on the X-axis. (A) Data taken from experiment represented by . (B) Data taken from experiment represented by .
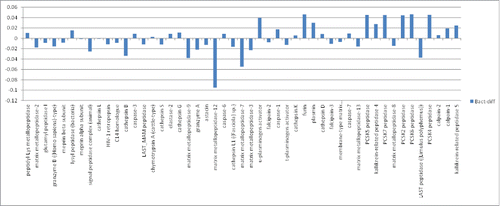
The above indicated connections between treatments with a wide variety of small molecules, cell death, and HLA-DR peptide occupancy suggests that a variety of proteases may have at least an indirect impact HLA-DR peptide occupancy, besides the canonical cathepsins, previously tied directly to antigen processing for MHC class II molecules. Recent work has indicated that HLA-DR epitope resistance correlates with protease expression levels.Citation3 Thus, to obtain a more global view of the sensitivity of HLA-DR epitopes to proteases, we developed an in silico algorithm, closely related to previously published, publicly available algorithms, for assessing protease sensitivity for a given epitope. Application of the scripted version of the algorithm to the entire set of immune epitope database (IEDB), HLA-DR bound bacterial epitopes indicated that, on average, many of these epitopes are resistant to a variety of proteases (), e.g., matrix metalloprotease-12 (MMP-12), expressed at a high level in macrophage, as well as MMP-7 and MMP-9.
Figure 4. Sensitivity of IEDB, HLA-DR bacterial epitopes to proteases. Proteases indicated along the X-axis; sensitivity along the Y-axis. Bars above zero represent proteases where the average sensitivity of all the IEDB bacterial epitopes is greater than the average sensitivity of all the epitopes for all the proteases. Bars below zero indicate proteases where the average sensitivity of all the IEDB bacterial epitopes is less than the average for the entire set of proteases. For example, most IEDB bacterial epitopes are relatively resistant to matrix metalloprotease-12 (MMP-12), consistent with high level MMP-12 expression in macrophage.Citation2,13 And, most IEDB bacterial epitopes are relatively resistant to Cathepsin L, B and S, consistent with the role of these proteases in antigen processing.Citation3,6,8,11
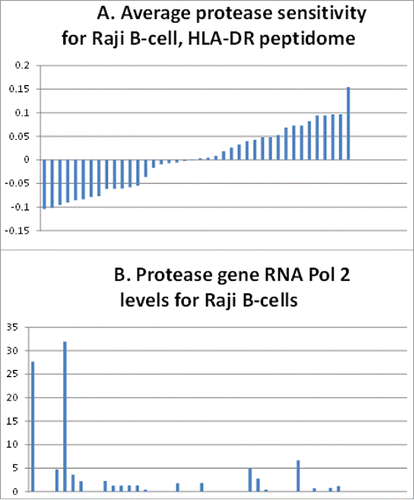
Finally, we obtained the average sensitivity of the entire HLA-DR peptidome of Raji B-cells to a wide variety of proteases, using the above, bioinformatics based algorithm. We then obtained the Raji RNA pol 2 values for each protease gene from the ENCODE database, which in turn are presumed to closely parallel the level of gene transcription. The level of RNA pol 2 for the protease genes is inversely proportional to the average sensitivity of the HLA-DR peptidome to the proteases (), again, consistent with the idea that many proteases indirectly impact the HLA-DR peptidome.
Figure 5. Correlation of Raji B-cell, protease gene, pol 2 levels with the HLA-DR peptidome epitopes' resistance to the proteases. (A) Sensitivity (above the average midline) or resistance (below the average midline) of epitopes of the Raji B-cell HLA-DR peptidome determined in ref.Citation3 (B) Levels of RNA polymerase 2 associated with the protease genes representing the above bars in the histogram of (A).
Discussion
The role of proteases in the formation of the MHC class II peptidome is at best poorly understood. While mouse models have indicated the role of several particular proteases in cleaving and generating peptides for MHC class II binding,Citation10 the studies have been highly limited. In particular, there is a casual assumption that all cells, and all cell states generated by various environmental stimuli, have the same process of MHC class II peptide loading. Furthermore, there is very little understanding of how cytokines or other environmental stimuli impact the protease effects on antigens that will ultimately become part of the MHC class II peptidome.
We recently developed a rapid assay for detecting HLA-DR peptidome variations, i.e., by observing changes in the ratio of surface CLIP to HLA-DR.Citation3 This assay was verified by eluting peptides from Raji B-cells in the presence and absence of the HDACI, Entinostat. Results from this approach indicated that Entinostat upregulated Cathepsin L1, and this in turn led to an increase in the Cathepsin L1 resistant peptides in the HLA-DR peptidome. While the CLIP/HLA-DR ratio does not indicate which peptides are present or absent in the HLA-DR peptidome, the change in the ratio is an indication of an overall alteration the peptidome constituents.
The results reported here indicate that this rapid assay can be used to determine whether numerous small molecules have an impact on the HLA-DR peptidome. In particular, the small molecules studied here have a range of biochemical effects, from HDAC and Cathepsin inhibition to tyrosine kinase inhibition. However, in all cases, or almost all cases, it is clear that the drugs could affect the peptidome. Futhermore, there was a correlation of cell death levels and the impact of the small molecule on the CLIP/HLA-DR ratio in Raji B-cells. This result, combined with the diverse set of drugs used, raises the question of how widespread the impact of various protease may be on the formation of the HLA-DR peptidome.
In untreated Raji B-cells, we established an inverse correlation between the RNA pol II levels of a wide variety of protease genes, available from ENCODE, and the overall sensitivity of the peptides of the peptidome to the indicated proteases. This correlation has only a modest strength but is statistically significant. Thus, the analysis is consistent with the results of the small molecule impacts on the CLIP/HLA-DR ratios, i.e., consistent with the idea that there are a number of other proteases, besides the canonical Cathepsin L1 and S, that have an impact on the HLA-DR peptidome.
Overall, the data and conclusions from this work raise the questions, how do small molecules, and potentially drugs received by patients, affect the HLA-DR mediated immune response; and can small molecules be used to generate a more diverse immune response for vaccines or generation of monoclonal antibodies?
Materials and methods
Cell culture, drug treatments, and CLIP and HLA-DR flow cytometry
Raji B-cells, and the other B-cell lines used in this report, were grown in culture as described.Citation3 Briefly, cells were grown in suspension in RPMI, with 10% fetal bovine serum, glutamine, penicillin, streptomycin and pyruvate. Cells were treated for 40 hours with indicated drugs (from stock solutions) in the following working concentrations: Entinostat (5 µM), Vorinostat (3 µM), Cathepsin L1 inhibitor, KD-1 (5 µM),Citation4 OPT-821 (6 µg/ml), Gleevec (3 µM), Vemurafenib (2.5 µM), and Genistein (100 µM). After 40 hours, the cells were recovered by centrifugation and stained for either CLIP or HLA-DR exactly as described.Citation3 Briefly, cells were resuspended at 4°C in PBS plus 2% human serum to block Fc receptors for 15 min. Cells were then treated with PE labeled anti-HLA-DR or FITC labeled anti-CLIP or isotype controls (in separate treatment reactions), to detect surface expression. Mean fluorescence values were obtained via flow cytometry as in ref.Citation3 Cell viability was determined with simultaneous DAPI staining as described.Citation3 The detailed collection of data representing , representing the flow cytometry analyses described above, are in the supporting online material (SOM).
Protease-related bioinformatics
The algorithms used for obtaining the relationship between protease sensitivity or resistance and RNA Pol2 levels associated with the protease genes in Raji B-cells, respectively, along with the scripted versions of the algorithm in Perl (version 5) code, are detailed in the SOM. Briefly, the publicly available MEROPS database's was used as a binding matrix for the protease sensitivity and resistance determinations, using 8 amino acid contacts for any given protease-substrate interaction. For measurements throughout an epitope, or entire protein, the 8 amino acid contact was shifted one amino acid at a time, to determine the highest level of sensitivity/resistance. The entire, publicly available Immune Epitope Database (IEDB) was downloaded from the IEDB website, and for the Raji B-cell epitopes, we used the data available from the SOM in ref.Citation3 The gene accession numbers in the ref.Citation3 SOM were used to download the reference sequence for each protein representing the Raji B-cell epitopes were given the same format as the IEDB data and copied to a MySQL database, which also contained the IEDB data. A non-parametric K-S test was used to assess statistical significance of the protease matrix binding scores, to establish the highest protease matrix binding score, when comparing 8-mers within a given length of epitope or protein. The algorithm and Perl code for obtaining the Raji B-cell Pol2 data from ENCODE aligns Pol2 regions (or anything in the *.narrowpeak format) with all the genes in the human genome, with genes defined by largest transcript, as in ref.Citation9
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
Authors would like to acknowledge the financial support of the taxpayers of the State of Florida.
References
- Accolla RS, Scarpellino L, Carra G, Guardiola J, Trans-acting element(s) operating across species barriers positively regulate expression of major histocompatibility complex class II genes. J Exp Med 1985; 162:1117-33; PMID:3862745; http://dx.doi.org/10.1084/jem.162.4.1117
- Belaaouaj A, Shipley JM, Kobayashi DK, Zimonjic DB, Popescu N, Silverman GA, Shapiro SD. Human macrophage metalloelastase. Genomic organization, chromosomal location, gene linkage, and tissue-specific expression. J Biol Chem 1995; 270:14568-75; PMID:7782320; http://dx.doi.org/10.1074/jbc.270.24.14568
- Cronin K, Escobar H, Szekeres K, Reyes-Vargas E, Rockwood AL, Lloyd MC, Delgado JC, Blanck G. Regulation of HLA-DR peptide occupancy by histone deacetylase inhibitors. Hum Vaccines Immunother 2013; 9:784-9; PMID:23328677; http://dx.doi.org/10.4161/hv.23085
- Dana D, De S, Rathod P, Davalos AR, Novoa DA, Paroly S, Torres VM, Afzal N, Lankalapalli RS, Rotenberg SA, Chang EJ, Subramaniam G, Kumar S. Development of a highly potent, selective, and cell-active inhibitor of cysteine cathepsin L-A hybrid design approach. Chem Commun 2014; 50:10875-8; PMID:25089379; http://dx.doi.org/10.1039/C4CC04037F
- Denzin LK, Cresswell P. HLA-DM induces CLIP dissociation from MHC class II alpha beta dimers and facilitates peptide loading. Cell 1995; 82:155-65; PMID:7606781; http://dx.doi.org/10.1016/0092-8674(95)90061-6
- Guncar G, Pungercic G, Klemencic I, Turk V, Turk D. Crystal structure of MHC class II-associated p41 Ii fragment bound to cathepsin L reveals the structural basis for differentiation between cathepsins L and S. EMBO J 1999; 18:793-803; PMID:10022822; http://dx.doi.org/10.1093/emboj/18.4.793
- Loosmore S, Gladstone P, Pious D, Jerry LM, Tamaoki T. Control of HLA-DR antigen gene expression at the pretranslational level: comparison of an HLA-DR-positive B lymphoblastoid cell line and its HLA-DR-negative variant. Immunogenetics 1982; 15:139-50; PMID:6800948; http://dx.doi.org/10.1007/BF00621947
- Matsunaga Y, Saibara T, Kido H, Katunuma N, Participation of cathepsin B in processing of antigen presentation to MHC class II. FEBS Lett 1993; 324:325-30; PMID:8405375; http://dx.doi.org/10.1016/0014-5793(93)80144-J
- Mauro JA, Blanck G. Functionally distinct gene classes as bigger or smaller transcription factor traps: a possible stochastic component to sequential gene expression programs in cancer. Gene 2014; 536:398-406; PMID:24291030; http://dx.doi.org/10.1016/j.gene.2013.11.013
- Pluger EB, Boes M, Alfonso C, Schroter CJ, Kalbacher H, Ploegh HL, Driessen C. Specific role for cathepsin S in the generation of antigenic peptides in vivo. Euro J Immunol 2002; 32:467-76; PMID:11813165; http://dx.doi.org/10.1002/1521-4141(200202)32:2%3c467::AID-IMMU467%3e3.0.CO;2-Y
- Riese RJ, Mitchell RN, Villadangos JA, Shi GP, Palmer JT, Karp ER, De Sanctis GT, Ploegh HL, Chapman HA. Cathepsin S activity regulates antigen presentation and immunity. J Clin Investigat 1998; 101:2351-63; PMID:9616206; http://dx.doi.org/10.1172/JCI1158
- Riese RJ, Wolf PR, Bromme D, Natkin LR, Villadangos JA, Ploegh HL, Chapman HA. Essential role for cathepsin S in MHC class II-associated invariant chain processing and peptide loading. Immunity 1996; 4:357-66; PMID:8612130; http://dx.doi.org/10.1016/S1074-7613(00)80249-6
- Shapiro SD, Kobayashi DK, Ley TJ. Cloning and characterization of a unique elastolytic metalloproteinase produced by human alveolar macrophages. J Biol Chem 1993; 268:23824-9; PMID:8226919
- Villadangos JA, Bryant RA, Deussing J, Driessen C, Lennon-Dumenil AM, Riese RJ, Roth W, Saftig P, Shi GP, Chapman HA, et al. Proteases involved in MHC class II antigen presentation. Immunol Rev 1999; 172 109-20; PMID:10631941; http://dx.doi.org/10.1111/j.1600-065X.1999.tb01360.x
