ABSTRACT
So far, the development of a human immunodeficiency virus (HIV) vaccine has been unsuccessful. However, recent progress in the field of broadly neutralizing antibodies (bNAbs) has reinvigorated the search for an HIV vaccine. bNAbs develop in a minority of HIV infected individuals and passive transfer of these bNAbs to non-human primates provides protection from HIV infection. Studies in a number of HIV infected individuals on bNAb maturation alongside viral evolution and escape have shed light on the features important for bNAb elicitation. Here we review the observations from these studies, and how they influence the rational design of HIV vaccines.
Introduction
The development of an effective human immunodeficiency virus (HIV) vaccine would represent a breakthrough in the battle against the ongoing HIV pandemic. However, after decades of research, an effective HIV vaccine remains elusive. Most effective antiviral vaccines currently used rely on the elicitation of neutralizing antibodies (NAbs) in the immunized individual.Citation1 These NAbs can prevent virus infection by direct neutralization of the virus upon entry. Although there have been 6 large scale HIV vaccine efficacy trials to date, none of them proved to be fully effective in preventing HIV transmission.Citation2-9 Nonetheless, modest partial protection was observed in the RV144 clinical trial, where non-NAbs were shown to be the correlate of protection, however the exact mechanism is still under investigation.Citation10-12 To enhance future vaccine efficacy improved vaccine immunogen design strategies should be explored, for instance those utilizing knowledge obtained from natural HIV infection.
One of the main features of HIV is its high sequence variability, which causes a great diversity in the circulating HIV strains.Citation13,14 As a result, the induction of strain-specific NAbs will not be adequate to prevent infection by all circulating HIV species and it is therefore highly desirable to elicit broadly neutralizing antibodies (bNAbs) by vaccination. These bNAbs are able to neutralize a broad range of heterologous HIV and several passive transfer studies have demonstrated that full protection against HIV acquisition can be achieved by bNAbs in rhesus macaques.Citation15-17 Therefore, bNAbs form important templates for HIV immunogen design. However, the question remains how to elicit these bNAbs in HIV naive, healthy individuals through vaccination.
Antibody responses during HIV infection
HIV infection is usually established by transmission of a single virus particle, the transmitted/founder (T/F) virus.Citation18 Several months post-seroconversion, NAbs will be produced, which can only neutralize autologous virus, making them strains-specific.Citation19-21 Nevertheless NAbs exert a selective pressure on the Env proteins of the circulating viruses, resulting in the appearance of HIV escape mutants. These escape viruses trigger new rounds of NAb affinity maturation, which can eventually lead to the development of heterologous NAbs that are capable of neutralizing a broader range of HIV viruses in approximately 20–30% of naturally HIV-1 infected individuals.Citation19,20,22-27 Usually these bNAbs arise after 2–3 y of persistent HIV infection and target more conserved areas on the HIV Env protein such as the CD4 binding site (CD4bs), V1V2 apex, gp120-gp41 interface, gp120 glycan patch, and the membrane proximal external region (MPER) of gp41.Citation19,25,28-36
In recent years, technologies and methods for isolating and characterizing Abs from infected individuals have become more efficient. This has resulted in a dramatic expansion of the number of bNAbs and their target epitopes that have become available for the HIV vaccine field.Citation25,33,34,37-40 It is becoming apparent that almost the entire HIV Env surface can serve as a target for various bNAbs.Citation41 Nevertheless some areas of the Env protein are targeted more often and therefore are designated as (super)sites of vulnerability. bNAbs possess a number of often-shared characteristics. For example, bNAbs are usually highly mutated. The mutation frequency in the variable region of the heavy chain (Vh) is 20–50%, which is 2–5 times more than typical Abs against other pathogens.Citation37,42 This suggests that multiple rounds of affinity maturation are necessary to acquire sufficient breadth. Second, bNAbs frequently have unusually long heavy chain complementary-determining region 3 regions (HCDR3s) of up to 38 amino acids.Citation43,44 Possessing longer HCDR3s is thought to provide bNAbs with the ability to penetrate the glycan shield that surrounds the conserved HIV Env protein domains.Citation45,46 Lastly, bNAbs are often auto- or polyreactive.Citation47-50 Auto- or polyreactivity could be advantageous to bNAbs since it might assist in increased virion binding through enhancing avidity.Citation51 However, autoreactivity would not be a desired property of bNAbs induced by vaccination, since this reactivity against self-antigens could potentially result in adverse effects. In general, all these characteristics are associated with negative selection during B cell development. Yet, in HIV infected individuals, Abs with either of these properties are not subjected to negative selection in early development or after maturation of the bNAbs. This is probably due to an impaired immune system in these individuals.Citation47,52,53 Therefore, it will be challenging to induce a bNAb response that displays these features in an uninfected individual with a fully functional immune system. Knowledge about the development of bNAbs in natural infection can guide rational vaccine design.Citation54
Co-evolution of virus and antibody
In general there are 2 different ways in which bNAbs can develop during natural infection. Both mechanisms depend on continuous virus evolution alongside Ab maturation. Either Abs evolve from the autologous neutralizing response, acquiring more breadth over time,Citation33,39 or they are established as an independent Ab lineage after escape from an early autologous neutralizing response.Citation25,34,38 An example of the former is the bNAb VRC26, directed to an N160 glycan dependent epitope at the V1/V2 apex. The germline of VRC26 was engaged by a superinfecting (SI) virus possessing the more commonly occurring N160 glycan, in comparison to the T/F virus that lacked this specific glycan.Citation39 Viral escape from early VRC26 Abs facilitated affinity maturation mainly in the CDRH3 of this lineage, increasing binding and neutralization properties. Recombination between the SI and T/F virus resulted in high diversity, especially in the V1/V2 apex, of circulating viruses further increasing Ab affinity maturation and thereby increasing breadth.
In the other process, selective pressure exerted by the autologous NAb response will result in the appearance of a new epitope capable of engaging the germline of an independent bNAb lineage.Citation34 Emergence of distinctive Ab lineages allows for the development of different bNAb waves during HIV infection. Combined, these bNAb waves might contribute to increased neutralization breadth and potency.Citation25,40,55 In one individual described by Wibmer et al.,Citation25 3 distinctive bNAb waves, targeting the V1V2 apex, CD4bs and a still undefined epitope, contributed to neutralization breadth. Proof that bNAbs complement each other in generating increased serum breadth came from a study by Bonsignori et al.Citation55 They demonstrated that serum neutralization could be recapitulated by combining 2 clones of distinct bNAb lineages, CH01-04 and CH30-34, targeting different epitopes. Indicating that combined, distinct bNAb lineages can complement each other to eventually create broad neutralization.Citation25,55 Similar observations were made in the individual that elicited the CH103 bNAb.Citation33 This individual also developed a second bNAb, CH235.Citation40,56 In contrast to the development of different Ab waves with multiple specificities, CH103 and CH235 target the same area on the Env protein (the CD4 binding site), permitting CH235 specific viral escape to influence the epitope for CH103. Whether interaction with a second (or third) NAb lineage, with similar or distinct epitopes, is crucial for development of fully mature bNAbs remains to be seen. If so, this might complicate vaccine design considerably.
Interestingly, these studies also reveal that clonal family members of bNAbs often show limited neutralization capacity even though they underwent similar levels of affinity maturation.Citation27,44,57 This underlines the importance to guide the Ab affinity maturation at different steps of the selection process. Very recently the developmental pathway was mapped for a bNAb response directed to the N332-glycan on the Env protein.Citation58 The Ab lineage diverged very early in development in response to Env escape mutants, overall the extend of SMH was limited and the N332-directed bNAbs contained few characteristics usually associated with negative selection during B-cell development. Suggesting a pathway of bNAb development that could more easily be achieved through vaccination.
Vaccine strategies
On HIV, the only target for NAbs is the Env protein, since this is the only protein readily accessible on the outside of the virus. Env is a trimeric protein comprised of proteolytically processed gp120 and gp41 subunits linked through a non-covalent interaction.Citation59-61 The Env protein is covered in a thick layer of glycans which conceal many of its epitopes.Citation61 During infection, Env can be present in different conformations. In the pre-fusion conformation, the Env protein has a compact or closed formation. This conformation is often referred to as the native form of Env and exposes the epitopes for bNAbs best. Binding to the CD4-receptor and subsequent conformational changes to facilitate membrane fusion, result in a more open conformation. In this state, epitopes for non-NAbs will be more easily accessible. Because these epitopes are generally immunodominant, an open Env conformation might divert the Ab elicitation away from the development of bNAbs. Therefore it is widely accepted that immunizations should contain Env proteins presented in the native-like conformation.
In clinical HIV vaccine trials to date monomeric, monovalent gp120 or gp140 soluble proteins have been used.Citation3,62-64 Although soluble gp120 elicits a weak NAb response against neutralization sensitive (Tier 1) viruses, it does not induce the desired bNAbs.Citation3,62-64 One possible reason is that many epitopes for non-NAbs are exposed that are usually obscured in a trimeric conformation and that might interfere with the induction of bNAbs, while conversely many bNAb epitopes, in particular those that depend on quaternary structure, are not present on gp120. Because autologous neutralization might be an important starting point for developing bNAbs as indicated by natural infection, first efforts were directed at eliciting such a response in immunized animals using improved immunogens. First attempts to increase NAb responses included the development of soluble gp140 Env proteins, truncated at gp41 to remove the transmembrane domain.Citation65,66 In the early generations of gp140 proteins cleavage between the gp120 and gp41 subunits was prevented, usually by mutation of the canonical furin cleavage site.Citation67 Furthermore, in many cases a trimerization motif was added to increase the trimerization propensity.Citation68,69 However, these uncleaved gp140 proteins assume a heterogeneous aberrant conformation and therefore do not resemble the native Env protein. Immunogenicity experiments with such gp140 proteins were disappointing, yielding only non-NAbs or NAbs against Tier 1 viruses.Citation64,70 Not even the autologous viruses could be neutralized. Therefore, attention has shifted to presenting the Env protein in a native-like conformation. Recently stable and correctly processed native-like HIV Env trimers have been generated, termed SOSIP proteins.Citation71,72 These Env trimers contain stabilizing mutations, keeping them in their closed, native conformation. In addition to SOSIP trimers, native flexibly linked trimers (NFL-trimers) form another line of research into native-like trimers.Citation73 In NFL constructs, the furin cleavage site is replaced by a flexible GS-linker, rendering the protein cleavage-independent while maintaining sufficient flexibility for the proteins to ensemble into native-like trimers. Other, more epitope-focused approaches using gp120 subunits, gp120 outer domains, or scaffolds are also being tested.Citation74-76 A recent study by Sanders et al.Citation77 showed that autologous neutralization could be induced by immunization with stable and native like BG505 SOSIP (subtype A) or B41 SOSIP (subtype B) proteins.Citation77 Autologous NAb responses were seen in both rabbits and macaques. However in spite of the high autologous NAb titers, heterologous neutralization was limited to Tier 1 viruses.Citation77
Because immune activation is relatively weak using single Env trimers, nanoparticles are being explored.Citation78 Clustering of Env proteins on the surface of a suitable nanoparticle, such as liposomes, ferritin or lumazine, might enhance immunogenicity through increased B-cell cross-linking.Citation78-80 Although important Env epitopes might be obscured by increased density. Immunizations with ferritin displayed SOSIP Envs increased the NAb titers in mice, as well as rabbits compared to soluble SOSIP Env trimers, however the responses were not broadened.Citation79 Although promising, SOSIP-, NFL-trimers, and nanoparticle display, therefore need to be developed further in order for them to elicit bNAb responses.
Activating Ab germline precursors that have the ability to evolve into bNAbs might be necessary to ensure development of broad neutralization activity.Citation81 However, the inferred germline versions of bNAbs do not usually bind with high affinity to native Env proteins or immunogens.Citation13 Thus, in order to activate desirable germline precursors, Env immunogens should be designed that are able to engage such germline precursors. Jardine et al.Citation82 designed an engineered gp120 outer domain (eOD) protein which was able to bind CD4bs directed bNAbs, as well as their inferred precursors, providing these immunogens with the ability to potentially engage germline bNAb lineages in humans, which was confirmed by the use of germline bNAb knock-in mice.Citation78,82 Although an Ab response was observed after immunogenicity experiments in these knock-in mice, there was no neutralizing activity. This confirms that additional affinity maturation of the Ab is necessary to gain neutralization activity.
Although it would be optimal if a single vaccination could induce an adequate protection against HIV infection, it is more likely that several booster vaccines are necessary to ensure full protection. From natural infection we know that Env diversity is positively correlated with the development of bNAbs later on in infection.Citation38,39,58,83 In addition, increased viral diversity is observed in HIV infected individuals directly preceding development of breadth. Based on this knowledge vaccination with a cocktail of different HIV variants could potentially be a way to induce bNAbs ().Citation84 Supporting the concept of a cocktail vaccine are co-evolution studies showing the importance of cooperation between different bNAb lineages with similar specificities.Citation40 In addition, the notion that multiple bNAb lineages with distinct specificities contribute to extended neutralization breadth in several HIV infected individuals, underlines the need for a diverse virus population able to engage several different bNAb lineages in a vaccination scheme.Citation25,55 Depending on the desired developmental pathway, the composition of the vaccine can vary from including several HIV subtypes, or immunogens specifically designed to expose a particular epitope, to only comprising virus variations from within the same HIV infected individual. In contrast, sequential immunizations with evolving Env proteins, of single or multiple subtypes, has been proposed as an alternative vaccine strategy ().Citation85 However, it should be noted that each bNAb lineage undergoes a tailored maturation pathway and the question remains if the same germline Ab can be activated in different individuals and if so, whether this would consequently give rise to the same Ab response.Citation86
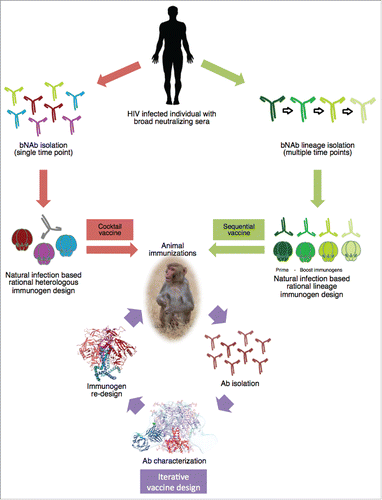
Figure 1. Schematic of rational HIV immunogen design based on natural infection. Env glycoprotein immunogens are rationally designed based on Abs isolated from an HIV infected individual that displays broad neutralization. Vaccine strategies entail cocktail vaccination to elicit bNAbs against a conserved site among different Env proteins. Or sequential vaccination with Env immunogens designed based on a developing bNAb lineage. Subsequent animal immunizations with the designed Env proteins and iterative vaccine design will guide optimization of the immunogen through isolation and characterization of NAbs from immunized animals, providing detailed knowledge on germline usage and target epitope. Which will in turn improve the Env protein immunogen design, improving it to elicit bNAbs.
Since B-cell interaction with T follicular helper cells (Tfh) is of great importance for the survival and maturation process of the B cell in the germinal center, the effect of a potential vaccine on these specific CD4+ T cells should be taken into account. Tfh cells assist follicular dendritic cells in selecting only high affinity B-cells for differentiation into plasma cells or long lived memory cells, and are important for induction of SHM.Citation87 During HIV infection an expansion of the amount of Tfh cells is observed, possibly due to persistent antigenic stimulation.Citation88 Resulting in increased stimulation of B cells in the germinal center, including B cells that would usually have been selected against, thus generating a higher Ab response. A positive correlation between HIV-specific Tfh cells and presence of HIV-specific NAbs has been observed.Citation89 However, there seems to be a fine balance in activating Tfh cells as another study suggests that continuous Tfh cell stimulation induces the upregulation of inhibitory molecules on B cells, affecting the functionality of the Tfh cell so that no adequate B cell help can be provided.Citation90 Even though Tfh cells are increasingly studied in relation to HIV infection, the current knowledge is still very limited.Citation91-93 And although the targeting of Tfh cells by vaccination is definitely something to consider, more research on this subject is required.
Conclusion
The relatively short time span in which significant steps have been made toward an HIV vaccine signifies the rapid progression of the HIV vaccine field. The studies describing the co-evolution of bNAbs and virus reviewed here show that there is not just one way of eliciting bNAbs and that it is very much a complex interplay of the humoral response and viral escape. These studies also highlight the importance of viral diversity and exposure of conserved epitopes on the Env proteins, as appearance of these features often directly precede bNAb development. It also seems likely that cooperation between several bNAb lineages is important since it generally enhances the neutralization breadth in HIV infected individuals. Whether single or multiple bNAb specificities are preferable remains unclear, as both can result in proper cross-neutralizing activity. In addition, important steps have been made with the development of stable, native-like trimers and the engagement of germline bNAbs by specifically designed Env trimers. Putting the knowledge from both natural infection and vaccine immunogen design together will further the development of improved Env immunogens and vaccine strategies toward a fully protective HIV vaccine.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Kathy West studios for providing us with the rhesus macaque photograph used in our figure.
Funding
MJG receives funding from the Aids Fonds Netherlands (grant #2012041). MMH receives funding from the national institutes of health (NIH) as part of the HIV research and design (HIVRAD) grant (#P01AI110657-01A1).
References
- Plotkin SA. Vaccines: Correlates of vaccine-induced immunity. Clin Infect Dis 2008; 47:401-9; PMID:18558875; http://dx.doi.org/10.1086/589862
- Flynn NM, Forthal DN, Harro CD, Judson FN, Mayer KH, Para MF, rgp120 HIV Vaccine Study Group. Placebo-controlled phase 3 trial of a recombinant glycoprotein 120 vaccine to prevent HIV-1 infection. J Infect Dis 2005; 191:654-65; PMID:15688278; http://dx.doi.org/10.1086/428404
- Pitisuttithum P, Gilbert P, Gurwith M, Heyward W, Martin M, van Griensven F, Hu D, Tappero JW, Choopanya K, Bangkok Vaccine Evaluation Group. Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in bangkok, thailand. J Infect Dis 2006; 194:1661-71; PMID:17109337; http://dx.doi.org/10.1086/508748
- Buchbinder SP, Mehrotra DV, Duerr A, Fitzgerald DW, Mogg R, Li D, Gilbert PB, Lama JR, Marmor M, Del Rio C, et al. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the step study): A double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet 2008; 372:1881-93; http://dx.doi.org/10.1016/S0140-6736(08)61591-3
- Gray G, Buchbinder S, Duerr A. Overview of STEP and phambili trial results: Two phase IIb test-of-concept studies investigating the efficacy of MRK adenovirus type 5 gag/pol/nef subtype B HIV vaccine. Curr Opin HIV AIDS 2010; 5:357-61; PMID:20978374; http://dx.doi.org/10.1097/COH.0b013e32833d2d2b
- Gray GE, Allen M, Moodie Z, Churchyard G, Bekker LG, Nchabeleng M, Mlisana K, Metch B, de Bruyn G, Latka MH, et al. Safety and efficacy of the HVTN 503/phambili study of a clade-B-based HIV-1 vaccine in south africa: A double-blind, randomised, placebo-controlled test-of-concept phase 2b study. Lancet Infect Dis 2011; 11:507-15; PMID:21570355; http://dx.doi.org/10.1016/S1473-3099(11)70098-6
- Hammer SM, Sobieszczyk ME, Janes H, Karuna ST, Mulligan MJ, Grove D, Koblin BA, Buchbinder SP, Keefer MC, Tomaras GD, et al. Efficacy trial of a DNA/rAd5 HIV-1 preventive vaccine. N Engl J Med 2013; 369:2083-92; PMID:24099601; http://dx.doi.org/10.1056/NEJMoa1310566
- Pitisuttithum P, Berman PW, Phonrat B, Suntharasamai P, Raktham S, Srisuwanvilai LO, Hirunras K, Kitayaporn D, Kaewkangwal J, Migasena S, et al. Phase I/II study of a candidate vaccine designed against the B and E subtypes of HIV-1. J Acquir Immune Defic Syndr 2004; 37:1160-5; PMID:15319676; http://dx.doi.org/10.1097/01.qai.0000136091.72955.4b
- Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in thailand. N Engl J Med 2009; 361:2209-20; PMID:19843557; http://dx.doi.org/10.1056/NEJMoa0908492
- Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med 2012; 366:1275-86; PMID:22475592; http://dx.doi.org/10.1056/NEJMoa1113425
- Karasavvas N, Billings E, Rao M, Williams C, Zolla-Pazner S, Bailer RT, Koup RA, Madnote S, Arworn D, Shen X, et al. The thai phase III HIV type 1 vaccine trial (RV144) regimen induces antibodies that target conserved regions within the V2 loop of gp120. AIDS Res Hum Retroviruses 2012; 28:1444-57; PMID:23035746; http://dx.doi.org/10.1089/aid.2012.0103
- Tomaras GD, Ferrari G, Shen X, Alam SM, Liao HX, Pollara J, Bonsignori M, Moody MA, Fong Y, Chen X, et al. Vaccine-induced plasma IgA specific for the C1 region of the HIV-1 envelope blocks binding and effector function of IgG. Proc Natl Acad Sci U S A 2013; 110:9019-24; PMID:23661056; http://dx.doi.org/10.1073/pnas.1301456110
- Bonsignori M, Alam SM, Liao HX, Verkoczy L, Tomaras GD, Haynes BF, Moody MA. HIV-1 antibodies from infection and vaccination: Insights for guiding vaccine design. Trends Microbiol 2012; 20:532-9; PMID:22981828; http://dx.doi.org/10.1016/j.tim.2012.08.011
- Taylor BS, Sobieszczyk ME, McCutchan FE, Hammer SM. The challenge of HIV-1 subtype diversity. N Engl J Med 2008; 358:1590-602; PMID:18403767; http://dx.doi.org/10.1056/NEJMra0706737
- Hessell AJ, Rakasz EG, Poignard P, Hangartner L, Landucci G, Forthal DN, Koff WC, Watkins DI, Burton DR. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Pathog 2009; 5:e1000433; PMID:19436712; http://dx.doi.org/10.1371/journal.ppat.1000433
- Burton DR, Hessell AJ, Keele BF, Klasse PJ, Ketas TA, Moldt B, Dunlop DC, Poignard P, Doyle LA, Cavacini L, et al. Limited or no protection by weakly or nonneutralizing antibodies against vaginal SHIV challenge of macaques compared with a strongly neutralizing antibody. Proc Natl Acad Sci U S A 2011; 108:11181-6; http://dx.doi.org/10.1073/pnas.1103012108
- Shingai M, Donau OK, Plishka RJ, Buckler-White A, Mascola JR, Nabel GJ, Nason MC, Montefiori D, Moldt B, Poignard P, et al. Passive transfer of modest titers of potent and broadly neutralizing anti-HIV monoclonal antibodies block SHIV infection in macaques. J Exp Med 2014; 211:2061-74; PMID:25155019; http://dx.doi.org/10.1084/jem.20132494
- Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A 2008; 105:7552-7; PMID:18490657; http://dx.doi.org/10.1073/pnas.0802203105
- Euler Z, van den Kerkhof TL, van Gils MJ, Burger JA, Edo-Matas D, Phung P, Wrin T, Schuitemaker H. Longitudinal analysis of early HIV-1-specific neutralizing activity in an elite neutralizer and in five patients who developed cross-reactive neutralizing activity. J Virol 2012; 86:2045-55; PMID:22156522; http://dx.doi.org/10.1128/JVI.06091-11
- Bunnik EM, Pisas L, van Nuenen AC, Schuitemaker H. Autologous neutralizing humoral immunity and evolution of the viral envelope in the course of subtype B human immunodeficiency virus type 1 infection. J Virol 2008; 82:7932-41; PMID:18524815; http://dx.doi.org/10.1128/JVI.00757-08
- Montefiori DC, Morris L, Ferrari G, Mascola JR. Neutralizing and other antiviral antibodies in HIV-1 infection and vaccination. Curr Opin HIV AIDS 2007; 2:169-76; PMID:19372883; http://dx.doi.org/10.1097/COH.0b013e3280ef691e
- Deeks SG, Schweighardt B, Wrin T, Galovich J, Hoh R, Sinclair E, Hunt P, McCune JM, Martin JN, Petropoulos CJ, et al. Neutralizing antibody responses against autologous and heterologous viruses in acute versus chronic human immunodeficiency virus (HIV) infection: Evidence for a constraint on the ability of HIV to completely evade neutralizing antibody responses. J Virol 2006; 80:6155-64; PMID:16731954; http://dx.doi.org/10.1128/JVI.00093-06
- Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, et al. Antibody neutralization and escape by HIV-1. Nature 2003; 422:307-12; PMID:12646921; http://dx.doi.org/10.1038/nature01470
- Moog C, Fleury HJ, Pellegrin I, Kirn A, Aubertin AM. Autologous and heterologous neutralizing antibody responses following initial seroconversion in human immunodeficiency virus type 1-infected individuals. J Virol 1997; 71:3734-41; PMID:9094648
- Wibmer CK, Bhiman JN, Gray ES, Tumba N, Abdool Karim SS, Williamson C, Morris L, Moore PL. Viral escape from HIV-1 neutralizing antibodies drives increased plasma neutralization breadth through sequential recognition of multiple epitopes and immunotypes. PLoS Pathog 2013; 9:e1003738; PMID:24204277; http://dx.doi.org/10.1371/journal.ppat.1003738
- van Gils MJ, Euler Z, Schweighardt B, Wrin T, Schuitemaker H. Prevalence of cross-reactive HIV-1-neutralizing activity in HIV-1-infected patients with rapid or slow disease progression. AIDS 2009; 23:2405-14; PMID:19770692; http://dx.doi.org/10.1097/QAD.0b013e32833243e7
- Bhiman JN, Anthony C, Doria-Rose NA, Karimanzira O, Schramm CA, Khoza T, Kitchin D, Botha G, Gorman J, Garrett NJ, et al. Viral variants that initiate and drive maturation of V1V2-directed HIV-1 broadly neutralizing antibodies. Nat Med 2015; 21:1332-6; PMID:26457756; http://dx.doi.org/10.1038/nm.3963
- Benjelloun F, Lawrence P, Verrier B, Genin C, Paul S. Role of human immunodeficiency virus type 1 envelope structure in the induction of broadly neutralizing antibodies. J Virol 2012; 86:13152-63; PMID:23015715; http://dx.doi.org/10.1128/JVI.01110-12
- Zolla-Pazner S, Cardozo T. Structure-function relationships of HIV-1 envelope sequence-variable regions refocus vaccine design. Nat Rev Immunol 2010; 10:527-35; PMID:20577269; http://dx.doi.org/10.1038/nri2801
- Blattner C, Lee JH, Sliepen K, Derking R, Falkowska E, de la Pena AT, Cupo A, Julien JP, van Gils M, Lee PS, et al. Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 env trimers. Immunity 2014; 40:669-80; PMID:24768348; http://dx.doi.org/10.1016/j.immuni.2014.04.008
- Falkowska E, Le KM, Ramos A, Doores KJ, Lee JH, Blattner C, Ramirez A, Derking R, van Gils MJ, Liang CH, et al. Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity 2014; 40:657-68; PMID:24768347; http://dx.doi.org/10.1016/j.immuni.2014.04.009
- van Gils MJ, Sanders RW. Broadly neutralizing antibodies against HIV-1: Templates for a vaccine. Virology 2013; 435:46-56; PMID:23217615; http://dx.doi.org/10.1016/j.virol.2012.10.004
- Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 2013; 496:469-76; PMID:23552890; http://dx.doi.org/10.1038/nature12053
- Moore PL, Gray ES, Wibmer CK, Bhiman JN, Nonyane M, Sheward DJ, Hermanus T, Bajimaya S, Tumba NL, Abrahams MR, et al. Evolution of an HIV glycan-dependent broadly neutralizing antibody epitope through immune escape. Nat Med 2012; 18:1688-92; PMID:23086475; http://dx.doi.org/10.1038/nm.2985
- Simek MD, Rida W, Priddy FH, Pung P, Carrow E, Laufer DS, Lehrman JK, Boaz M, Tarragona-Fiol T, Miiro G, et al. Human immunodeficiency virus type 1 elite neutralizers: Individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J Virol 2009; 83:7337-48; PMID:19439467; http://dx.doi.org/10.1128/JVI.00110-09
- Saphire EO, Parren PW, Barbas C, Burton DR, Wilson IA. Crystallization and preliminary structure determination of an intact human immunoglobulin, b12: An antibody that broadly neutralizes primary isolates of HIV-1. Acta Crystallographica Section D: Biological Crystallography 2001; 57:168-71; http://dx.doi.org/10.1107/S0907444900017376
- Klein F, Diskin R, Scheid JF, Gaebler C, Mouquet H, Georgiev IS, Pancera M, Zhou T, Incesu RB, Fu BZ, et al. Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell 2013; 153:126-38; PMID:23540694; http://dx.doi.org/10.1016/j.cell.2013.03.018
- Sather DN, Carbonetti S, Malherbe D, Pissani F, Stuart AB, Hessell AJ, Gray MD, Mikell I, Kalams SA, Haigwood NL, et al. Emergence of broadly neutralizing antibodies and viral co-evolution in two subjects during the early stages of infection with the human immunodeficiency virus type 1. J Virol 2014; 88(22):12968-81; PMID:25122781
- Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, DeKosky BJ, Ernandes MJ, Georgiev IS, Kim HJ, Pancera M, et al. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature 2014; 509:55-62; PMID:24590074; http://dx.doi.org/10.1038/nature13036
- Gao F, Bonsignori M, Liao HX, Kumar A, Xia SM, Lu X, Cai F, Hwang KK, Song H, Zhou T, et al. Cooperation of B cell lineages in induction of HIV-1-broadly neutralizing antibodies. Cell 2014; 158:481-91; PMID:25065977; http://dx.doi.org/10.1016/j.cell.2014.06.022
- Derking R, Ozorowski G, Sliepen K, Yasmeen A, Cupo A, Torres JL, Julien JP, Lee JH, van Montfort T, de Taeye SW, et al. Comprehensive antigenic map of a cleaved soluble HIV-1 envelope trimer. PLoS Pathog 2015; 11:e1004767; PMID:25807248; http://dx.doi.org/10.1371/journal.ppat.1004767
- Verkoczy L, Kelsoe G, Moody MA, Haynes BF. Role of immune mechanisms in induction of HIV-1 broadly neutralizing antibodies. Curr Opin Immunol 2011; 23:383-90; PMID:21524897; http://dx.doi.org/10.1016/j.coi.2011.04.003
- Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 2011; 477:466-70; PMID:21849977; http://dx.doi.org/10.1038/nature10373
- Doria-Rose NA, Bhiman JN, Roark RS, Schramm CA, Gorman J, Chuang GY, Pancera M, Cale EM, Ernandes MJ, Louder MK, et al. New member of the V1V2-directed CAP256-VRC26 lineage that shows increased breadth and exceptional potency. J Virol 2015; 90:76-91; PMID:26468542; http://dx.doi.org/10.1128/JVI.01791-15
- Julien JP, Sok D, Khayat R, Lee JH, Doores KJ, Walker LM, Ramos A, Diwanji DC, Pejchal R, Cupo A, et al. Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans. PLoS Pathog 2013; 9:e1003342; PMID:23658524; http://dx.doi.org/10.1371/journal.ppat.1003342
- Pejchal R, Doores KJ, Walker LM, Khayat R, Huang PS, Wang SK, Stanfield RL, Julien JP, Ramos A, Crispin M, et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science 2011; 334:1097-103; PMID:21998254; http://dx.doi.org/10.1126/science.1213256
- Verkoczy L, Diaz M. Autoreactivity in HIV-1 broadly neutralizing antibodies: Implications for their function and induction by vaccination. Curr Opin HIV AIDS 2014; 9:224-34; PMID:24714565; http://dx.doi.org/10.1097/COH.0000000000000049
- Ofek G, Tang M, Sambor A, Katinger H, Mascola JR, Wyatt R, Kwong PD. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J Virol 2004; 78:10724-37; PMID:15367639; http://dx.doi.org/10.1128/JVI.78.19.10724-10737.2004
- Cardoso RM, Zwick MB, Stanfield RL, Kunert R, Binley JM, Katinger H, Burton DR, Wilson IA. Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity 2005; 22:163-73; PMID:15723805; http://dx.doi.org/10.1016/j.immuni.2004.12.011
- Haynes BF, Moody MA, Verkoczy L, Kelsoe G, Alam SM. Antibody polyspecificity and neutralization of HIV-1: A hypothesis. Hum Antibodies 2005; 14:59-67; PMID:16720975
- Liu M, Yang G, Wiehe K, Nicely NI, Vandergrift NA, Rountree W, Bonsignori M, Alam SM, Gao J, Haynes BF, et al. Polyreactivity and autoreactivity among HIV-1 antibodies. J Virol 2015; 89:784-98; PMID:25355869; http://dx.doi.org/10.1128/JVI.02378-14
- Mouquet H, Nussenzweig MC. Polyreactive antibodies in adaptive immune responses to viruses. Cell Mol Life Sci 2012; 69:1435-45; PMID:22045557; http://dx.doi.org/10.1007/s00018-011-0872-6
- Moir S, Fauci AS. B cells in HIV infection and disease. Nat Rev Immunol 2009; 9:235-45; PMID:19319142; http://dx.doi.org/10.1038/nri2524
- Doria-Rose NA, Mascola JR. HIV immunology goes out on a limb. Immunity 2016; 44:1088-90; PMID:27192575; http://dx.doi.org/10.1016/j.immuni.2016.05.004
- Bonsignori M, Hwang KK, Chen X, Tsao CY, Morris L, Gray E, Marshall DJ, Crump JA, Kapiga SH, Sam NE, et al. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J Virol 2011; 85:9998-10009; PMID:21795340; http://dx.doi.org/10.1128/JVI.05045-11
- Bonsignori M, Zhou T, Sheng Z, Chen L, Gao F, Joyce MG, Ozorowski G, Chuang GY, Schramm CA, Wiehe K, et al. Maturation pathway from germline to broad HIV-1 neutralizer of a CD4-mimic antibody. Cell 2016; 165:449-63; PMID:26949186; http://dx.doi.org/10.1016/j.cell.2016.02.022
- Sok D, Laserson U, Laserson J, Liu Y, Vigneault F, Julien JP, Briney B, Ramos A, Saye KF, Le K, et al. The effects of somatic hypermutation on neutralization and binding in the PGT121 family of broadly neutralizing HIV antibodies. PLoS Pathog 2013; 9:e1003754; PMID:24278016
- MacLeod DT, Choi NM, Briney B, Garces F, Ver LS, Landais E, Murrell B, Wrin T, Kilembe W, Liang CH, et al. Early antibody lineage diversification and independent limb maturation lead to broad HIV-1 neutralization targeting the env high-mannose patch. Immunity 2016; 44:1215-26; PMID:27192579; http://dx.doi.org/10.1016/j.immuni.2016.04.016
- Ward AB, Wilson IA. Insights into the trimeric HIV-1 envelope glycoprotein structure. Trends Biochem Sci 2015; 40:101-7; PMID:25600289; http://dx.doi.org/10.1016/j.tibs.2014.12.006
- Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, et al. Structure and immune recognition of trimeric pre-fusion HIV-1 env. Nature 2014; 514:455-61; PMID:25296255; http://dx.doi.org/10.1038/nature13808
- Wang W, Nie J, Prochnow C, Truong C, Jia Z, Wang S, Chen XS, Wang Y. A systematic study of the N-glycosylation sites of HIV-1 envelope protein on infectivity and antibody-mediated neutralization. Retrovirology 2013; 10:14; PMID:23384254; http://dx.doi.org/10.1186/1742-4690-10-14
- Mascola JR, Montefiori DC. The role of antibodies in HIV vaccines. Annu Rev Immunol 2010; 28:413-44; PMID:20192810; http://dx.doi.org/10.1146/annurev-immunol-030409-101256
- Gilbert P, Wang M, Wrin T, Petropoulos C, Gurwith M, Sinangil F, D'Souza P, Rodriguez-Chavez IR, DeCamp A, Giganti M, et al. Magnitude and breadth of a nonprotective neutralizing antibody response in an efficacy trial of a candidate HIV-1 gp120 vaccine. J Infect Dis 2010; 202:595-605; PMID:20608874; http://dx.doi.org/10.1086/654816
- Kovacs JM, Nkolola JP, Peng H, Cheung A, Perry J, Miller CA, Seaman MS, Barouch DH, Chen B. HIV-1 envelope trimer elicits more potent neutralizing antibody responses than monomeric gp120. Proc Natl Acad Sci U S A 2012; 109:12111-6; PMID:22773820; http://dx.doi.org/10.1073/pnas.1204533109
- Earl PL, Sugiura W, Montefiori DC, Broder CC, Lee SA, Wild C, Lifson J, Moss B. Immunogenicity and protective efficacy of oligomeric human immunodeficiency virus type 1 gp140. J Virol 2001; 75:645-53; PMID:11134278; http://dx.doi.org/10.1128/JVI.75.2.645-653.2001
- Stamatatos L, Lim M, Cheng-Mayer C. Generation and structural analysis of soluble oligomeric gp140 envelope proteins derived from neutralization-resistant and neutralization-susceptible primary HIV type 1 isolates. AIDS Res Hum Retroviruses 2000; 16:981-94; PMID:10890360; http://dx.doi.org/10.1089/08892220050058407
- Schulke N, Vesanen MS, Sanders RW, Zhu P, Lu M, Anselma DJ, Villa AR, Parren PW, Binley JM, Roux KH, et al. Oligomeric and conformational properties of a proteolytically mature, disulfide-stabilized human immunodeficiency virus type 1 gp140 envelope glycoprotein. J Virol 2002; 76:7760-76; PMID:12097589; http://dx.doi.org/10.1128/JVI.76.15.7760-7776.2002
- Yang X, Farzan M, Wyatt R, Sodroski J. Characterization of stable, soluble trimers containing complete ectodomains of human immunodeficiency virus type 1 envelope glycoproteins. J Virol 2000; 74:5716-25; PMID:10823881; http://dx.doi.org/10.1128/JVI.74.12.5716-5725.2000
- Yang X, Lee J, Mahony EM, Kwong PD, Wyatt R, Sodroski J. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J Virol 2002; 76:4634-42; PMID:11932429; http://dx.doi.org/10.1128/JVI.76.9.4634-4642.2002
- Spearman P, Lally MA, Elizaga M, Montefiori D, Tomaras GD, McElrath MJ, Hural J, De Rosa SC, Sato A, Huang Y, et al. A trimeric, V2-deleted HIV-1 envelope glycoprotein vaccine elicits potent neutralizing antibodies but limited breadth of neutralization in human volunteers. J Infect Dis 2011; 203:1165-73; PMID:21451004; http://dx.doi.org/10.1093/infdis/jiq175
- Sanders RW, Vesanen M, Schuelke N, Master A, Schiffner L, Kalyanaraman R, Paluch M, Berkhout B, Maddon PJ, Olson WC, et al. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol 2002; 76:8875-89; PMID:12163607; http://dx.doi.org/10.1128/JVI.76.17.8875-8889.2002
- Sanders RW, Derking R, Cupo A, Julien J, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Peña A T, Korzun J. A next-generation cleaved, soluble HIV-1 env trimer, BG505 SOSIP. 664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS pathogens 2013; 9:e1003618; PMID:24068931; http://dx.doi.org/10.1371/journal.ppat.1003618
- Sharma SK, de Val N, Bale S, Guenaga J, Tran K, Feng Y, Dubrovskaya V, Ward AB, Wyatt RT. Cleavage-independent HIV-1 env trimers engineered as soluble native spike mimetics for vaccine design. Cell Rep 2015; 11:539-50; PMID:25892233; http://dx.doi.org/10.1016/j.celrep.2015.03.047
- Barouch DH, Alter G, Broge T, Linde C, Ackerman ME, Brown EP, Borducchi EN, Smith KM, Nkolola JP, Liu J, et al. Protective efficacy of adenovirus/protein vaccines against SIV challenges in rhesus monkeys. Science 2015; 349:320-4; PMID:26138104; http://dx.doi.org/10.1126/science.aab3886
- Hulot SL, Korber B, Giorgi EE, Vandergrift N, Saunders KO, Balachandran H, Mach LV, Lifton MA, Pantaleo G, Tartaglia J, et al. Comparison of immunogenicity in rhesus macaques of transmitted-founder, HIV-1 group M consensus, and trivalent mosaic envelope vaccines formulated as a DNA prime, NYVAC, and envelope protein boost. J Virol 2015; 89:6462-80; PMID:25855741; http://dx.doi.org/10.1128/JVI.00383-15
- Sliepen K, Sanders RW. HIV-1 envelope glycoprotein immunogens to induce broadly neutralizing antibodies. Expert Rev Vaccines 2016; 15:349-65; PMID:26654478
- Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, Ozorowski G, Cupo A, Simonich C, Goo L, et al. HIV-1 VACCINES. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science 2015; 349:aac4223; PMID:26089353; http://dx.doi.org/10.1126/science.aac4223
- Dosenovic P, von Boehmer L, Escolano A, Jardine J, Freund NT, Gitlin AD, McGuire AT, Kulp DW, Oliveira T, Scharf L, et al. Immunization for HIV-1 broadly neutralizing antibodies in human ig knockin mice. Cell 2015; 161:1505-15; PMID:26091035; http://dx.doi.org/10.1016/j.cell.2015.06.003
- Sliepen K, Ozorowski G, Burger JA, van Montfort T, Stunnenberg M, LaBranche C, Montefiori DC, Moore JP, Ward AB, Sanders RW. Presenting native-like HIV-1 envelope trimers on ferritin nanoparticles improves their immunogenicity. Retrovirology 2015; 12:82; PMID:26410741; http://dx.doi.org/10.1186/s12977-015-0210-4
- Ingale J, Stano A, Guenaga J, Sharma SK, Nemazee D, Zwick MB, Wyatt RT. High-density array of well-ordered HIV-1 spikes on synthetic liposomal nanoparticles efficiently activate B cells. Cell Rep 2016; 15:1986-99; PMID:27210756; http://dx.doi.org/10.1016/j.celrep.2016.04.078
- Jardine J, Julien JP, Menis S, Ota T, Kalyuzhniy O, McGuire A, Sok D, Huang PS, MacPherson S, Jones M, et al. Rational HIV immunogen design to target specific germline B cell receptors. Science 2013; 340:711-6; PMID:23539181; http://dx.doi.org/10.1126/science.1234150
- Jardine JG, Ota T, Sok D, Pauthner M, Kulp DW, Kalyuzhniy O, Skog PD, Thinnes TC, Bhullar D, Briney B, et al. HIV-1 VACCINES. priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science 2015; 349:156-61; PMID:26089355; http://dx.doi.org/10.1126/science.aac5894
- Liao HX, Tsao CY, Alam SM, Muldoon M, Vandergrift N, Ma BJ, Lu X, Sutherland LL, Scearce RM, Bowman C, et al. Antigenicity and immunogenicity of transmitted/founder, consensus, and chronic envelope glycoproteins of human immunodeficiency virus type 1. J Virol 2013; 87:4185-201; PMID:23365441; http://dx.doi.org/10.1128/JVI.02297-12
- Bricault CA, Kovacs JM, Nkolola JP, Yusim K, Giorgi EE, Shields JL, Perry J, Lavine CL, Cheung A, Ellingson-Strouss K, et al. A multivalent clade C HIV-1 env trimer cocktail elicits a higher magnitude of neutralizing antibodies than any individual component. J Virol 2015; 89:2507-19; PMID:25540368; http://dx.doi.org/10.1128/JVI.03331-14
- Malherbe DC, Doria-Rose NA, Misher L, Beckett T, Puryear WB, Schuman JT, Kraft Z, O'Malley J, Mori M, Srivastava I, et al. Sequential immunization with a subtype B HIV-1 envelope quasispecies partially mimics the in vivo development of neutralizing antibodies. J Virol 2011; 85:5262-74; PMID:21430056; http://dx.doi.org/10.1128/JVI.02419-10
- Haynes BF, Kelsoe G, Harrison SC, Kepler TB. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nat Biotechnol 2012; 30:423-33; PMID:22565972; http://dx.doi.org/10.1038/nbt.2197
- Crotty S. Follicular helper CD4 T cells (TFH). Annu Rev Immunol 2011; 29:621-63; PMID:21314428; http://dx.doi.org/10.1146/annurev-immunol-031210-101400
- Lindqvist M, van Lunzen J, Soghoian DZ, Kuhl BD, Ranasinghe S, Kranias G, Flanders MD, Cutler S, Yudanin N, Muller MI, et al. Expansion of HIV-specific T follicular helper cells in chronic HIV infection. J Clin Invest 2012; 122:3271-80; PMID:22922259; http://dx.doi.org/10.1172/JCI64314
- Yamamoto T, Lynch RM, Gautam R, Matus-Nicodemos R, Schmidt SD, Boswell KL, Darko S, Wong P, Sheng Z, Petrovas C, et al. Quality and quantity of TFH cells are critical for broad antibody development in SHIVAD8 infection. Sci Transl Med 2015; 7:298ra120; PMID:26223303; http://dx.doi.org/10.1126/scitranslmed.aab3964
- Cubas RA, Mudd JC, Savoye AL, Perreau M, van Grevenynghe J, Metcalf T, Connick E, Meditz A, Freeman GJ, Abesada-Terk G, Jr, et al. Inadequate T follicular cell help impairs B cell immunity during HIV infection. Nat Med 2013; 19:494-9; PMID:23475201; http://dx.doi.org/10.1038/nm.3109
- Cubas R, Perreau M. The dysfunction of T follicular helper cells. Curr Opin HIV AIDS 2014; 9:485-91; PMID:25023620; http://dx.doi.org/; http://dx.doi.org/10.1097/COH.0000000000000095
- de Bree GJ, Lynch RM. B cells in HIV pathogenesis. Curr Opin Infect Dis 2016; 29:23-30; PMID:26658653; http://dx.doi.org/10.1097/QCO.0000000000000225
- Locci M, Havenar-Daughton C, Landais E, Wu J, Kroenke MA, Arlehamn CL, Su LF, Cubas R, Davis MM, Sette A, et al. Human circulating PD-1+CXCR3-CXCR5+ memory tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses. Immunity 2013; 39:758-69; PMID:24035365; http://dx.doi.org/10.1016/j.immuni.2013.08.031
