ABSTRACT
Thirty-three infants aged ∼2 months had serial stool samples collected after receipt of Rotarix® vaccine dose 1, and were assessed for shedding of porcine circovirus type 1 DNA and Rotavirus group A RNA by molecular methods. We did not find strong evidence that porcine circovirus type 1 replication occurred. Porcine circovirus type 1 genome with the same sequence as that in Rotarix® was detected in a few infants as late as day ≥ 13; while this timing could suggest there may have been replication and not just transient passage through the gastrointestinal tract, the lack of increase in copy number in any infant supports transient passage and there are inherent limitations to the results. We found that 21% of infants did not shed Rotarix® RVA RNA beyond the day 3 sample, which may suggest lack of vaccine virus replication. Of the infants in whom Rotarix RVA RNA shedding continued, peak copy numbers were reached on days 3–5 for ∼40%, and after day 5 in ∼60%, and shedding can be prolonged (≥ 45 days).
Introduction
In 2010, Victoria et al. reported the identification of the full genome of porcine circovirus type 1 (PCV-1) in Rotarix® vaccine.Citation1 Rotarix® (GlaxoSmithKline Biologicals, GSK), a rotavirus vaccine that contains a live attenuated human rotavirus strain, was added to the U.S. national immunization program in April 2008 as one of the 2 available rotavirus vaccines recommended to be given to all infants.Citation2 Rotarix® is a 2-dose series, with doses recommended for infants at ages 2 and 4 months. The initial PCV-1 report and subsequent investigations led the Food and Drug Administration (FDA) to recommend a temporary suspension of Rotarix® in the United States on March 22, 2010, while further assessing the issue. On May 14, 2010, the FDA lifted the suspension, concluding that, while live PCV-1 was detected in the vaccine, there was no evidence that PCV-1 (nor porcine circovirus type 2 [PCV-2]; short DNA segments of both PCV-1 and PCV-2 had been detected in the rotavirus vaccine, RotaTeq® [Merck and Co., Inc.]) poses a safety risk in humans; neither porcine circovirus was known to cause infection or illness in humans.Citation3-5
As part of the manufacturer's evaluation of PCV-1 in Rotarix® vaccine, testing for PCV-1 was performed in stool samples from 10 vaccinated infants following dose 1 in each of 4 earlier clinical trials; the samples were limited to those that had been collected as part of the trial.Citation6 GSK reported that, among these samples, PCV-1 with sequence identical to that in Rotarix® was detected in 40% (2/5) of the samples collected from infants on post-vaccination day 3, 5% (2/38) on day 7 (with 2 additional samples of the 40 tested having inconclusive results), and 0% on days 10, 15, 22, 30, and 45 (number of infants with samples tested from these days were 5, 40, 10, 5 and 5, respectively).Citation6 GSK concluded that these data were consistent with transient passage of DNA without replication.
In order to more fully examine the shedding of PCV-1 DNA following the receipt of Rotarix® (Rotarix® PCV-1 DNA), we collected and tested stool samples from 33 infants after receiving their first dose at about age 2 months. We also measured the shedding of Rotarix® Rotavirus group A RNA (Rotarix® RVA RNA) to better understand the replication of the rotavirus vaccine virus in vivo.
Results
Thirty-three infants were enrolled after parents gave consent, and provided ≥ 1 stool sample following Rotarix® vaccination. The median age at receipt was 9.8 weeks; interquartile range [IQR] 9.1–10.5; range 8.0–14.7 weeks). All but one infant (32/33, 97%) were non-Hispanic/non-Latino; 28 (85%) were Black, 1 (3%) was White, 3 (9%) were reported to be bi-racial, and 1 (3%) infant's race was unknown. At the time of vaccine receipt, 18 (55%) were reported to be receiving only formula as their milk source, 6 (18%) were receiving only breastmilk, and 9 (27%) were receiving both. Only 1 infant (breastfed) was receiving solid food. Twenty-three were born after ≥ 37 week gestation and 10 had been born before 37 weeks gestation (range 28–36 weeks). Eighteen full-term and 10 premature infants submitted all (or missed only 1) of the 10 samples desired during the first 15 d post-vaccination ().
Table 1. Copy numbers per mL of stool of Rotarix® RVA RNA and Rotarix® PCV-1 DNA, by infant and sample post-vaccination collection day. In each cell, the first number indicates Rotarix® RVA RNA copy number/mL of stool (highlighted in yellow if Rotarix® RVA RNA was detected). The second number in each cell indicates Rotarix® PCV-1 DNA copy number/mL of stool (highlighted in blue if Rotarix® PCV-1 DNA was detected). Cells highlighted both yellow and blue indicate that Rotarix® RVA RNA and Rotarix® PCV-1 DNA were detected. Negative sign (−) indicates that sample was collected and tested and was negative. Empty cell indicates that the sample was not collected. Among samples indicated as collected during days 5–15, 5 infants had 1 or 2 specimens that were collected 1 day later than the specific day indicated on the table. Samples indicated as day 30 were collected on post-vaccination days 28–33 and those indicated as day 45 were collected on post-vaccination days 43–47
Of the 271 samples extracted with MagNA Pure, 17 were positive only for Rotarix® PCV-1 DNA, 79 were positive only for Rotarix® RVA RNA, and 18 samples were positive for both viruses. Of the 271 samples extracted with Master Pure™, 6 were only positive for Rotarix® PCV-1 DNA, 121 were positive only for Rotarix® RVA RNA, and 29 samples were positive for both viruses. Because each extraction method had limitations (with MagNA Pure platform–the possibility of competition between viruses [Rotarix® RVA RNA and Rotarix® PCV-1 DNA] for the limited surface binding area on the magnetic beads during the extraction; with MasterPure–the prolonged storage of stool dilutions may have resulted in degradation of viral nucleic acid), we combined the results from both extractions and reported the result as positive if either method yielded a positive result.
Rotarix® PCV-1 DNA detection
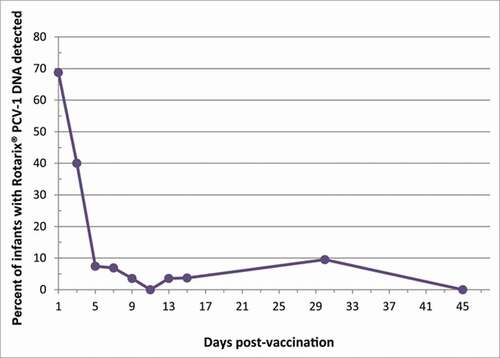
Out of a total of 271 samples tested, 43 (16%) were found to contain Rotarix® PCV-1 DNA. Overall, 76% (25/33) subjects that submitted ≥ 1 stool sample had Rotarix® PCV-1 DNA detected, and 24% (8/33) never had Rotarix® PCV-1 DNA detected. The proportion of samples with Rotarix® PCV-1 DNA detected decreased from 69% on ∼post-vaccination day 1, to 40% on day 3 (, ). Rotarix® PCV-1 DNA was then detected in 17% (5/29) of infants who submitted samples on day 5 or later: in one subject each, detected on day 5 and 7 (and negative thereafter; infant #23, breastfed), day 8, 10 and 13 (with 1 intervening negative sample; infant #19, breastfed), day 5 and 15 (with 4 intervening negative samples; infant #6, formula-fed), day 31 (earlier positive on day 1 only; infant #9, breastfed), day 36 (earlier positives on day 1 and 3 only, infant #15, formula-fed). Three of 10 (30%) premature infants had Rotarix® PCV-1 DNA detection on day ≥ 5, as did 2/19 (11%) full-term infants (2-sided Fisher's exact p = 0.31).
Table 2. Number of samples tested and proportion with Rotarix® PCV-1 DNA detected, and with Rotarix® RVA RNA detected
Rotarix® PCV-1 DNA copy number
For Rotarix® stock, Rotarix® PCV-1 DNA was detected at 1.1 × 107 copies/vial (1 mL) of Rotarix® vaccine. In stool samples, Rotarix® PCV-1 DNA was detected in the range of 8.1 × 101 to 1.0 × 105 copies per mL of stool.
Of those with Rotarix® PCV-1 DNA detected on day 1, copy number on that day was in the range described above, with median of 4.7×103 copies per mL of stool (). In all infants, the copy number detected in a later Rotarix® PCV-1 DNA-positive sample was lower than or at approximately the same level as the previous copy number detected: the maximum increase in copy number was in infant #9, with a 3-fold increase in copy number from samples collected on day 1 (5.9×103 copies per mL) and day 31 (1.8×104 copies per mL).
Rotarix® RVA RNA detection
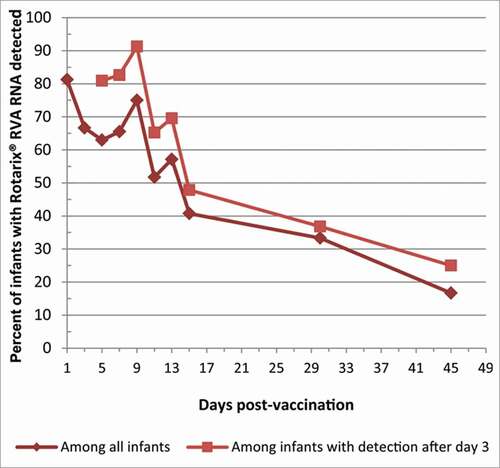
Overall, 94% (31/33) subjects that submitted at least one stool sample had Rotarix® RVA RNA ever detected and 2 subjects (who each only submitted a sample on day 1) never had Rotarix® RVA RNA detected (). The overall Rotarix® RVA RNA detection rate was 81%, 67%, 63%, 63%, and 75% on day 1, 3, 5, 7, and 9 respectively, and then decreased over time to 17% detection on day 45 (, ). Of the 29 infants who submitted at least the 4 samples in the first week, and including pt#10 who submitted off schedule, 10% (3/29) did not have Rotarix® RVA RNA detected beyond the day 1 sample, suggesting lack of replication, and 3 more infants (for total 6/29, 21%) did not have Rotarix® RVA RNA detected beyond the day 3 sample. Examining the detection rate among only those who did have detection beyond day 3, the Rotarix® RVA RNA detection rate was about 10–15 percentage points higher at each time point (e.g, 81%, 91%, and 25% on days 5, 9 and 45, respectively) than with all infants combined ().
Rotarix® RVA RNA copy number
For Rotarix® stock, Rotarix® RVA RNA was detected at 2.2 × 107 copies/vial (1 mL) of Rotarix® vaccine. In stool samples, Rotarix® RVA RNA was detected in the range of 1.2 × 102 to 1.3 × 1010 copies per mL of stool.
For the 3 infants described above who had Rotarix® RVA RNA detected only in samples from days 1 and 3, the copy numbers were similar (within one log) in both of their samples. To inform possible timing of peak replication, copy numbers were examined over time in the 21 infants who submitted all 8 requested samples during the first 15 d and had Rotarix® RVA RNA detected beyond day 3. In 43% (9/21) of these subjects, the highest Rotarix® RVA RNA copy number had been reached on days 3 or 5 (i.e., copy numbers in later samples were lower or no more than 1 log higher). In 57% (12/21), a higher copy number (by ≥ 100-fold, or reaching a copy number of ≥ 102 if previously undetectable) was detected in at least 1 sample beyond day 5. Of the 12 in the latter group, 5 had no Rotarix® RVA RNA detected on day 3, 2 had none detected on day 5, and 2 had none detected on days 3 and 5. Using the highest copy number detected for each of the 21 infants on day ≥ 3, the median value was 5.2×106 copies/mL (IQR 1.4×105 to 1.0×108; range 377 to 1.3×1010)
Among the 7 infants with Rotarix® RVA RNA detected on day∼30, the median copy number was 1.2×105/mL. On day ∼45, the 3 subjects with Rotarix® RVA RNA detected had 2.4×103, 6.4×103, and 1.5×105 copies/mL.
Discussion
We believe this is the first study in the scientific literature evaluating the shedding of Rotarix® PCV-1 DNA following Rotarix® vaccination in infants. We used 2 extraction methods to minimize the impact of extraction and prolonged storage on viral detection. We were able to detect both Rotarix® PCV-1 RNA and Rotarix® RVA RNA in the same samples, and assessed shedding in samples collected up to 45 d post-vaccination. In our sample of infants who received dose 1 of Rotarix®, we did not find strong evidence that Rotarix® PCV-1 DNA replication occurred. We found that 21% of infants did not shed Rotarix® RVA RNA beyond the day 3 sample, which may indicate lack of vaccine virus replication. Of the infants in whom shedding continued, peak copy numbers frequently were reached after day 5, and there was evidence of prolonged shedding (≥ 45 days) in some infants.
In their limited number of samples tested, GSK concluded that their results, including Rotarix® PCV-1 DNA detection on day 7, were consistent with transient passage of DNA without replication.Citation6 It is not known with certainty how long Rotarix® PCV-1 DNA can be detected in the stool of vaccinated infants and be consistent with only transient passage. RotaTeq® vaccine was found to contain only short segments of PCV-2 DNA (RotaTeq® PCV-2 DNA) and was not shown to be infectious in vitroCitation4; in a study examining shedding in young US infants following receipt of RotaTeq® vaccine, RotaTeq® PCV-2 DNA was detected in some infants at the latest sample collected which was on day 9 post-vaccination.Citation7 In our study, Rotarix® PCV-1 DNA was detected in one infant each as late as day 7, 13, 15, 31 and 36. In the last 3 infants, several earlier samples did not have Rotarix® PCV-1 DNA detected. It is not clear if this indicates that shedding can be very intermittent and prolonged even if there is only passage and not replication, that Rotarix® PCV-1 DNA is unevenly distributed throughout a sample so that any one aliquot tested may not be representative, that there may have been a labeling or laboratory testing error (although multiple procedures were in place to prevent these), or that infants may have been re-exposed to Rotarix® PCV-1 DNA. To the last possibility, records indicated that none of these infants had received a second dose of Rotarix® until ≥ 3 weeks after the samples had all been collected; in the sample with Rotarix® PCV-1 DNA detected from day 36 (infant #15), Rotarix® RVA RNA was not detected, but in the sample with Rotarix® PCV-1 DNA detected from day 31 (infant #9), Rotarix® RVA RNA was detected in high copy number. Our overall finding that, among infants with Rotarix® PCV-1 DNA detected, copy number in later samples were about the same or lower than in previous samples where Rotarix® PCV-1 DNA had been detected is more supportive of passage rather than replication.
Following vaccination with live-attenuated rotavirus vaccines like Rotarix®, vaccine RVA virus is expected to replicate in the upper gastrointestinal tract and be shed into fecal matter. In our population of almost exclusively non-Hispanic infants, and using techniques expected to detect even small quantities of viral RNA, we did not detect Rotarix® RVA RNA beyond the day 1 sample in 10% of infants, and not beyond the day 3 sample in 21%, cumulatively. This suggests that in at least 10% (and perhaps even 21%) of our infants, vaccine virus may not have replicated. Interestingly, this is the approximate proportion (25–26%) of non-Hispanic US infants found in a control group to be FUT2 (α [1,2] fucosyltransferase 2) nonsecretors.Citation8,9 FUT2 nonsecretors may be protected against infection with P[8] rotavirus such as the vaccine virus contained in Rotarix®.Citation8,10-12 However, we did not perform serologic assessments to more fully determine which infants likely had “vaccine-take,” and, as described earlier, the duration of shedding from just passive transit of vaccine virus is not known. In studies evaluating a rotavirus vaccine consisting of a human neonatal strain, in which stool samples from infants aged ∼2 months were collected daily for the first week after vaccination, investigators considered detection of vaccine virus by RT-PCR in any stool sample collected on post-vaccination day 3 or later as indicative of vaccine virus replication.Citation13,14
Although several previous studies describing the timing of shedding of Rotarix® RVA RNA in vaccinees used enzyme immunoassay (EIA) to detect rotavirus antigen in stool samples (and the EIA-positive samples may have been subsequently tested by RT-PCR to confirm presence of vaccine strain), we used quantitative real-time RT-PCR (qRT-PCR) on freshly extracted samples to detect Rotarix® RVA RNA.Citation15,16 Recently, we compared RVA detection rates by EIA and qRT-PCR among children with and without acute gastroenteritis.Citation17 The rate of RVA detection was 10% and 5% higher by qRT-PCR when compared with EIA for children with and without acute gastroenteritis, respectively. Only Hsieh et.al. previously reported on Rotarix® RVA RNA shedding after vaccination of infants (35 Taiwanese infants, similar in age to our infants), over time, using qRT-PCR; these investigators also assessed the shedding by EIA.Citation18 Our protocol was very similar to that of Hsieh and colleagues. We believe our choice of positive control, dsRNA transcript vs. Hsieh's plasmid dsDNA, allowed us to more precisely calculate copy numbers. Additionally, the sensitivity and specificity of the assay we used for RVA RNA detection should be higher due to our alterations of the probe and the enzyme used during qRT-PCR. Hsieh et al. detected Rotarix® RVA RNA by qRT-PCR in as high as 90% and 88% of stool samples collected on days 4–5, and days 6–7, respectively, which are higher than our results among all infants but more similar to our results among only those with shedding beyond day 3 (81% and 83% on day 5 and day 7, respectively). Their detection rate at last sample (40% [2/5] on days 22–28) is similar to our rates of 33–37% from our larger sample size measured on day ∼30. Hsieh et al demonstrated the much higher detection rate (∼4-fold higher at each time point) when samples were examined by qRT-PCR vs. EIA, and also provided valuable information on shedding following dose 2 (lower detection rate than after dose 1).Citation18
Four limitations of this study should be noted. It is possible that we did not detect or detected at lower copy number, Rotarix® PCV-1 DNA and/or Rotarix® RVA RNA, given that each of our testing approaches had limitations; we combined results to try and minimize the impact of each limitation. Second, we used only one SNP difference to identify Rotarix® PCV-1 DNA, though this SNP appears to be specific for the Rotarix® PCV-1 strain when compared with wild-type strain sequences deposited in GeneBank. Third, identification of the Rotarix® PCV-1 DNA, in the stool of our infants does not absolutely prove that the Rotarix® vaccine was the only possible source of the PCV-1 DNA in each of the samples. Fourth, although we quantified PCV-1 DNA by molecular methods, we did not attempt to ascertain if the samples contained live PCV-1 virus.
In conclusion, using molecular methods, in our sample of young infants who received dose 1 of Rotarix®, we did not find strong evidence that PCV-1 replication occurred. We found that 21% of infants did not shed Rotarix® RVA RNA beyond the day 3 sample. Of the infants in whom Rotarix RVA RNA shedding continued, peak copy numbers frequently were reached after day 5, and shedding can be prolonged (≥ 45 days).
Participants, materials and methods
Participants
Our goal was to enroll 20 healthy full-term infants and 10 preterm (gestational age < 37 weeks) infants who provided almost all the samples during the first 15 d post-vaccination with dose 1 of Rotarix®. Study participants were recruited from the Pediatric Ambulatory Clinic at Hughes Spalding Children's Hospital in Atlanta, Georgia. Parents of infants were approached for enrollment on the day their infant was to receive the first dose of Rotarix®, which was administered as part of routine care by their primary healthcare provider (typically given concomitantly with pentavalent vaccine [DTaP-IPV-Hib] and pneumococcal conjugate vaccine). Parents who agreed to have their infant participate provided written informed consent. Parents were asked to collect, label, and freeze (if delivery of the sample to the hospital was not possible within 2 hours of collection) a stool sample on days 1, 3, 5, 7, 9, 11, 13, 15, ∼30 and ∼45 following receipt of the first Rotarix® dose. Reminders for stool collection were made by telephone calls or text messaging to the caregivers. Most samples were brought by the parent to the hospital or were picked up from the home by a courier service on the same day the sample was collected from the infant; parents sometimes kept ≥ 1 sample frozen at home until delivery to the hospital.
This study was approved by the governing institutional review boards of the hospital, the academic medical center, and the Centers for Disease Control and Prevention.
Sample collection
Between August 28, 2012 and December 28, 2013, 271 stool samples were collected from 33 infants.
Sample storage
Diapers were kept frozen during transport and until laboratory tests were performed. Diapers were identified only by a unique sample number, and laboratory staff were blinded to date of specimen collection, sample order, and specific infant. Diapers were thawed by placing them at 4°C overnight (approximately 12 hours) prior to extraction of total nucleic acid (TNA).
Standard controls
Rotavirus NSP3 dsRNA transcript was used to generate the standard curve for calculating the copy number of all rotavirus positive samples.Citation15 Commercially available Rotarix® vaccine (Lot #:A41FB315A, GlaxoSmithKline) was used as the positive control for qRT-PCR assays targeting the Rotarix® strain. Rotarix® vaccine used in the qRT-PCR experiments was a lyophilized pellet and was reconstituted following manufacturer's guidelines. PCV-1 single-stranded DNA oligonucleotide positive control standard was used to generate the standard curve used for calculating the copy numbers of PCV-1.Citation7
Testing from MagNA Pure TNA extracts
Fecal material from each diaper was eluted into 1000 μL of phosphate buffered saline (PBS). Total nucleic acid (TNA) from each of the 271 specimens was extracted using the MagNA Pure Compact instrument and the Total Nucleic Acid Isolation Kit I (catalog #:03731146001 and 03730964001, Roche Applied Science). To avoid any possible cross-contamination of samples with Rotarix® vaccine stock, the vaccine was extracted in separate run. All extractions were performed according to the manufacturer's instructions. TNA extracts were analyzed immediately by real-time PCR (qPCR) for PCV-1 DNACitation7 and for Rotavirus group A by a duplex qRT-PCR targeting the NSP3 gene of RVA and using Xeno® RNA as an internal control.Citation15 Remaining TNA was stored at −80°C until further testing. Rotavirus positive samples were further tested by qRT-PCR targeting the NSP2 and VP4 genes of the Rotarix® vaccine strain, as previously described.Citation16 For Rotarix® RVA positive samples, the NSP2 gene was sequenced using published primers.Citation19 The qPCR assay, targeting PCV-1 strains found either in Rotarix® vaccine or as a wild-type, was run under previously described conditions.Citation7 All PCV-1 qPCR positive samples were sequenced using a previously published PCR assay.Citation7 During data analysis, we speculated that there might be competition between the viruses, RVA and PCV-1, for the limited surface area of the magnetic bead used during the MagNA Pure extraction, as this was observed during a similar study for RotaTeq® vaccinees.Citation7 The decision to re-extract and re-analyze the samples was further supported by comparing the results of 50 randomly selected samples from the current study, for which we found an increase of 19% and 2% for Rotarix® RVA RNA and Rotarix® PCV-1 DNA detection, respectively, for samples extracted using the Master Pure kit (data not shown). We choose to re-extract the samples with the Master Pure™ kit because this approach is based on total nucleic acid precipitation which is not limited by the binding surface of the bead-based method.
Testing from MasterPure™ Complete RNA and DNA Purification extracts
Stool aliquots previously prepared for MagnaPure TNA extraction (which were kept in 4°C) were used for extraction with MasterPure™. Total nucleic acid was manually extracted using MasterPure™ Complete RNA and DNA Purification (catalog #:MC85200, Epicentre® (An Illumina Company)). All extractions were performed according to the manufacturer's instructions. TNA extracts were analyzed immediately by all real-time PCR assays described above. Samples that were previously sequenced for all targets using MagnaPure TNA extracts were not re-sequenced. All other Rotarix® RVA positive samples were sequence confirmed by obtaining NSP2 and VP6 genes using published primers.Citation19
Conventional hemi-nested PCR assay for PCV-1
PCV-1 positive samples extracted with MagNA Pure were sequence confirmed with published primers designed when there were few PCV-1 sequences. During the study we noticed increased numbers of PCV-1 sequences published in the GenBank database. After comparing our sequences to the newly published PCV-1 sequences, we found a single nucleotide polymorphism (SNP), base 1163 (reference sequence GenBank accession number HM143844), which differentiates all wild-type PCV-1 strains in GenBank from the PCV-1 strain associated with Rotarix®. All PCV-1 positive samples were sequenced using the following protocol. The PCV-1 forward primer spanned nucleotides 889 to 913 (PCV-1PCV-1F-889: 5′-GGC CGA TTT GAA GCA GTG GAC CCA-3′); the PCV-1 reverse primer spanned nucleotides 1406–1383 (PCV-1PCV-1R-1406: 5′-CCC CTA CCT TTC CAA TAC CG-3′); and the forward primer for the hemi-nested PCR spanned nucleotides 1060 to 1080 (PCV-1PCV-1F-1060: 5′-GGG CTG TGG CTG CAT TTT GG-3′). The oligonucleotide primers were designed to amplify 494 bp and 303 bp regions of open reading frame 1 (ORF1) in the first round and hemi-nested second round reactions, respectively. The extracted TNAs from the vaccine and stool samples were used as PCR templates in the first reaction, and 2 μL of the amplified product was used for second round of amplification. The hemi-nested amplification was performed in a 50 μL reaction mixture containing 1 × PCR buffer, 0.2 mM of each dNTP, 1.25 mM MgCl2, 1 μM of each primer, and 2.5 U of Platinum Taq DNA polymerase (catalog #:10966026, ThermoFisher Scientific). Both first and hemi-nested PCR reactions were run on a GeneAmp PCR System 9600 Thermal cycler (catalog #:4339386, ThermoFisher Scientific) for 35 cycles at 95°C for 1 min (denaturation), 65°C for 1 min (for primer annealing), and 72°C for 1 min (extension) with a final extension step at 72°C for 10 min. Electrophoresis of amplified products was accomplished on a 3% agarose gel stained with GelRed™ (catalog #: 41003, Biotium, Inc.). Second hemi-nested round products were visualized on a UV transilluminator, excised and purified using a QIAquick Gel Extraction kit (catalog #: 28704, Qiagen, Inc.) following the manufacturer's instructions. DNA cycle sequencing was accomplished using a Big Dye Terminator Cycle Sequencing kit v1.1 (catalog #: 4337450, ThermoFisher Scientific) with individual primers from the second round reaction, PCV-1F-1060 and PCV-1R-1406. The cycle sequenced product was purifiedCitation20 and resulting amplicons were sequenced on ABI Prism 3130XL automated sequencer (catalog #: 4474242, ThermoFisher Scientific).
Data presented
Data from this study were presented at the 11th International Rotavirus Symposium in New Delhi, India on September 4th, 2014.
Disclaimer
The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the Centers for Disease Control and Prevention. Names of specific vendors, manufacturers, or products are included for public health and informational purposes; inclusion does not imply endorsement of the vendors, manufacturers, or products by the Centers for Disease Control and Prevention or the US Department of Health and Human Services.
Abbreviations
| RVA | = | Rotavirus group A |
| Rotarix® | = | Rotarix® vaccine (GlaxoSmithKline Biologicals) |
| Rotarix® RVA | = | rotavirus strain found in Rotarix® vaccine |
| Rotarix® PCV-1 | = | porcine circovirus type 1 strain found in Rotarix® vaccine |
| Rotarix® RVA RNA | = | RNA of rotavirus strain found in Rotarix® vaccine |
| Rotarix® PCV-1 DNA | = | DNA of porcine circovirus type 1 strain found in Rotarix® vaccine |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to acknowledge Jon R. Gentsch (CDC, Atlanta, GA) for his scientific guidance. We would like to thank the clinic staff, including nurses and providers at the Pediatric Ambulatory Clinic, Hughes Spalding Children's Hospital. We are especially grateful to Ms. Gloria Smykle, Ms. Teresa Pilgram, and Dr. Joy Smith for their assistance to help recruit and enroll the infants. We want to thank the microbiology laboratory staff at the Clinical Microbiology Laboratory, Children's Healthcare of Atlanta at Egleston for assistance in sample receipt and storage. Finally, we thank the families of the infants enrolled in the study for their careful attention to the collection of the specimens.
Funding
This work was funded through the CDC-funded Emerging Infections Program grant number U50CK000196–02. Dr. Immergluck received funding support from National Center for Advancing Translations Sciences, NIH, Clinical Research and Education Career Development Program, Grant#8R25MD007589–10.
References
- Victoria JG, Wang C, Jones MS, Jaing C, McLoughlin K, Gardner S, Delwart EL. Viral nucleic acids in live-attenuated vaccines: detection of minority variants and an adventitious virus. J Virol 2010; 84:6033-40; PMID:20375174; https://doi.org/http://dx.doi.org/10.1128/JVI.02690-09
- Cortese MM, Parashar UD. Prevention of rotavirus gastroenteritis among infants and children: recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR Recomm Rep 2009; 58:1-25; PMID:19194371
- US Food and Drug Administration. Information for parents and caregivers. 2010. Available at: http://www.fda.gov/BiologicsBloodVaccines/Vaccines/ApprovedProducts/ucm205547.htm. Accessed 05 April 2016.
- McClenahan SD, Krause PR, Uhlenhaut C. Molecular and infectivity studies of porcine circovirus in vaccines. Vaccine 2011; 29:4745-53; PMID:21569811; https://doi.org/http://dx.doi.org/10.1016/j.vaccine.2011.04.087
- Baylis SA, Finsterbusch T, Bannert N, Blumel J, Mankertz A. Analysis of porcine circovirus type 1 detected in Rotarix vaccine. Vaccine 2011; 29:690-7; PMID:21093497; https://doi.org/http://dx.doi.org/10.1016/j.vaccine.2010.11.028
- Howe B. Vaccines and related biological products advisory committee meeting: ROTARIX (Rotavirus Vaccine, Live Oral): GSK's PCV1 Investigation (PPTX - 2.4MB). 2010. Available at: http://www.fda.gov/AdvisoryCommittees/CommitteesMeetingMaterials/BloodVaccinesandOtherBiologics/VaccinesandRelatedBiologicalProductsAdvisoryCommittee/ucm211828.htm. Accessed 05 April 2016.
- Esona MD, Mijatovic-Rustempasic S, Yen C, Parashar UD, Gentsch JR, Bowen MD, LaRussa P. Detection of PCV-2 DNA in stool samples from infants vaccinated with RotaTeq®. Hum Vaccin Immunother 2014; 10:25-32; PMID:24104203; https://doi.org/http://dx.doi.org/10.4161/hv.26731
- Payne DC, Currier RL, Staat MA, Sahni LC, Selvarangan R, Halasa NB, Englund JA, Weinberg GA, Boom JA, Szilagyi PG, et al. Epidemiologic association between FUT2 secretor status and severe rotavirus gastroenteritis in children in the United States. JAMA Pediatr 2015; 169:1040-5; PMID:26389824; https://doi.org/http://dx.doi.org/10.1001/jamapediatrics.2015.2002
- Currier RL, Payne DC, Staat MA, Selvarangan R, Shirley SH, Halasa N, Boom JA, Englund JA, Szilagyi PG, Harrison CJ, et al. Innate susceptibility to norovirus infections influenced by FUT2 genotype in a United States pediatric population. Clinical Infect Dis 2015; 60:1631-8; https://doi.org/http://dx.doi.org/10.1093/cid/civ165
- Kambhampati A, Payne DC, Costantini V, Lopman BA. Host genetic susceptibility to enteric viruses: A systematic review and metaanalysis. Clinical Infect Dis 2016; 62:11-8; https://doi.org/http://dx.doi.org/10.1093/cid/civ873
- Nordgren J, Sharma S, Bucardo F, Nasir W, Günaydın G, Ouermi D, Nitiema LW, Becker-Dreps S, Simpore J, Hammarström L, et al. Both Lewis and secretor status mediate susceptibility to rotavirus infections in a rotavirus genotype-dependent manner. Clinical Infect Dis 2014; 59:1567-73; https://doi.org/http://dx.doi.org/10.1093/cid/ciu633
- Imbert-Marcille BM, Barbé L, Dupé M, Le Moullac-Vaidye B, Besse B, Peltier C, Ruvoën-Clouet N, Le Pendu J. A FUT2 gene common polymorphism determines resistance to rotavirus A of the P[8]genotype. J Infect Dis 2014; 209:1227-30; PMID:24277741; https://doi.org/http://dx.doi.org/10.1093/infdis/jit655
- Bines JE, Danchin M, Jackson P, Handley A, Watts E, Lee KJ, West A, Cowley D, Chen MY, Barnes GL, et al. Safety and immunogenicity of RV3-BB human neonatal rotavirus vaccine administered at birth or in infancy: a randomised, double-blind, placebo-controlled trial. Lancet Infect Dis 2015; 15:1389-97; PMID:26318715; https://doi.org/http://dx.doi.org/10.1016/S1473-3099(15)00227-3
- Danchin M, Kirkwood CD, Lee KJ, Bishop RF, Watts E, Justice FA, Clifford V, Cowley D, Buttery JP, Bines JE, et al. Phase I trial of RV3-BB rotavirus vaccine: a human neonatal rotavirus vaccine. Vaccine 2013; 31:2610-6; PMID:23597719; https://doi.org/http://dx.doi.org/10.1016/j.vaccine.2013.04.008
- Mijatovic-Rustempasic S, Tam KI, Kerin TK, Lewis JM, Gautam R, Quaye O, Gentsch JR, Bowen MD. Sensitive and specific quantitative detection of rotavirus a by one-step real-time reverse transcription PCR assay without antecedent dsRNA denaturation. J Clin Microbiol 2013; 9:3047-54; https://doi.org/http://dx.doi.org/10.1128/JCM.01192-13
- Gautam R, Esona MD, Mijatovic-Rustempasic S, Ian Tam K, Gentsch JR, Bowen MD. Real-time RT-PCR assays to differentiate wild-type group A rotavirus strains from Rotarix and RotaTeq vaccine strains in stool samples. Hum Vaccin Immunother 2014; 10:767-77; PMID:24342877; https://doi.org/http://dx.doi.org/10.4161/hv.27388
- Tate JE, Mijatovic-Rustempasic S, Tam KI, Lyde FC, Payne DC, Szilagyi P, Edwards K, Staat MA, Weinberg GA, Hall CB, et al. Comparison of 2 assays for diagnosing rotavirus and evaluating vaccine effectiveness in children with gastroenteritis. Emerg Infect Dis 2013; 19:1245-52; PMID:23876518; https://doi.org/http://dx.doi.org/10.3201/eid1908.130461
- Hsieh YC, Wu FT, Hsiung CA, Wu HS, Chang KY, Huang YC. Comparison of virus shedding after lived attenuated and pentavalent reassortant rotavirus vaccine. Vaccine 2014; 32:1199-204; PMID:24076325; https://doi.org/http://dx.doi.org/10.1016/j.vaccine.2013.08.041
- Mijatovic-Rustempasic S, Banyai K, Esona MD, Foytich K, Bowen MD, Gentsch JR. Genome sequence based molecular epidemiology of unusual US Rotavirus A G9 strains isolated from Omaha, USA between 1997 and 2000. Infect Genet Evol 2011; 11:522-7; PMID:21130184; https://doi.org/http://dx.doi.org/10.1016/j.meegid.2010.11.012
- Mijatovic-Rustempasic S, Frace MA, Bowen MD. Cost-effective paramagnetic bead technique for purification of cycle sequencing products. Sequencing 2012; 2012:4; https://doi.org/http://dx.doi.org/10.1155/2012/767959